

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC140022.13 + phase: 0
(213 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
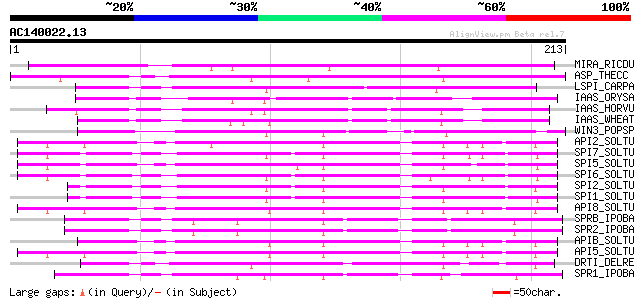
Score E
Sequences producing significant alignments: (bits) Value
MIRA_RICDU (P13087) Miraculin precursor (MIR) 176 4e-44
ASP_THECC (P32765) 21 kDa seed protein precursor 136 3e-32
LSPI_CARPA (P80691) Latex serine proteinase inhibitor 96 5e-20
IAAS_ORYSA (P29421) Alpha-amylase/subtilisin inhibitor (RASI) 76 7e-14
IAAS_HORVU (P07596) Alpha-amylase/subtilisin inhibitor precursor... 68 1e-11
IAAS_WHEAT (P16347) Endogenous alpha-amylase/subtilisin inhibito... 67 3e-11
WIN3_POPSP (P16335) Wound responsive GWIN3 protein precursor 66 5e-11
API2_SOLTU (Q43646) Aspartic protease inhibitor 2 precursor (Cat... 65 2e-10
SPI7_SOLTU (P30941) Serine protease inhibitor 7 precursor (PIG) ... 64 3e-10
SPI5_SOLTU (Q41484) Serine protease inhibitor 5 precursor (gCDI-B1) 64 3e-10
SPI6_SOLTU (Q41433) Probable serine protease inhibitor 6 precurs... 63 4e-10
SPI2_SOLTU (P58515) Serine protease inhibitor 2 (PSPI-21) (PSPI-... 63 4e-10
SPI1_SOLTU (P58514) Serine protease inhibitor 1 (PSPI-21) (PSPI-... 63 4e-10
API8_SOLTU (P17979) Aspartic protease inhibitor 8 precursor (pi8... 63 4e-10
SPRB_IPOBA (P10965) Sporamin B precursor 62 1e-09
SPR2_IPOBA (P14716) Sporamin B precursor (Clone PIM0535) 62 1e-09
APIB_SOLTU (P16348) Aspartic protease inhibitor 11 (Cathepsin D ... 60 3e-09
API5_SOLTU (P58519) Aspartic protease inhibitor 5 precursor (pi1... 60 4e-09
DRTI_DELRE (P83667) Kunitz-type serine protease inhibitor DrTI 59 8e-09
SPR1_IPOBA (P14715) Sporamin A precursor (Clone PIM0335) 57 2e-08
>MIRA_RICDU (P13087) Miraculin precursor (MIR)
Length = 220
Score = 176 bits (446), Expect = 4e-44
Identities = 96/210 (45%), Positives = 126/210 (59%), Gaps = 17/210 (8%)
Query: 8 LAVLFALSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFV 67
++ L A + PLL AD++P V+D +G+K+R G +YYI PV G G
Sbjct: 14 VSALLAAAANPLLSAADSAPNPVLDIDGEKLRTGTNYYIVPVLRDH---------GGGLT 64
Query: 68 LIARSPNET--CPLNVVVV--EGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTSCE-- 121
+ A +PN T CP VV E + + F P NPK+ V+RVSTDLNI S C
Sbjct: 65 VSATTPNGTFVCPPRVVQTRKEVDHDRPLAFFPENPKEDVVRVSTDLNINFSAFMPCRWT 124
Query: 122 ESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKY--EDDYKFVFCPTVCNFC 179
ST+W LD +D STGQ+FVT GGV GNPG +T+ +WFKIE++ YK VFCPTVC C
Sbjct: 125 SSTVWRLDKYDESTGQYFVTIGGVKGNPGPETISSWFKIEEFCGSGFYKLVFCPTVCGSC 184
Query: 180 KVMCRNVGIFRDSNGNQRVALTDVPYKVRF 209
KV C +VGI+ D G +R+AL+D P+ F
Sbjct: 185 KVKCGDVGIYIDQKGRRRLALSDKPFAFEF 214
>ASP_THECC (P32765) 21 kDa seed protein precursor
Length = 221
