

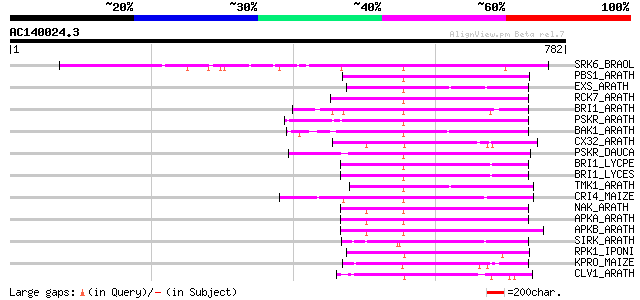
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.3 + phase: 0 /pseudo
(782 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 402 e-111
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 199 2e-50
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 194 1e-48
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 191 6e-48
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 185 5e-46
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 183 1e-45
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 181 9e-45
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 180 1e-44
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 179 2e-44
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 178 4e-44
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 178 4e-44
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 169 2e-41
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 167 8e-41
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 167 1e-40
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 167 1e-40
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 163 2e-39
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 162 2e-39
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 161 7e-39
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 142 3e-33
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 138 5e-32
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 402 bits (1034), Expect = e-111
Identities = 268/769 (34%), Positives = 398/769 (50%), Gaps = 93/769 (12%)
Query: 71 DGAVVWMYDRNQPIAIDSAVLSLDYSGVLKIEFQNRNVPIIIYYSPQPTNDTVATMLDTG 130
D VW+ +R+ P++ L + + ++ ++ N+ V + VA +L G
Sbjct: 79 DRTYVWVANRDNPLSNAIGTLKISGNNLVLLDHSNKPVWWTNLTRGNERSPVVAELLANG 138
Query: 131 NFVLQQLHPNGTKSILWQSFDSPVDTLLPTMKLGVNRKTGHNWSLVSRLAHSLPTPGELS 190
NFV++ N LWQSFD P DTLLP MKLG N KTG N L S + P+ G S
Sbjct: 139 NFVMRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYNLKTGLNRFLTSWRSSDDPSSGNFS 198
Query: 191 LEWEPKE-GELNIRKSGKVHWKSGKLKSNGM-FENIPAKVQRIYQ-YIIVSNKDEDSFAF 247
+ E + E + + +SG NG+ F IP + Y Y + N +E ++ F
Sbjct: 199 YKLETQSLPEFYLSRENFPMHRSGPW--NGIRFSGIPEDQKLSYMVYNFIENNEEVAYTF 256
Query: 248 E------------VKDGKFIRWFISPKGRLISDAGSTSNADMC--------YGYKSDEGC 287
+ +G F R P R+ + S+ C Y Y C
Sbjct: 257 RMTNNSFYSRLTLISEGYFQRLTWYPSIRIWNRFWSSPVDPQCDTYIMCGPYAY-----C 311
Query: 288 QVANADMC---YGYNSD-----------GGCQKWEEIPNCREPGEVFRKMVGRPNKDNAT 333
V + +C G+N GGC + ++ +C G K + P AT
Sbjct: 312 DVNTSPVCNCIQGFNPRNIQQWDQRVWAGGCIRRTQL-SCSGDGFTRMKKMKLPETTMAT 370
Query: 334 TDEPANGYDDCKMRCWRNCNCYGFE--ELYSNFTGCIYYSWNSTQDV-----DLDKKNNF 386
D G +CK RC +CNC F ++ + +GC+ ++ +D+ D +
Sbjct: 371 VDRSI-GVKECKKRCISDCNCTAFANADIRNGGSGCVIWT-ERLEDIRNYATDAIDGQDL 428
Query: 387 YALVKPTKSPPNSHGKRRIWIGAAIATALLILCPLILF-LAKKKQKYALQGKKSKRKEGK 445
Y + + +I I + ++L+L LI+F L K+KQK A S +
Sbjct: 429 YVRLAAADIAKKRNASGKI-ISLTVGVSVLLL--LIMFCLWKRKQKRAKASAISIANTQR 485
Query: 446 MKDLAESYDIKDLENDFKGH------DIKVFNFTSILEATMDFSSENKLGQGGYGPVYKG 499
++L + + + +F G ++ + ++++AT +FSS NKLGQGG+G VYKG
Sbjct: 486 NQNLPMNEMVLSSKREFSGEYKFEELELPLIEMETVVKATENFSSCNKLGQGGFGIVYKG 545
Query: 500 ILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------ 553
L G+E+AVKRLSKTS QG EF NE+ LI LQH NLVQ+LGCCI +E++L
Sbjct: 546 RLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHINLVQVLGCCIEGDEKMLIYEYLE 605
Query: 554 ----------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENL 597
L+W +R +I G+++GLLYLH+ SR +IIHRDLK SNILLD+N+
Sbjct: 606 NLSLDSYLFGKTRRSKLNWNERFDITNGVARGLLYLHQDSRFRIIHRDLKVSNILLDKNM 665
Query: 598 NPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIIC 657
PKISDFGMAR+F + E+ NT ++VGTYGYMSPEYAM GI S KSDV+SFGV++LEI+
