

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140024.11 + phase: 0
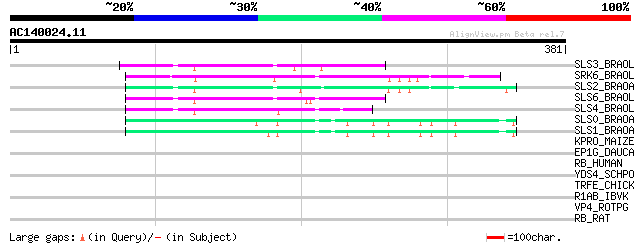
(381 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SLS3_BRAOL (P17840) S-locus-specific glycoprotein S13 precursor ... 77 6e-14
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 71 4e-12
SLS2_BRAOA (P22553) S-locus-specific glycoprotein BS29-2 precursor 71 5e-12
SLS6_BRAOL (P07761) S-locus-specific glycoprotein S6 precursor (... 69 3e-11
SLS4_BRAOL (P17841) S-locus-specific glycoprotein S14 precursor ... 60 1e-08
SLS0_BRAOA (P22551) S-locus-specific glycoprotein precursor 59 2e-08
SLS1_BRAOA (P22552) S-locus-specific glycoprotein BS29-1 precursor 57 8e-08
KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1 precu... 36 0.14
EP1G_DAUCA (Q39688) Epidermis-specific secreted glycoprotein EP1... 35 0.41
RB_HUMAN (P06400) Retinoblastoma-associated protein (PP110) (P10... 33 1.2
YDS4_SCHPO (O14180) Hypothetical protein C4F8.04 in chromosome I 32 2.1
TRFE_CHICK (P02789) Ovotransferrin precursor (Conalbumin) (Aller... 32 3.5
R1AB_IBVK (P12723) Replicase polyprotein 1ab (pp1ab) (ORF1ab pol... 31 4.6
VP4_ROTPG (P23045) Outer capsid protein VP4 (Hemagglutinin) (Out... 30 7.8
RB_RAT (P33568) Retinoblastoma-associated protein (PP105) (RB) (... 30 7.8
>SLS3_BRAOL (P17840) S-locus-specific glycoprotein S13 precursor
(SLSG-13) (Fragment)
Length = 434
Score = 77.4 bits (189), Expect = 6e-14
Identities = 58/200 (29%), Positives = 98/200 (49%), Gaps = 22/200 (11%)
Query: 76 YGKVVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN----TVATML 131
Y VWV + ++ + + L + + ++ ++ N+ ++S+ N VA +L
Sbjct: 79 YRTYVWVANRDNPLSNDIGTLKISGNNLVLLDHSNKS---VWSTNVTRGNERSPVVAELL 135
Query: 132 DAGNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPK 191
D GNFV++ N + LWQSFDYP+D L+P MKLG + KTG N L S + +P
Sbjct: 136 DNGNFVMRDSNSNNASQFLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTS--WRSSDDPS 193
Query: 192 QGE----LNIKKSGKVYWKSGKLKS------NGL-FENIPANVQSRYQ-YIIVSNKDEDS 239
G+ L +++ + Y SG + NG IP + + Y Y N +E +
Sbjct: 194 SGDYSYKLELRRLPEFYLSSGSFRLHRSGPWNGFRISGIPEDQKLSYMVYNFTENSEEAA 253
Query: 240 FTFEVKDGKF-AQWELSSKG 258
+TF + + F ++ +SS G
Sbjct: 254 YTFLMTNNSFYSRLTISSTG 273
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 71.2 bits (173), Expect = 4e-12
Identities = 79/318 (24%), Positives = 130/318 (40%), Gaps = 67/318 (21%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNT--VATMLDAGNFV 137
VWV + ++ + L + + ++ ++ N KP+ + + + VA +L GNFV
Sbjct: 83 VWVANRDNPLSNAIGTLKISGNNLVLLDHSN-KPVWWTNLTRGNERSPVVAELLANGNFV 141
Query: 138 LQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLV---------SDKFNLEW 188
++ N + LWQSFDYP+D L+P MKLG N KTG N L S F+ +
Sbjct: 142 MRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYNLKTGLNRFLTSWRSSDDPSSGNFSYKL 201