Score = 136 bits (343), Expect = 3e-32
Identities = 79/219 (36%), Positives = 127/219 (57%), Gaps = 15/219 (6%)
Query: 1 MKSICILLAVLFALSTQP-LLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
MK+ ++ +LFA +++ G A+A+ V+DT+G +++ GV YY+ + G
Sbjct: 1 MKTATAVVLLLFAFTSKSYFFGVANAANSPVLDTDGDELQTGVQYYV----LSSISG--- 53
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQG--VTFTPVNPKKGVIRVSTDLNIK--TS 115
G G + + R+ ++CP VV G V F+ + K V+RVSTD+NI+
Sbjct: 54 --AGGGGLALGRATGQSCPEIVVQRRSDLDNGTPVIFSNADSKDDVVRVSTDVNIEFVPI 111
Query: 116 LNTSCEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPT 174
+ C ST+W LD++D+S G+W+VTT GV G PG +T+ +WFKIEK YKF FCP+
Sbjct: 112 RDRLCSTSTVWRLDNYDNSAGKWWVTTDGVKGEPGPNTLCSWFKIEKAGVLGYKFRFCPS 171
Query: 175 VCNFCKVMCRNVGIFRDSNGNQRVALTDVPYKVRFQPSA 213
VC+ C +C ++G D +G R+AL+D + F+ ++
Sbjct: 172 VCDSCTTLCSDIGRHSDDDGQIRLALSDNEWAWMFKKAS 210
>LSPI_CARPA (P80691) Latex serine proteinase inhibitor
Length = 184
Score = 96.3 bits (238), Expect = 5e-20
Identities = 61/180 (33%), Positives = 92/180 (50%), Gaps = 12/180 (6%)
Query: 26 SPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVE 85
+P+ +VD +GK V GVDY++ + G G G G + + CPL+VV
Sbjct: 2 APKPIVDIDGKPVLYGVDYFV----VSAIWGAG----GGGLTVYGPGNKKKCPLSVVQDP 53
Query: 86 GFRGQGVTFTPV-NPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGG 144
G+ + F+ + N K ++ S DLN+K ++ +C E+T W +D F G W VT GG
Sbjct: 54 FDNGEPIIFSAIKNVKDNIVFESVDLNVKFNITINCNETTAWKVDRFPGVIG-WTVTLGG 112
Query: 145 VLGNPGKDTVDNWFKIEK--YEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVALTD 202
G G ++ + FKI+K YKF FCP+ + C NV IF D +R+ LT+
Sbjct: 113 EKGYHGFESTHSMFKIKKAGLPFSYKFHFCPSYPRTRLIPCNNVDIFFDKYRIRRLILTN 172
>IAAS_ORYSA (P29421) Alpha-amylase/subtilisin inhibitor (RASI)
Length = 176
Score = 75.9 bits (185), Expect = 7e-14
Identities = 59/193 (30%), Positives = 93/193 (47%), Gaps = 31/193 (16%)
Query: 26 SPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVV- 84
+P V DTEG ++ A YY+ +P +P G G + +P P ++V
Sbjct: 1 APPPVYDTEGHELSADGSYYV--LPASPGHGGG----------LTMAPRRVIPCPLLVAQ 48
Query: 85 ---EGFRGQGVTFTP----VNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQ 137
E +G V FTP +P+ +RVSTD+ I+ + T C +ST W + D + TG
Sbjct: 49 ETDERRKGFPVRFTPWGGAASPR--TLRVSTDVRIRFNAATLCVQSTEWHVGD-EPLTGA 105
Query: 138 WFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQR 197
V TG ++G P +N F++EKY YK V C+ C+++G+ RD G
Sbjct: 106 RRVVTGPLIG-PSPSGRENAFRVEKYGGGYKLV-------SCRDSCQDLGVSRDGAGAAW 157
Query: 198 VALTDVPYKVRFQ 210
+ + P+ V F+
Sbjct: 158 IGASQPPHVVVFK 170
>IAAS_HORVU (P07596) Alpha-amylase/subtilisin inhibitor precursor
(BASI)
Length = 203
Score = 68.2 bits (165), Expect = 1e-11