Sbjct: 666 IPKISDFGMARIFERDETEANTMKVVGTYGYMSPEYAMYGIFSEKSDVFSFGVIVLEIVS 725
Query: 658 GRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDT-------FVPDEVQRCIHVG 710
G+KN F+++D +L+ + W W +G L+++DP + D+ F P EV +CI +G
Sbjct: 726 GKKNRGFYNLDYENDLLSYVWSRWKEGRALEIVDPVIVDSLSSQPSIFQPQEVLKCIQIG 785
Query: 711 LLCVQQYANDRPTMSDVISMLTNKYKLTTLPRRPAFYIRREIYDGETTS 759
LLCVQ+ A RP MS V+ M ++ P+ P + +RR Y+ + +S
Sbjct: 786 LLCVQELAEHRPAMSSVVWMFGSEATEIPQPKPPGYCVRRSPYELDPSS 834
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 199 bits (506), Expect = 2e-50
Identities = 114/288 (39%), Positives = 167/288 (57%), Gaps = 25/288 (8%)
Query: 470 FNFTSILEATMDFSSENKLGQGGYGPVYKGIL-ATGQEVAVKRLSKTSGQGIVEFRNELA 528
F F + ATM+F + LG+GG+G VYKG L +TGQ VAVK+L + QG EF E+
Sbjct: 74 FAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQGNREFLVEVL 133
Query: 529 LICELQHTNLVQLLGCCIHEEERIL-----------------------LDWKKRLNIIEG 565
++ L H NLV L+G C ++R+L LDW R+ I G
Sbjct: 134 MLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDWNMRMKIAAG 193
Query: 566 ISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGT 625
++GL +LH + +I+RD K+SNILLDE +PK+SDFG+A++ + + R++GT
Sbjct: 194 AAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKSHVSTRVMGT 253
Query: 626 YGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWND-G 684
YGY +PEYAM G + KSDVYSFGV+ LE+I GRK NL+ A L+ND
Sbjct: 254 YGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHGEQNLVAWARPLFNDRR 313
Query: 685 EYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLT 732
++++L DP L F + + + V +C+Q+ A RP ++DV++ L+
Sbjct: 314 KFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALS 361
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 194 bits (492), Expect = 1e-48
Identities = 113/282 (40%), Positives = 162/282 (57%), Gaps = 27/282 (9%)
Query: 475 ILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQ 534
I+EAT FS +N +G GG+G VYK L + VAVK+LS+ QG EF E+ + +++
Sbjct: 910 IVEATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVK 969
Query: 535 HTNLVQLLGCCIHEEERIL-----------------------LDWKKRLNIIEGISQGLL 571
H NLV LLG C EE++L LDW KRL I G ++GL
Sbjct: 970 HPNLVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGLA 1029
Query: 572 YLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSP 631
+LH IIHRD+KASNILLD + PK++DFG+AR+ + ES V+T I GT+GY+ P
Sbjct: 1030 FLHHGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVST-VIAGTFGYIPP 1088
Query: 632 EYAMEGICSTKSDVYSFGVLLLEIICGRK--NNSFHDVDRPLNLIGHAWELWNDGEYLQL 689
EY +TK DVYSFGV+LLE++ G++ F + + NL+G A + N G+ + +
Sbjct: 1089 EYGQSARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGG-NLVGWAIQKINQGKAVDV 1147
Query: 690 LDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
+DP L + + R + + +LC+ + RP M DV+ L
Sbjct: 1148 IDPLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKAL 1189
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 191 bits (485), Expect = 6e-48
Identities = 113/307 (36%), Positives = 170/307 (54%), Gaps = 27/307 (8%)
Query: 452 SYDIK--DLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILAT-GQEVA 508
S D+K +L + G + F F + EAT +F S+ LG+GG+G V+KG + Q VA
Sbjct: 71 SLDVKGLNLNDQVTGKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVA 130
Query: 509 VKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL--------------- 553
+K+L + QGI EF E+ + H NLV+L+G C ++R+L
Sbjct: 131 IKQLDRNGVQGIREFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLH 190
Query: 554 --------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFG 605
LDW R+ I G ++GL YLH +I+RDLK SNILL E+ PK+SDFG
Sbjct: 191 VLPSGKKPLDWNTRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFG 250
Query: 606 MARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFH 665
+A++ + + R++GTYGY +P+YAM G + KSD+YSFGV+LLE+I GRK