Query: 189 EPKQ-GELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQ-YIIVSNKDEDSFTFEVK 245
E + E + + +SG NG+ F IP + + Y Y + N +E ++TF +
Sbjct: 202 ETQSLPEFYLSRENFPMHRSGPW--NGIRFSGIPEDQKLSYMVYNFIENNEEVAYTFRMT 259
Query: 246 DGKF-AQWELSSKGK-----------------------------LVGDDGY--IANADMC 273
+ F ++ L S+G + G Y + + +C
Sbjct: 260 NNSFYSRLTLISEGYFQRLTWYPSIRIWNRFWSSPVDPQCDTYIMCGPYAYCDVNTSPVC 319
Query: 274 ---YGYNS-----------DGGCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFDVTY 319
G+N GGC + + +C G KK P +T D +
Sbjct: 320 NCIQGFNPRNIQQWDQRVWAGGCIRRTQL-SCSGDGFTRMKKMKLPETTMATV---DRSI 375
Query: 320 SYSDCKIRCWKNCSCNGF 337
+CK RC +C+C F
Sbjct: 376 GVKECKKRCISDCNCTAF 393
>SLS2_BRAOA (P22553) S-locus-specific glycoprotein BS29-2 precursor
Length = 435
Score = 70.9 bits (172), Expect = 5e-12
Identities = 79/334 (23%), Positives = 134/334 (39%), Gaps = 75/334 (22%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN----TVATMLDAGN 135
VWV + ++ + + L + ++ ++ N+ ++S+ N VA +L GN
Sbjct: 83 VWVANRDNPLSCSIGTLKISNMNLVLLDHSNKS---LWSTNHTRGNERSPVVAELLANGN 139
Query: 136 FVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGEL 195
FVL+ N LWQSFDYP+D L+P MKLG + +TG N L S + +P G+
Sbjct: 140 FVLRDSNKNDRSGFLWQSFDYPTDTLLPEMKLGYDLRTGLNRFLTS--WRSSDDPSSGDF 197
Query: 196 NIK------------KSGKVYWKSGKLKSNGLFENIPANVQSRYQ-YIIVSNKDEDSFTF 242
+ K K + +SG G F +P + + Y Y N +E ++TF
Sbjct: 198 SYKLQTRRLPEFYLFKDDFLVHRSGPWNGVG-FSGMPEDQKLSYMVYNFTQNSEEVAYTF 256
Query: 243 EVKDGK-FAQWELSSKG-----------------------------KLVGDDGY--IANA 270
+ + +++ +SS G K+ G Y + +
Sbjct: 257 LMTNNSIYSRLTISSSGYFERLTWTPSSGMWNVFWSSPEDFQCDVYKICGAYSYCDVNTS 316
Query: 271 DMC-------------YGYNS-DGGCQKWEDIPTCREPGEMFQKKAGRPSIDNSTTYEFD 316
+C +G + GGC++ + +C G KK P +T D
Sbjct: 317 PVCNCIQRFDPSNVQEWGLRAWSGGCRRRTRL-SCSGDGFTRMKKMKLP---ETTMAIVD 372
Query: 317 VTYSYSDCKIRCWKNCSCNGFQL--YYSNMTGCV 348
+ +C+ RC +C+C F + TGCV
Sbjct: 373 RSIGLKECEKRCLSDCNCTAFANADIRNGGTGCV 406
>SLS6_BRAOL (P07761) S-locus-specific glycoprotein S6 precursor
(SLSG-6)
Length = 436
Score = 68.6 bits (166), Expect = 3e-11
Identities = 57/198 (28%), Positives = 95/198 (47%), Gaps = 26/198 (13%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN----TVATMLDAGN 135
VWV + ++ + L + + ++ + N+ ++S+ N VA +L GN
Sbjct: 84 VWVANRDNPLSNAIGTLKISGNNLVLLGHTNKS---VWSTNLTRGNERLPVVAELLSNGN 140
Query: 136 FVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDKFNLEWEPKQGEL 195
FV++ N + LWQSFDYP+D L+P MKLG + KTG N L S + +P G+
Sbjct: 141 FVMRDSSNNDASEYLWQSFDYPTDTLLPEMKLGYDLKTGLNRFLTS--WRSSDDPSSGDF 198
Query: 196 NIKKSGK-----VYW-------KSGKLKSNGL-FENIPANVQSRYQ-YIIVSNKDEDSFT 241
+ K + W +SG NG+ F IP + + Y Y N +E ++T
Sbjct: 199 SYKLETRSLPEFYLWHGIFPMHRSGPW--NGVRFSGIPEDQKLSYMVYNFTENSEEVAYT 256