Identities = 63/205 (30%), Positives = 91/205 (43%), Gaps = 31/205 (15%)
Query: 15 STQPLLGEAD----ASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIA 70
S+ PL A A P V DT+G ++RA +YY+ + G G + +A
Sbjct: 9 SSSPLFWPAPPSRAADPPPVHDTDGHELRADANYYV--LSANRAHGGG--------LTMA 58
Query: 71 RSPNETCPLNVVVVEGFRGQG--VTFTP--VNPKKGVIRVSTDLNIKTSLNTSCEESTIW 126
CPL V + G V TP V P +IR+STD+ I T+C +ST W
Sbjct: 59 PGHGRHCPLFVSQDPNGQHDGFPVRITPYGVAPSDKIIRLSTDVRISFRAYTTCLQSTEW 118
Query: 127 TLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYE----DDYKFVFCPTVCNFCKVM 182
+D + + G+ V TG V +P +N F+IEKY +YK + C
Sbjct: 119 HIDS-ELAAGRRHVITGPV-KDPSPSGRENAFRIEKYSGAEVHEYKLMSCGD-------W 169
Query: 183 CRNVGIFRDSNGNQRVALTDVPYKV 207
C+++G+FRD G PY V
Sbjct: 170 CQDLGVFRDLKGGAWFLGATEPYHV 194
>IAAS_WHEAT (P16347) Endogenous alpha-amylase/subtilisin inhibitor
(WASI)
Length = 180
Score = 67.0 bits (162), Expect = 3e-11
Identities = 59/189 (31%), Positives = 86/189 (45%), Gaps = 27/189 (14%)
Query: 27 PEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVV-VE 85
P V DT+G ++RA +YY+ +P G G + +A CPL V +
Sbjct: 2 PPPVHDTDGNELRADANYYV--LPANRAHGGG--------LTMAPGHGRRCPLFVSQEAD 51
Query: 86 GFR-GQGVTFTPVN--PKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTT 142
G R G V P P +IR+STD+ I T+C +ST W +D + +G+ V T
Sbjct: 52 GQRDGLPVRIAPHGGAPSDKIIRLSTDVRISFRAYTTCVQSTEWHIDS-ELVSGRRHVIT 110
Query: 143 GGVLGNPGKDTVDNWFKIEKYE----DDYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRV 198
G V +P +N F+IEKY +YK + C C+++G+FRD G
Sbjct: 111 GPV-RDPSPSGRENAFRIEKYSGAEVHEYKLMACGD-------SCQDLGVFRDLKGGAWF 162
Query: 199 ALTDVPYKV 207
PY V
Sbjct: 163 LGATEPYHV 171
>WIN3_POPSP (P16335) Wound responsive GWIN3 protein precursor
Length = 200
Score = 66.2 bits (160), Expect = 5e-11
Identities = 59/193 (30%), Positives = 86/193 (43%), Gaps = 33/193 (17%)
Query: 27 PEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEG 86
P V+D G++V+AG Y I F ++ + N C +V++ G
Sbjct: 30 PAAVLDFYGREVQAGASYLIDQ---------------EDFRVVNATINPICNSDVILSTG 74
Query: 87 FRGQGVTFTPV-NPKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDFDSSTGQWFVTTG 143
G VTF+PV N GVIR T + + +T + +W + F+S+ + VTTG
Sbjct: 75 IEGLPVTFSPVINSTDGVIREGTLITVSFDASTCGMAGVTPMWKIG-FNSTAKGYIVTTG 133
Query: 144 GVLGNPGKDTVDNWFKIEKYEDD---YKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVAL 200
GV D + N FKI K+E D Y+ +CP FC+ C VG D V+
Sbjct: 134 GV------DRL-NLFKITKFESDSSFYQLSYCPNSEPFCECPCVPVGANSDKYLAPNVSY 186
Query: 201 TDVPYKVRFQPSA 213
D RF+P A
Sbjct: 187 AD----FRFKPDA 195
>API2_SOLTU (Q43646) Aspartic protease inhibitor 2 precursor
(Cathepsin D inhibitor) (CathDinh) (CathIhn)
Length = 220
Score = 64.7 bits (156), Expect = 2e-10