Sbjct: 251 LAKVGPSGDKTHVSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNT 310
Query: 666 DVDRPLNLIGHAWELWND-GEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTM 724
+ NL+G A L+ D + +++DP L + + + + + +CVQ+ RP +
Sbjct: 311 KTRKDQNLVGWARPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVV 370
Query: 725 SDVISML 731
SDV+ L
Sbjct: 371 SDVVLAL 377
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 185 bits (469), Expect = 5e-46
Identities = 128/382 (33%), Positives = 196/382 (50%), Gaps = 61/382 (15%)
Query: 399 SHGKRRIWIGAAIATALLI--LCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAESY--- 453
SHG+R + ++A LL +C L L ++ + K+ ++KE +++ AE +
Sbjct: 784 SHGRRPASLAGSVAMGLLFSFVCIFGLILVGREMR-----KRRRKKEAELEMYAEGHGNS 838
Query: 454 -DIKDLENDFKGHDIKV---------------FNFTSILEATMDFSSENKLGQGGYGPVY 497
D ++K +K F +L+AT F +++ +G GG+G VY
Sbjct: 839 GDRTANNTNWKLTGVKEALSINLAAFEKPLRKLTFADLLQATNGFHNDSLIGSGGFGDVY 898
Query: 498 KGILATGQEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---- 553
K IL G VA+K+L SGQG EF E+ I +++H NLV LLG C +ER+L
Sbjct: 899 KAILKDGSAVAIKKLIHVSGQGDREFMAEMETIGKIKHRNLVPLLGYCKVGDERLLVYEF 958
Query: 554 -------------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLD 594
L+W R I G ++GL +LH IIHRD+K+SN+LLD
Sbjct: 959 MKYGSLEDVLHDPKKAGVKLNWSTRRKIAIGSARGLAFLHHNCSPHIIHRDMKSSNVLLD 1018
Query: 595 ENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLE 654
ENL ++SDFGMAR+ + ++ ++ + + GT GY+ PEY CSTK DVYS+GV+LLE
Sbjct: 1019 ENLEARVSDFGMARLMSAMDTHLSVSTLAGTPGYVPPEYYQSFRCSTKGDVYSYGVVLLE 1078
Query: 655 IICGRKNNSFHDVDRPLNLIG----HAWELWNDGEYLQLLDPSLC--DTFVPDEVQRCIH 708
++ G++ D NL+G HA +D + DP L D + E+ + +
Sbjct: 1079 LLTGKRPTDSPDFG-DNNLVGWVKQHAKLRISD-----VFDPELMKEDPALEIELLQHLK 1132
Query: 709 VGLLCVQQYANDRPTMSDVISM 730
V + C+ A RPTM V++M
Sbjct: 1133 VAVACLDDRAWRRPTMVQVMAM 1154
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 183 bits (465), Expect = 1e-45
Identities = 117/367 (31%), Positives = 191/367 (51%), Gaps = 29/367 (7%)
Query: 388 ALVKPTKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMK 447
AL+K +S + G + IG A + L+ ++ L +++ + + + + K
Sbjct: 645 ALIK--RSRRSRGGDIGMAIGIAFGSVFLLTLLSLIVLRARRRSGEVDPEIEESESMNRK 702
Query: 448 DLAESYDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEV 507
+L E L F+ +D K ++ +L++T F N +G GG+G VYK L G++V
Sbjct: 703 ELGEIGS--KLVVLFQSND-KELSYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKV 759
Query: 508 AVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL-------------- 553
A+K+LS GQ EF E+ + QH NLV L G C ++ +R+L
Sbjct: 760 AIKKLSGDCGQIEREFEAEVETLSRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWL 819
Query: 554 ---------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDF 604
L WK RL I +G ++GLLYLH+ I+HRD+K+SNILLDEN N ++DF
Sbjct: 820 HERNDGPALLKWKTRLRIAQGAAKGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADF 879
Query: 605 GMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSF 664
G+AR+ + E+ V+T+ +VGT GY+ PEY + + K DVYSFGV+LLE++ ++
Sbjct: 880 GLARLMSPYETHVSTD-LVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDM 938
Query: 665 HDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTM 724
+LI ++ ++ ++ DP + E+ R + + LC+ + RPT
Sbjct: 939 CKPKGCRDLISWVVKMKHESRASEVFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTT 998
Query: 725 SDVISML 731
++S L
Sbjct: 999 QQLVSWL 1005
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 181 bits (458), Expect = 9e-45
Identities = 130/374 (34%), Positives = 192/374 (50%), Gaps = 52/374 (13%)
Query: 390 VKPTKSPPNSHGKRRIWI----GAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGK 445
+ PT PP+ G RI G A ALL P I AL + K+ +
Sbjct: 210 ISPT--PPSPAGSNRITGAIAGGVAAGAALLFAVPAI----------ALAWWRRKKPQDH 257