Query: 242 FEVKDGK-FAQWELSSKG 258
F + + +++ LSS+G
Sbjct: 257 FRMTNNSIYSRLTLSSEG 274
>SLS4_BRAOL (P17841) S-locus-specific glycoprotein S14 precursor
(SLSG-14) (Fragment)
Length = 434
Score = 59.7 bits (143), Expect = 1e-08
Identities = 48/185 (25%), Positives = 88/185 (46%), Gaps = 21/185 (11%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNN----TVATMLDAGN 135
VWV + ++ + + L + + ++ N+ ++S+ N VA +L GN
Sbjct: 81 VWVANRDNPLSSSIGTLKISGNNPCHLDHSNKS---VWSTNLTRGNERSPVVADVLANGN 137
Query: 136 FVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLVSDK---------FNL 186
FV++ N + LWQSFD+P+D L+P MKL + KTG N L S + F+
Sbjct: 138 FVMRDSNNNDASGFLWQSFDFPTDTLLPEMKLSYDLKTGLNRFLTSRRSSDDPSSGDFSY 197
Query: 187 EWEPKQ-GELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQYIIVSNKDEDSFTFEV 244
+ EP++ E + + ++SG NG+ F +P + + Y + +S ++ F +
Sbjct: 198 KLEPRRLPEFYLSSGVFLLYRSGPW--NGIRFSGLPDDQKLSY-LVYISQDMRVAYKFRM 254
Query: 245 KDGKF 249
+ F
Sbjct: 255 TNNSF 259
>SLS0_BRAOA (P22551) S-locus-specific glycoprotein precursor
Length = 444
Score = 59.3 bits (142), Expect = 2e-08
Identities = 75/334 (22%), Positives = 132/334 (39%), Gaps = 73/334 (21%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQ 139
VWV + ++++ + L + ++ ++ ++ N + A +L GNFVL+
Sbjct: 88 VWVANRDNALHNSMGTLKISHASLVLLDHSNTPVWSTNFTGVAHLPVTAELLANGNFVLR 147
Query: 140 QFLPNGSMSVLWQSFDYPSDVLIPMMKLG------VNRKTGHNWSLVSD------KFNLE 187
N +WQSFDYP D L+P MKLG N K +W +D F LE
Sbjct: 148 DSKTNDLDRFMWQSFDYPVDTLLPEMKLGRNLIGSENEKILTSWKSPTDPSSGDFSFILE 207
Query: 188 WEPKQGELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQYI---IVSNKDEDSFTFE 243
E E + K+ +++G NG+ F IP + YI + N +E +++F+
Sbjct: 208 TEGFLHEFYLLKNEFKVYRTGPW--NGVRFNGIPK--MQNWSYIDNSFIDNNEEVAYSFQ 263
Query: 244 VKDGK--FAQWELSSKG--KLVGDDGYIANADMCYGYNSDG-------GCQKWEDI---P 289
V + ++ +SS G +++ + +M + + D G + D+ P
Sbjct: 264 VNNNHNIHTRFRMSSTGYLQVITWTKTVPQRNMFWSFPEDTCDLYKVCGPYAYCDMHTSP 323
Query: 290 TCREPGEMFQKKAGR-----------------------------PSIDNSTTYEFDVTYS 320
TC K AGR + ++ D
Sbjct: 324 TCNCIKGFVPKNAGRWDLRDMSGGCVRSSKLSCGEGDGFLRMSQMKLPETSEAVVDKRIG 383
Query: 321 YSDCKIRCWKNCSCNGFQLYYSNM------TGCV 348
+C+ +C ++C+C G Y+NM +GCV
Sbjct: 384 LKECREKCVRDCNCTG----YANMDIMNGGSGCV 413
>SLS1_BRAOA (P22552) S-locus-specific glycoprotein BS29-1 precursor
Length = 444
Score = 57.0 bits (136), Expect = 8e-08
Identities = 74/334 (22%), Positives = 129/334 (38%), Gaps = 73/334 (21%)
Query: 80 VWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVLQ 139
VWV + ++++ + L + ++ ++ ++ N + A +L GNFVL+
Sbjct: 88 VWVANRDNALHNSMGTLKISHASLVLLDHSNTPVWSTNFTGVAHLPVTAELLANGNFVLR 147
Query: 140 QFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHN------WSLVSD------KFNLE 187
+WQSFDYP D L+P MKLG NR N W +D F LE