Identities = 66/223 (29%), Positives = 100/223 (44%), Gaps = 30/223 (13%)
Query: 4 ICILLAVLFA--LSTQPLLGEADASP--EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
+C+L V+F+ ++Q L+ SP + V+DT GK++ Y I + GRG
Sbjct: 10 LCLLPIVVFSSTFTSQNLIDLPSESPLPKPVLDTNGKELNPNSSYRIISI------GRGA 63
Query: 60 CVVGSGFVLIARSPNET--CPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTDLNIKTSLN 117
G V + +SPN CP V G TF P G I LNI+ ++
Sbjct: 64 L---GGDVYLGKSPNSDGPCPDGVFRYNSDVGPSGTFVRFIPLSGGIFEDQLLNIQFNIA 120
Query: 118 TS--CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPT 174
T C TIW + + ++ + TGG +G ++FKI K + Y ++CP
Sbjct: 121 TVKLCVSYTIWKVGNLNAYFRTMLLETGGTIGQAD----SSYFKIVKLSNFGYNLLYCPI 176
Query: 175 ----VCNFCK--VMCRNVGIFRDSNGNQRVAL-TDVPYKVRFQ 210
+C FC+ C VG+ NG +R+AL + P V FQ
Sbjct: 177 TPPFLCPFCRDDNFCAKVGVV-IQNGKRRLALVNENPLDVLFQ 218
>SPI7_SOLTU (P30941) Serine protease inhibitor 7 precursor (PIG)
(PIGEN1) (Allergen Sola t 4) (STPIB) (STPIA) (pKEN14-28)
(pF4)
Length = 221
Score = 63.5 bits (153), Expect = 3e-10
Identities = 66/227 (29%), Positives = 103/227 (45%), Gaps = 37/227 (16%)
Query: 4 ICILLAVLFA---LSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
+C++ V+F+ S P+ +DA+P V+D GK++ + + Y I +T G
Sbjct: 9 LCLVPIVVFSSTFTSKNPINLPSDATP--VLDVAGKELDSRLSYRII---STFWGALG-- 61
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPV-------NPKKGVIRVSTDLNIK 113
G V + +SPN P + G + TPV + +G+ LNI+
Sbjct: 62 ----GDVYLGKSPNSDAPCANGIFRYNSDVGPSGTPVRFIGSSSHFGQGIFENEL-LNIQ 116
Query: 114 TSLNTS--CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFV 170
+++TS C TIW + D+D+S G + TGG +G +WFKI K Y +
Sbjct: 117 FAISTSKLCVSYTIWKVGDYDASLGTMLLETGGTIGQAD----SSWFKIVKSSQFGYNLL 172
Query: 171 FCPTV----CNFCK--VMCRNVGIFRDSNGNQRVALT-DVPYKVRFQ 210
+CP C F C VG+ NG +R+AL D P V F+
Sbjct: 173 YCPVTSTMSCPFSSDDQFCLKVGVVH-QNGKRRLALVKDNPLDVSFK 218
>SPI5_SOLTU (Q41484) Serine protease inhibitor 5 precursor (gCDI-B1)
Length = 213
Score = 63.5 bits (153), Expect = 3e-10
Identities = 64/220 (29%), Positives = 96/220 (43%), Gaps = 31/220 (14%)
Query: 4 ICILLAVLFA---LSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
+C++ V+F+ S P+ +DA+P V+D GK++ + Y I + GRG
Sbjct: 9 LCLVPIVVFSSTFTSQNPINLPSDATP--VLDVTGKELDPRLSYRIISI------GRGAL 60
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVNPKKGVIRVSTD------LNIKT 114
G V + +SPN P V G + TPV LNI+
Sbjct: 61 ---GGDVYLGKSPNSDAPCANGVFRFNSDVGPSGTPVRFIGSSSHFGPHIFEGELLNIQF 117
Query: 115 SLNTS--CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVF 171
++T C TIW + D+D+S G + TGG +G +WFKI K Y ++
Sbjct: 118 DISTVKLCVSYTIWKVGDYDASLGTMLLETGGTIGQAD----SSWFKIVKSSQLGYNLLY 173
Query: 172 CPTVCNFCKVMCRNVGIFRDSNGNQRVALT-DVPYKVRFQ 210