Query: 446 MKDLAESYDIKDLENDFKGH--DIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILAT 503
D+ E D + H +K F+ + A+ +FS++N LG+GG+G VYKG LA
Sbjct: 258 FFDVPA-------EEDPEVHLGQLKRFSLRELQVASDNFSNKNILGRGGFGKVYKGRLAD 310
Query: 504 GQEVAVKRLSKTSGQG-IVEFRNELALICELQHTNLVQLLGCCIHEEERIL--------- 553
G VAVKRL + QG ++F+ E+ +I H NL++L G C+ ER+L
Sbjct: 311 GTLVAVKRLKEERTQGGELQFQTEVEMISMAVHRNLLRLRGFCMTPTERLLVYPYMANGS 370
Query: 554 --------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNP 599
LDW KR I G ++GL YLH + KIIHRD+KA+NILLDE
Sbjct: 371 VASCLRERPESQPPLDWPKRQRIALGSARGLAYLHDHCDPKIIHRDVKAANILLDEEFEA 430
Query: 600 KISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGR 659
+ DFG+A++ +++ V T + GT G+++PEY G S K+DV+ +GV+LLE+I G+
Sbjct: 431 VVGDFGLAKLMDYKDTHV-TTAVRGTIGHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQ 489
Query: 660 KNNSFHDV--DRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQY 717
+ + D + L+ L + + L+D L + +EV++ I V LLC Q
Sbjct: 490 RAFDLARLANDDDVMLLDWVKGLLKEKKLEALVDVDLQGNYKDEEVEQLIQVALLCTQSS 549
Query: 718 ANDRPTMSDVISML 731
+RP MS+V+ ML
Sbjct: 550 PMERPKMSEVVRML 563
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 180 bits (457), Expect = 1e-44
Identities = 113/325 (34%), Positives = 172/325 (52%), Gaps = 42/325 (12%)
Query: 455 IKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGIL----------ATG 504
I D + ++KV+NF + AT +F ++ LGQGG+G VY+G + +G
Sbjct: 59 ISDSGKLLESPNLKVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSG 118
Query: 505 QEVAVKRLSKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERILL---------- 554
VA+KRL+ S QG E+R+E+ + L H NLV+LLG C ++E +L+
Sbjct: 119 MIVAIKRLNSESVQGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLE 178
Query: 555 ----------DWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDF 604
W R+ I+ G ++GL +LH R ++I+RD KASNILLD N + K+SDF
Sbjct: 179 SHLFRRNDPFPWDLRIKIVIGAARGLAFLHSLQR-EVIYRDFKASNILLDSNYDAKLSDF 237
Query: 605 GMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSF 664
G+A++ E T RI+GTYGY +PEY G KSDV++FGV+LLEI+ G +
Sbjct: 238 GLAKLGPADEKSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGL---TA 294
Query: 665 HDVDRPL---NLIGHAW---ELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYA 718
H+ RP +L+ W EL N Q++D + + + L C++
Sbjct: 295 HNTKRPRGQESLVD--WLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDP 352
Query: 719 NDRPTMSDVISMLTNKYKLTTLPRR 743
+RP M +V+ +L + L +P R
Sbjct: 353 KNRPHMKEVVEVLEHIQGLNVVPNR 377
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 179 bits (455), Expect = 2e-44
Identities = 114/364 (31%), Positives = 180/364 (49%), Gaps = 33/364 (9%)
Query: 394 KSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSKRKEGKMKDLAES- 452
KS N + +G + T L+ L++ L + KK+ E ++ +
Sbjct: 662 KSKKNIRKIVAVAVGTGLGTVFLLTVTLLIILRTTSRGEVDPEKKADADEIELGSRSVVL 721
Query: 453 YDIKDLENDFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRL 512
+ KD N+ DI L++T F+ N +G GG+G VYK L G +VA+KRL
Sbjct: 722 FHNKDSNNELSLDDI--------LKSTSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRL 773
Query: 513 SKTSGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL------------------- 553
S +GQ EF+ E+ + QH NLV LLG C ++ +++L
Sbjct: 774 SGDTGQMDREFQAEVETLSRAQHPNLVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVD 833
Query: 554 ----LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARM 609
LDWK RL I G ++GL YLH+ I+HRD+K+SNILL + ++DFG+AR+
Sbjct: 834 GPPSLDWKTRLRIARGAAEGLAYLHQSCEPHILHRDIKSSNILLSDTFVAHLADFGLARL 893
Query: 610 FTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDR 669
++ V T+ +VGT GY+ PEY + + K DVYSFGV+LLE++ GR+
Sbjct: 894 ILPYDTHVTTD-LVGTLGYIPPEYGQASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRG 952