Sbjct: 148 DSKTTALDRFMWQSFDYPVDTLLPEMKLGRNRNGSGNEKILTSWKSPTDPSSGDYSFILE 207
Query: 188 WEPKQGELNIKKSGKVYWKSGKLKSNGL-FENIPANVQSRYQYI---IVSNKDEDSFTFE 243
E E + + +++G NG+ F IP + YI + N E +++F+
Sbjct: 208 TEGFLHEFYLLNNEFKVYRTGPW--NGVRFNGIPK--MQNWSYIDNSFIDNNKEVAYSFQ 263
Query: 244 VKDGK--FAQWELSSKG--KLVGDDGYIANADMCYGYNSDG-------GCQKWEDI---P 289
V + ++ +SS G +++ + +M + + D G + D+ P
Sbjct: 264 VNNNHNIHTRFRMSSTGYLQVITWTKTVPQRNMFWSFPEDTCDLYKVCGPYAYCDMHTSP 323
Query: 290 TCREPGEMFQKKAGR-----------------------------PSIDNSTTYEFDVTYS 320
TC K AGR + ++ D
Sbjct: 324 TCNCIKGFVPKNAGRWDLRDMSGGCVRSSKLSCGEGDGFLRMSQMKLPETSEAVVDKRIG 383
Query: 321 YSDCKIRCWKNCSCNGFQLYYSNM------TGCV 348
+C+ +C ++C+C G Y+NM +GCV
Sbjct: 384 LKECREKCVRDCNCTG----YANMDIMNGGSGCV 413
>KPRO_MAIZE (P17801) Putative receptor protein kinase ZmPK1
precursor (EC 2.7.1.37)
Length = 817
Score = 36.2 bits (82), Expect = 0.14
Identities = 21/85 (24%), Positives = 38/85 (44%), Gaps = 4/85 (4%)
Query: 79 VVWVYDINHSIDFNTSVLSLDYSGVLKIESQNRKPIIIYSSPQPTNNTVATMLDAGNFVL 138
+VW + + + S L+L G + + + + T A +LD GN V+
Sbjct: 86 IVWSANPDRPVHARRSALTLQKDGNMVLTDYDGAAVWRADGNNFTGVQRARLLDTGNLVI 145
Query: 139 QQFLPNGSMSVLWQSFDYPSDVLIP 163
+ + + +WQSFD P+D +P
Sbjct: 146 E----DSGGNTVWQSFDSPTDTFLP 166
>EP1G_DAUCA (Q39688) Epidermis-specific secreted glycoprotein EP1
precursor (52/54-kDa medium protein)
Length = 389
Score = 34.7 bits (78), Expect = 0.41
Identities = 39/137 (28%), Positives = 57/137 (41%), Gaps = 22/137 (16%)
Query: 123 TNNTVATMLDAGN--FVLQQFLPNGSMSV-------LWQSFDYPSDVLI--PMMKLGVNR 171
+N VA N V + LPNG+M + LWQSFD P+D L+ +K+G
Sbjct: 119 SNGQVAWQTSTANKGVVGLKILPNGNMVLYDSKGKFLWQSFDTPTDTLLVGQSLKMGAVT 178
Query: 172 K------TGHNWSLVSDKFNLEWEPKQGELNIKKSGKVYWKSGKLKSNGLFENIPANVQS 225
K G N V+ ++L EPK L K + K + S LF + N
Sbjct: 179 KLVSRASPGEN---VNGPYSLVMEPKGLHLYYKPTTSP--KPIRYYSFSLFTKLNKNESL 233
Query: 226 RYQYIIVSNKDEDSFTF 242
+ N+++ F F
Sbjct: 234 QNVTFEFENENDQGFAF 250
>RB_HUMAN (P06400) Retinoblastoma-associated protein (PP110)
(P105-RB) (RB)
Length = 928
Score = 33.1 bits (74), Expect = 1.2
Identities = 28/83 (33%), Positives = 39/83 (46%), Gaps = 10/83 (12%)
Query: 168 GVNRKTGHNWSLVSDKFNLEWEPKQGELNIKKSGKVYWKS--------GKLKSNGLFENI 219
G NR L +D +E K+ E NI + VY+K+ G + SNGL E
Sbjct: 256 GQNRSARIAKQLENDTRIIEVLCKEHECNIDEVKNVYFKNFIPFMNSLGLVTSNGLPE-- 313
Query: 220 PANVQSRYQYIIVSNKDEDSFTF 242
N+ RY+ I + NKD D+ F
Sbjct: 314 VENLSKRYEEIYLKNKDLDARLF 336
>YDS4_SCHPO (O14180) Hypothetical protein C4F8.04 in chromosome I
Length = 306
Score = 32.3 bits (72), Expect = 2.1
Identities = 18/57 (31%), Positives = 31/57 (53%), Gaps = 2/57 (3%)
Query: 198 KKSGKVYWKSGKLKSNGLFENIPANVQSR--YQYIIVSNKDEDSFTFEVKDGKFAQW 252
K+ K K + K L ENIPA ++S+ Y I+ +K ++ E+KD +F+ +
Sbjct: 30 KERAKEEEKDPEKKRLRLSENIPATIESKRVYDETIIEDKPDEELQAELKDDEFSAY 86