CP + C VG+ NG +R+AL D P V F+
Sbjct: 174 CPFSSD--DQFCLKVGVVH-QNGKRRLALVKDNPLDVSFK 210
>SPI6_SOLTU (Q41433) Probable serine protease inhibitor 6 precursor
(AM66)
Length = 221
Score = 63.2 bits (152), Expect = 4e-10
Identities = 65/227 (28%), Positives = 103/227 (44%), Gaps = 37/227 (16%)
Query: 4 ICILLAVLFA---LSTQPLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPC 60
+C+ V+F+ S P+ +DA+P V+D GK++ + Y+I +T G
Sbjct: 9 LCLFPIVVFSSTFTSQNPINLPSDATP--VLDVTGKELDPRLSYHII---STFWGALG-- 61
Query: 61 VVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPV-------NPKKGVIRVSTDLNIK 113
G V + +SPN P + G + TPV + +G+ LNI+
Sbjct: 62 ----GDVYLGKSPNSDAPCANGIFRYNSDVGPSGTPVRFIGSSSHFGQGIFENEL-LNIQ 116
Query: 114 TSLNTS--CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKI-EKYEDDYKFV 170
+++TS C TIW + D+D+S G + TGG +G +WFKI + + Y +
Sbjct: 117 FAISTSKLCVSYTIWKVGDYDASLGTMLLETGGTIGQAD----SSWFKIVQSSQFGYNLL 172
Query: 171 FCPTV----CNFCK--VMCRNVGIFRDSNGNQRVALT-DVPYKVRFQ 210
+CP C F C VG+ NG +R+AL D P V F+
Sbjct: 173 YCPVTSTMSCPFSSDDQFCLKVGVVH-QNGKRRLALVKDNPLDVSFK 218
>SPI2_SOLTU (P58515) Serine protease inhibitor 2 (PSPI-21)
(PSPI-21-5.2)
Length = 186
Score = 63.2 bits (152), Expect = 4e-10
Identities = 59/199 (29%), Positives = 91/199 (45%), Gaps = 28/199 (14%)
Query: 23 ADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVV 82
+DA+P V+D GK++ + + Y I +T G G V + +SPN P
Sbjct: 3 SDATP--VLDVTGKELDSRLSYRII---STFWGALG------GDVYLGKSPNSDAPCANG 51
Query: 83 VVEGFRGQGVTFTPV-------NPKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDFDS 133
+ G + TPV + +G+ LNI+ +++TS C TIW + D+D+
Sbjct: 52 IFRYNSDVGPSGTPVRFIGSSSHFGQGIFENEL-LNIQFAISTSKLCVSYTIWKVGDYDA 110
Query: 134 STGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCKVMCRNVGIFRDS 192
S G + TGG +G +WFKI K Y ++CP + C VG+
Sbjct: 111 SLGTMLLETGGTIGQAD----SSWFKIVKSSQLGYNLLYCPVTSSSDDQFCSKVGVVH-Q 165
Query: 193 NGNQRVAL-TDVPYKVRFQ 210
NG +R+AL + P V FQ
Sbjct: 166 NGKRRLALVNENPLDVLFQ 184
>SPI1_SOLTU (P58514) Serine protease inhibitor 1 (PSPI-21)
(PSPI-21-6.3)
Length = 187
Score = 63.2 bits (152), Expect = 4e-10
Identities = 59/199 (29%), Positives = 91/199 (45%), Gaps = 28/199 (14%)
Query: 23 ADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVV 82
+DA+P V+D GK++ + + Y I +T G G V + +SPN P
Sbjct: 3 SDATP--VLDVTGKELDSRLSYRII---STFWGALG------GDVYLGKSPNSDAPCANG 51
Query: 83 VVEGFRGQGVTFTPV-------NPKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDFDS 133
V G + TPV + +G+ LNI+ +++TS C TIW + D+D+
Sbjct: 52 VFRYNSDVGPSGTPVRFIGSSSHFGQGIFENEL-LNIQFAISTSKLCVSYTIWKVGDYDA 110
Query: 134 STGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCKVMCRNVGIFRDS 192
S G + TGG +G +WFKI K Y ++CP + C VG+