Query: 670 PLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVIS 729
+LI ++ + ++ DP + D +E+ + + C+ + RPT ++S
Sbjct: 953 SRDLISWVLQMKTEKRESEIFDPFIYDKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVS 1012
Query: 730 MLTN 733
L N
Sbjct: 1013 WLEN 1016
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 178 bits (452), Expect = 4e-44
Identities = 108/289 (37%), Positives = 158/289 (54%), Gaps = 27/289 (9%)
Query: 467 IKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNE 526
++ F +LEAT F +++ +G GG+G VYK L G VA+K+L SGQG EF E
Sbjct: 873 LRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAE 932
Query: 527 LALICELQHTNLVQLLGCCIHEEERIL-----------------------LDWKKRLNII 563
+ I +++H NLV LLG C EER+L L+W R I
Sbjct: 933 METIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIA 992
Query: 564 EGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIV 623
G ++GL +LH IIHRD+K+SN+LLDENL ++SDFGMAR+ + ++ ++ + +
Sbjct: 993 IGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLA 1052
Query: 624 GTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWND 683
GT GY+ PEY CSTK DVYS+GV+LLE++ G++ D NL+G +L
Sbjct: 1053 GTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFG-DNNLVGWV-KLHAK 1110
Query: 684 GEYLQLLDPSLC--DTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISM 730
G+ + D L D + E+ + + V C+ RPTM V++M
Sbjct: 1111 GKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 178 bits (452), Expect = 4e-44
Identities = 108/289 (37%), Positives = 158/289 (54%), Gaps = 27/289 (9%)
Query: 467 IKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNE 526
++ F +LEAT F +++ +G GG+G VYK L G VA+K+L SGQG EF E
Sbjct: 873 LRKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAE 932
Query: 527 LALICELQHTNLVQLLGCCIHEEERIL-----------------------LDWKKRLNII 563
+ I +++H NLV LLG C EER+L L+W R I
Sbjct: 933 METIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIA 992
Query: 564 EGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIV 623
G ++GL +LH IIHRD+K+SN+LLDENL ++SDFGMAR+ + ++ ++ + +
Sbjct: 993 IGAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLA 1052
Query: 624 GTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWND 683
GT GY+ PEY CSTK DVYS+GV+LLE++ G++ D NL+G +L
Sbjct: 1053 GTPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFG-DNNLVGWV-KLHAK 1110
Query: 684 GEYLQLLDPSLC--DTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISM 730
G+ + D L D + E+ + + V C+ RPTM V++M
Sbjct: 1111 GKITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAM 1159
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 169 bits (429), Expect = 2e-41
Identities = 97/288 (33%), Positives = 163/288 (55%), Gaps = 30/288 (10%)
Query: 479 TMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSK--TSGQGIVEFRNELALICELQHT 536
T +FSS+N LG GG+G VYKG L G ++AVKR+ +G+G EF++E+A++ +++H
Sbjct: 585 TNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSEIAVLTKVRHR 644
Query: 537 NLVQLLGCCIHEEERIL------------------------LDWKKRLNIIEGISQGLLY 572
+LV LLG C+ E++L L WK+RL + +++G+ Y
Sbjct: 645 HLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTLALDVARGVEY 704
Query: 573 LHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPE 632
LH + IHRDLK SNILL +++ K++DFG+ R+ + + + T RI GT+GY++PE
Sbjct: 705 LHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIET-RIAGTFGYLAPE 763
Query: 633 YAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELW--NDGEYLQLL 690
YA+ G +TK DVYSFGV+L+E+I GRK+ + ++L+ ++ + + + +
Sbjct: 764 YAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYINKEASFKKAI 823
Query: 691 DPSL-CDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLTNKYKL 737
D ++ D V + C + RP M +++L++ +L
Sbjct: 824 DTTIDLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNILSSLVEL 871