>TRFE_CHICK (P02789) Ovotransferrin precursor (Conalbumin) (Allergen
Gal d 3) (Gal d III) (Serum transferrin)
Length = 705
Score = 31.6 bits (70), Expect = 3.5
Identities = 12/36 (33%), Positives = 22/36 (60%)
Query: 339 LYYSNMTGCVFLSWNSTQYVDMVPDKFYTLVKTTKS 374
L + ++T C+F T Y + + DKFYT++ + K+
Sbjct: 654 LLFKDLTKCLFKVREGTTYKEFLGDKFYTVISSLKT 689
>R1AB_IBVK (P12723) Replicase polyprotein 1ab (pp1ab) (ORF1ab
polyprotein) [Includes: Replicase polyprotein 1a (pp1a)
(ORF1a)] [Contains: p39; p35] (Fragment)
Length = 521
Score = 31.2 bits (69), Expect = 4.6
Identities = 17/47 (36%), Positives = 26/47 (55%)
Query: 134 GNFVLQQFLPNGSMSVLWQSFDYPSDVLIPMMKLGVNRKTGHNWSLV 180
G+ VL+Q+LP G++ V DY SD + ++ KT H + LV
Sbjct: 298 GSTVLKQWLPEGTLLVDTDIVDYVSDAHVSVLSDCNKYKTEHKFDLV 344
>VP4_ROTPG (P23045) Outer capsid protein VP4 (Hemagglutinin) (Outer
layer protein VP4) [Contains: Outer capsid protein VP8;
Outer capsid protein VP5]
Length = 775
Score = 30.4 bits (67), Expect = 7.8
Identities = 47/167 (28%), Positives = 69/167 (41%), Gaps = 35/167 (20%)
Query: 48 SKQGKFCLQFGNNSNSDFQCLFISVNAD--------YGKVVWVYD---INHSIDFNTSVL 96
S + KF F NN+N DFQ L + +D +G VW ++ + + D++T+
Sbjct: 134 SDKWKFFEMFRNNANIDFQ-LQRPLTSDTKLAGFLTHGGRVWTFNGETPHATTDYSTTSN 192
Query: 97 SLDYSGVLKIE------SQNRK--PIIIYSSP--QPTNNTVATMLDAGNFVLQQFLPNG- 145
D V+ E SQ K I P Q T N V L + + Q+ N
Sbjct: 193 LPDVEVVIHTEFYIIPRSQESKCNEYINTGLPPMQNTRNVVPVALSSRSITYQRAQVNED 252
Query: 146 ---SMSVLWQSFDYPSDVLI------PMMKLGVNRKTGHNWSLVSDK 183
S + LW+ Y D+ I ++KLG G+ WS VS K
Sbjct: 253 IIISKTSLWKEMQYNRDITIRFKFGNSIVKLG---GLGYKWSEVSFK 296
>RB_RAT (P33568) Retinoblastoma-associated protein (PP105) (RB)
(Fragment)
Length = 899
Score = 30.4 bits (67), Expect = 7.8
Identities = 25/83 (30%), Positives = 39/83 (46%), Gaps = 10/83 (12%)
Query: 168 GVNRKTGHNWSLVSDKFNLEWEPKQGELNIKKSGKVYWKS--------GKLKSNGLFENI 219
G NR L SD +E K+ E N+ + VY+K+ G + SNGL E
Sbjct: 228 GQNRSARIAKQLESDTRTIEVLCKEHECNVDEVKNVYFKNFIPFISSLGIVSSNGLPE-- 285
Query: 220 PANVQSRYQYIIVSNKDEDSFTF 242
++ RY+ + + +KD D+ F
Sbjct: 286 LESLSKRYEEVYLKSKDLDARLF 308
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.134 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 49,409,788
Number of Sequences: 164201
Number of extensions: 2233660
Number of successful extensions: 4186
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 9
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 4167
Number of HSP's gapped (non-prelim): 25
length of query: 381
length of database: 59,974,054
effective HSP length: 112
effective length of query: 269
effective length of database: 41,583,542
effective search space: 11185972798
effective search space used: 11185972798
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC140024.11