Sbjct: 111 SLGTMLLETGGTIGQAD----SSWFKIVKSSQLGYNLLYCPVTSSSDDQFCLKVGVVH-Q 165
Query: 193 NGNQRVALT-DVPYKVRFQ 210
NG +R+AL D P + F+
Sbjct: 166 NGKRRLALVKDNPLDISFK 184
>API8_SOLTU (P17979) Aspartic protease inhibitor 8 precursor (pi8)
(PI-8) (API) (API-8) (Cathepsin D inhibitor)
Length = 220
Score = 63.2 bits (152), Expect = 4e-10
Identities = 66/223 (29%), Positives = 101/223 (44%), Gaps = 30/223 (13%)
Query: 4 ICILLAVLFA--LSTQPLLGEADASP--EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
+C+L V+F+ ++Q L+ SP + V+DT GK++ Y I + GRG
Sbjct: 10 LCLLPIVVFSSTFTSQNLIDLPSESPLPKPVLDTNGKELNPDSSYRIISI------GRGA 63
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVN--PKKGVIRVSTDLNIKTSLN 117
G V + +SPN P V G + TPV P G I LNI+ ++
Sbjct: 64 L---GGDVYLGKSPNSDAPCPDGVFRYNSDVGPSGTPVRFIPLSGGIFEDQLLNIQFNIP 120
Query: 118 TS--CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPT 174
T C TIW + + ++ + TGG +G ++FKI K + Y ++CP
Sbjct: 121 TVKLCVSYTIWKVGNLNAYFRTMLLETGGTIGQAD----SSYFKIVKLSNFGYNLLYCPI 176
Query: 175 ----VCNFCK--VMCRNVGIFRDSNGNQRVAL-TDVPYKVRFQ 210
+C FC+ C VG+ NG +R+AL + P V FQ
Sbjct: 177 TPPFLCPFCRDDNFCAKVGVV-IQNGKRRLALVNENPLDVLFQ 218
>SPRB_IPOBA (P10965) Sporamin B precursor
Length = 216
Score = 61.6 bits (148), Expect = 1e-09
Identities = 57/197 (28%), Positives = 90/197 (44%), Gaps = 21/197 (10%)
Query: 22 EADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI-ARSPNETCPLN 80
E +S V+D G +VRAG +YYI + G G G G L+ S + C +
Sbjct: 33 ETASSETPVLDINGDEVRAGENYYI----VSAIWGAG----GGGLRLVRLDSSSNECASD 84
Query: 81 VVVVEG--FRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDFDSSTG 136
V+V G +T TP +P+ V+ ST + ++ T+ C + W + DS +G
Sbjct: 85 VIVSRSDFDNGDPITITPADPEATVVMPSTYQTFRFNIATNKLCVNNVNWGI-KHDSESG 143
Query: 137 QWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDS-NGN 195
Q+FV G + + N FKIE D+ + + C F C NVG + D
Sbjct: 144 QYFVKAGEFVSDN-----SNQFKIEVVNDNLN-AYKISYCQFGTEKCFNVGRYYDPLTRA 197
Query: 196 QRVALTDVPYKVRFQPS 212
R+AL++ P+ +P+
Sbjct: 198 TRLALSNTPFVFVIKPT 214
>SPR2_IPOBA (P14716) Sporamin B precursor (Clone PIM0535)
Length = 216
Score = 61.6 bits (148), Expect = 1e-09
Identities = 56/197 (28%), Positives = 91/197 (45%), Gaps = 21/197 (10%)
Query: 22 EADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLI-ARSPNETCPLN 80
E +S V+D G +VRAG +YY+ + G G G G L+ S + C +
Sbjct: 33 ETASSETPVLDINGDEVRAGENYYM----VSAIWGAG----GGGLRLVRLDSSSNECASD 84
Query: 81 VVVVEG--FRGQGVTFTPVNPKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDFDSSTG 136
V+V G +T TP +P+ V+ ST + ++ T+ C + W + DS +G
Sbjct: 85 VIVSRSDFDNGDPITITPADPEATVVMPSTYQTFRFNIATNKLCVNNVNWGI-KHDSESG 143
Query: 137 QWFVTTGGVLGNPGKDTVDNWFKIEKYEDDYKFVFCPTVCNFCKVMCRNVGIFRDS-NGN 195