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 167 bits (424), Expect = 8e-41
Identities = 120/388 (30%), Positives = 190/388 (48%), Gaps = 34/388 (8%)
Query: 381 DKKNNFYALVKPTKSPPNSHGKRRIWIGAAIATALLILCPLILFLAKKKQKYALQGKKSK 440
D+ +F K TKS + RI++ + +L+L + + K + S
Sbjct: 400 DECLSFCLSQKRTKSRKLMAFQMRIFVAEIVFAVVLVLSVSVTTCLYVRHKLR-HCQCSN 458
Query: 441 RKEGKMKDLAESYDIKDLENDFKGHDIKV-----FNFTSILEATMDFSSENKLGQGGYGP 495
R+ K A S+ +++ D+K+ F++ + +AT FS ++++G+G +
Sbjct: 459 RELRLAKSTAYSFRKDNMKIQPDMEDLKIRRAQEFSYEELEQATGGFSEDSQVGKGSFSC 518
Query: 496 VYKGILATGQEVAVKRLSKTSG--QGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL 553
V+KGIL G VAVKR K S + EF NEL L+ L H +L+ LLG C ER+L
Sbjct: 519 VFKGILRDGTVVAVKRAIKASDVKKSSKEFHNELDLLSRLNHAHLLNLLGYCEDGSERLL 578
Query: 554 ------------------------LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKAS 589
L+W +R+ I ++G+ YLH Y+ +IHRD+K+S
Sbjct: 579 VYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIAVQAARGIEYLHGYACPPVIHRDIKSS 638
Query: 590 NILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFG 649
NIL+DE+ N +++DFG++ + + GT GY+ PEY +TKSDVYSFG
Sbjct: 639 NILIDEDHNARVADFGLSILGPADSGTPLSELPAGTLGYLDPEYYRLHYLTTKSDVYSFG 698
Query: 650 VLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHV 709
V+LLEI+ GRK + N++ A L G+ +LDP L + +++ V
Sbjct: 699 VVLLEILSGRKAIDMQFEEG--NIVEWAVPLIKAGDIFAILDPVLSPPSDLEALKKIASV 756
Query: 710 GLLCVQQYANDRPTMSDVISMLTNKYKL 737
CV+ DRP+M V + L + L
Sbjct: 757 ACKCVRMRGKDRPSMDKVTTALEHALAL 784
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 167 bits (423), Expect = 1e-40
Identities = 103/300 (34%), Positives = 163/300 (54%), Gaps = 35/300 (11%)
Query: 466 DIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGIL----------ATGQEVAVKRLSKT 515
++K F+ + + AT +F ++ +G+GG+G V+KG + TG +AVKRL++
Sbjct: 52 NLKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLNQE 111
Query: 516 SGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------------------- 553
QG E+ E+ + +L H NLV+L+G C+ EE R+L
Sbjct: 112 GFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTFYQ 171
Query: 554 -LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
L W R+ + G ++GL +LH ++ ++I+RD KASNILLD N N K+SDFG+AR
Sbjct: 172 PLSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKASNILLDSNYNAKLSDFGLARDGPM 230
Query: 613 QESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLN 672
++ + R++GT GY +PEY G S KSDVYSFGV+LLE++ GR+ + N
Sbjct: 231 GDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGEHN 290
Query: 673 LIGHAWE-LWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
L+ A L N L+++DP L + + + L C+ A RPTM++++ +
Sbjct: 291 LVDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVKTM 350
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 167 bits (422), Expect = 1e-40
Identities = 103/300 (34%), Positives = 162/300 (53%), Gaps = 35/300 (11%)
Query: 466 DIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGIL----------ATGQEVAVKRLSKT 515
++K F+F + AT +F ++ LG+GG+G V+KG + TG +AVK+L++
Sbjct: 52 NLKSFSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQD 111
Query: 516 SGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------------------- 553
QG E+ E+ + + H +LV+L+G C+ +E R+L
Sbjct: 112 GWQGHQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQ 171
Query: 554 -LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
L WK RL + G ++GL +LH S ++I+RD K SNILLD N K+SDFG+A+
Sbjct: 172 PLSWKLRLKVALGAAKGLAFLHS-SETRVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPI 230
Query: 613 QESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLN 672
+ + R++GT+GY +PEY G +TKSDVYSFGV+LLE++ GR+ + N
Sbjct: 231 GDKSHVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERN 290
Query: 673 LIGHAWE-LWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
L+ A L N + +++D L D + +E + + L C+ RP MS+V+S L
Sbjct: 291 LVEWAKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHL 350
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 163 bits (412), Expect = 2e-39
Identities = 104/321 (32%), Positives = 165/321 (51%), Gaps = 35/321 (10%)
Query: 466 DIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGIL----------ATGQEVAVKRLSKT 515
++K F F + AT +F ++ LG+GG+G V+KG + TG +AVK+L++
Sbjct: 53 NLKSFTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQD 112
Query: 516 SGQGIVEFRNELALICELQHTNLVQLLGCCIHEEERIL---------------------- 553
QG E+ E+ + + H NLV+L+G C+ +E R+L
Sbjct: 113 GWQGHQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQ 172
Query: 554 -LDWKKRLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQ 612
L W RL + G ++GL +LH + +I+RD K SNILLD N K+SDFG+A+
Sbjct: 173 PLSWTLRLKVALGAAKGLAFLHN-AETSVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPT 231
Query: 613 QESIVNTNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLN 672
+ + RI+GTYGY +PEY G +TKSDVYS+GV+LLE++ GR+ +
Sbjct: 232 GDKSHVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQK 291
Query: 673 LIGHAWELW-NDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
L+ A L N + +++D L D + +E + + L C+ RP M++V+S L
Sbjct: 292 LVEWARPLLANKRKLFRVIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHL 351
Query: 732 TNKYKLTTLPRRPAFYIRREI 752
+ L R ++R +
Sbjct: 352 EHIQTLNEAGGRNIDMVQRRM 372
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 162 bits (411), Expect = 2e-39
Identities = 96/285 (33%), Positives = 157/285 (54%), Gaps = 25/285 (8%)
Query: 468 KVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNEL 527
+ F ++ ++ T +F E +G+GG+G VY G++ G++VAVK LS+ S QG EFR E+
Sbjct: 562 RYFKYSEVVNITNNF--ERVIGKGGFGKVYHGVI-NGEQVAVKVLSEESAQGYKEFRAEV 618
Query: 528 ALICELQHTNLVQLLGCC--------IHE-------------EERILLDWKKRLNIIEGI 566
L+ + HTNL L+G C I+E + +L W++RL I
Sbjct: 619 DLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRSFILSWEERLKISLDA 678
Query: 567 SQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTY 626
+QGL YLH + I+HRD+K +NILL+E L K++DFG++R F+ + S + + G+
Sbjct: 679 AQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQISTVVAGSI 738
Query: 627 GYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWNDGEY 686
GY+ PEY + KSDVYS GV+LLE+I G+ + ++ +++ H + +G+
Sbjct: 739 GYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTEK-VHISDHVRSILANGDI 797
Query: 687 LQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
++D L + + + + L C + + RPTMS V+ L
Sbjct: 798 RGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMEL 842
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 161 bits (407), Expect = 7e-39
Identities = 99/286 (34%), Positives = 154/286 (53%), Gaps = 29/286 (10%)
Query: 475 ILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTS-GQGIVEFRNELALICEL 533
+LEAT + + + +G+G +G +YK L+ + AVK+L T G V E+ I ++
Sbjct: 809 VLEATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKV 868
Query: 534 QHTNLVQLLGCCIHEEERILL----------------------DWKKRLNIIEGISQGLL 571
+H NL++L + +E ++L DW R NI G + GL
Sbjct: 869 RHRNLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPKPLDWSTRHNIAVGTAHGLA 928
Query: 572 YLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYGYMSP 631
YLH I+HRD+K NILLD +L P ISDFG+A++ Q + + +N + GT GYM+P
Sbjct: 929 YLHFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAP 988
Query: 632 EYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHAWELWND-GEYLQLL 690
E A + S +SDVYS+GV+LLE+I RK + +++G +W GE +++
Sbjct: 989 ENAFTTVKSRESDVYSYGVVLLELIT-RKKALDPSFNGETDIVGWVRSVWTQTGEIQKIV 1047
Query: 691 DPS----LCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISMLT 732
DPS L D+ V ++V + + L C ++ + RPTM DV+ LT
Sbjct: 1048 DPSLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQLT 1093
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 142 bits (359), Expect = 3e-33
Identities = 97/297 (32%), Positives = 148/297 (49%), Gaps = 43/297 (14%)
Query: 470 FNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRLSKTSGQGIVEFRNELAL 529
+++ +++AT F E LG+G G VYKG+L + VAVK+L QG F+ EL++
Sbjct: 524 YSYRELVKATRKFKVE--LGRGESGTVYKGVLEDDRHVAVKKLENVR-QGKEVFQAELSV 580
Query: 530 ICELQHTNLVQLLGCCIHEEER----------------------ILLDWKKRLNIIEGIS 567
I + H NLV++ G C R ILLDW+ R NI G++
Sbjct: 581 IGRINHMNLVRIWGFCSEGSHRLLVSEYVENGSLANILFSEGGNILLDWEGRFNIALGVA 640
Query: 568 QGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVNTNRIVGTYG 627
+GL YLH +IH D+K NILLD+ PKI+DFG+ ++ + S N + + GT G
Sbjct: 641 KGLAYLHHECLEWVIHCDVKPENILLDQAFEPKITDFGLVKLLNRGGSTQNVSHVRGTLG 700
Query: 628 YMSPEYAMEGICSTKSDVYSFGVLLLEIICGRK-------NNSFHDVDRPL------NLI 674
Y++PE+ + K DVYS+GV+LLE++ G + + H + R L L
Sbjct: 701 YIAPEWVSSLPITAKVDVYSYGVVLLELLTGTRVSELVGGTDEVHSMLRKLVRMLSAKLE 760
Query: 675 GHAWELWNDGEYLQLLDPSLCDTFVPDEVQRCIHVGLLCVQQYANDRPTMSDVISML 731
G + W DG LD L + + I + + C+++ + RPTM + L
Sbjct: 761 GEE-QSWIDG----YLDSKLNRPVNYVQARTLIKLAVSCLEEDRSKRPTMEHAVQTL 812
Score = 43.1 bits (100), Expect = 0.003
Identities = 33/117 (28%), Positives = 50/117 (42%), Gaps = 27/117 (23%)
Query: 53 FCLYFDSEEAHLVVSSGVDGAVVWMYDRNQPIAIDSAVLSL---------DYSGVLKIEF 103
F +++ EA ++ + +VW + ++P+ + L+L DY G
Sbjct: 69 FSVWYSKTEA----AAANNKTIVWSANPDRPVHARRSALTLQKDGNMVLTDYDGAAVWRA 124
Query: 104 QNRNVPIIIYYSPQPTNDTVATMLDTGNFVLQQLHPNGTKSILWQSFDSPVDTLLPT 160
N T A +LDTGN V++ N +WQSFDSP DT LPT
Sbjct: 125 DGNNF----------TGVQRARLLDTGNLVIEDSGGN----TVWQSFDSPTDTFLPT 167
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 138 bits (348), Expect = 5e-32
Identities = 98/311 (31%), Positives = 152/311 (48%), Gaps = 55/311 (17%)
Query: 461 DFKGHDIKVFNFTSILEATMDFSSENKLGQGGYGPVYKGILATGQEVAVKRL-SKTSGQG 519
DFK D+ LE + EN +G+GG G VY+G + +VA+KRL + +G+
Sbjct: 682 DFKSEDV--------LECLKE---ENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRS 730
Query: 520 IVEFRNELALICELQHTNLVQLLGCCIHEEERILL---------------------DWKK 558
F E+ + ++H ++V+LLG +++ +LL W+
Sbjct: 731 DHGFTAEIQTLGRIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGHLQWET 790
Query: 559 RLNIIEGISQGLLYLHKYSRLKIIHRDLKASNILLDENLNPKISDFGMARMFTQQESIVN 618
R + ++GL YLH I+HRD+K++NILLD + ++DFG+A+ +
Sbjct: 791 RHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASEC 850
Query: 619 TNRIVGTYGYMSPEYAMEGICSTKSDVYSFGVLLLEIICGRKNNSFHDVDRPLNLIGHA- 677
+ I G+YGY++PEYA KSDVYSFGV+LLE+I G+K P+ G
Sbjct: 851 MSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKK---------PVGEFGEGV 901
Query: 678 ----WELWNDGEYLQLLDPSLCDTFVPDE-----VQRCIHV---GLLCVQQYANDRPTMS 725
W + E Q D ++ V + IHV ++CV++ A RPTM
Sbjct: 902 DIVRWVRNTEEEITQPSDAAIVVAIVDPRLTGYPLTSVIHVFKIAMMCVEEEAAARPTMR 961
Query: 726 DVISMLTNKYK 736
+V+ MLTN K
Sbjct: 962 EVVHMLTNPPK 972
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 100,763,540
Number of Sequences: 164201
Number of extensions: 4656197
Number of successful extensions: 13732
Number of sequences better than 10.0: 1633
Number of HSP's better than 10.0 without gapping: 1122
Number of HSP's successfully gapped in prelim test: 512
Number of HSP's that attempted gapping in prelim test: 10346
Number of HSP's gapped (non-prelim): 2119
length of query: 782
length of database: 59,974,054
effective HSP length: 118
effective length of query: 664
effective length of database: 40,598,336
effective search space: 26957295104
effective search space used: 26957295104
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 70 (31.6 bits)
Medicago: description of AC140024.3