Q+FV G + + N FKIE D+ + + C F C NVG + D
Sbjct: 144 QYFVKAGEFVSDN-----SNQFKIEVVNDNLN-AYKISYCQFGTEKCFNVGRYYDPLTRA 197
Query: 196 QRVALTDVPYKVRFQPS 212
R+AL+++P+ +P+
Sbjct: 198 TRLALSNIPFVFVIKPT 214
>APIB_SOLTU (P16348) Aspartic protease inhibitor 11 (Cathepsin D
inhibitor PDI) (Allergen Sola t 2)
Length = 188
Score = 60.5 bits (145), Expect = 3e-09
Identities = 59/196 (30%), Positives = 87/196 (44%), Gaps = 26/196 (13%)
Query: 27 PEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEG 86
P+ V+DT GK++ Y I + GRG G V + +SPN P V
Sbjct: 5 PKPVLDTNGKELNPNSSYRIISI------GRGAL---GGDVYLGKSPNSDAPCPDGVFRY 55
Query: 87 FRGQGVTFTPVN--PKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDFDSSTGQWFVTT 142
G + TPV P G I LNI+ ++ T C TIW + + ++ + T
Sbjct: 56 NSDVGPSGTPVRFIPLSGGIFEDQLLNIQFNIATVKLCVSYTIWKVGNLNAYFRTMLLET 115
Query: 143 GGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPT----VCNFCK--VMCRNVGIFRDSNGN 195
GG +G ++FKI K + Y ++CP +C FC+ C VG+ NG
Sbjct: 116 GGTIGQAD----SSYFKIVKLSNFGYNLLYCPITPPFLCPFCRDDNFCAKVGVV-IQNGK 170
Query: 196 QRVAL-TDVPYKVRFQ 210
+R+AL + P V FQ
Sbjct: 171 RRLALVNENPLDVLFQ 186
>API5_SOLTU (P58519) Aspartic protease inhibitor 5 precursor (pi13)
(PI-13)
Length = 220
Score = 60.1 bits (144), Expect = 4e-09
Identities = 65/223 (29%), Positives = 99/223 (44%), Gaps = 30/223 (13%)
Query: 4 ICILLAVLFA--LSTQPLLGEADASP--EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGP 59
+C+L V+F+ ++Q L+ SP + V+DT GK++ Y I + GRG
Sbjct: 10 LCLLPIVVFSSTFTSQNLIDLPSESPVPKPVLDTNGKELNPNSSYRIISI------GRGA 63
Query: 60 CVVGSGFVLIARSPNETCPLNVVVVEGFRGQGVTFTPVN--PKKGVIRVSTDLNIKTSLN 117
G V + +SPN P V G + TPV P I LNI+ ++
Sbjct: 64 L---GGDVYLGKSPNSDAPCPDGVFRYNSDVGPSGTPVRFIPLSTNIFEDQLLNIQFNIP 120
Query: 118 TS--CEESTIWTLDDFDSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYED-DYKFVFCPT 174
T C TIW + + ++ + TGG +G ++FKI K Y ++CP
Sbjct: 121 TVKLCVSYTIWKVGNLNAHLRTMLLETGGTIGQAD----SSYFKIVKSSKFGYNLLYCPI 176
Query: 175 ----VCNFCK--VMCRNVGIFRDSNGNQRVAL-TDVPYKVRFQ 210
+C FC+ C VG+ NG +R+AL + P V FQ
Sbjct: 177 TRHFLCPFCRDDNFCAKVGVV-IQNGKRRLALVNENPLDVLFQ 218
>DRTI_DELRE (P83667) Kunitz-type serine protease inhibitor DrTI
Length = 185
Score = 58.9 bits (141), Expect = 8e-09
Identities = 55/188 (29%), Positives = 82/188 (43%), Gaps = 23/188 (12%)
Query: 28 EQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETCPLNVVVVEGF 87
E+V D EG V G +YYI + G G G V R+ CP++++ +
Sbjct: 4 EKVYDIEGYPVFLGSEYYI----VSAIIG-----AGGGGVRPGRTRGSMCPMSIIQEQSD 54
Query: 88 RGQG--VTFTPVNPKKGVIRVSTDLNIKTSLNTSCEESTIWTLDDFDSSTGQWFVTTGGV 145
G V F+ +G I T+L I+ C ES+ W + +G+ V GG
Sbjct: 55 LQMGLPVRFSSPEESQGKIYTDTELEIEFVEKPDCAESSKWVI---VKDSGEARVAIGGS 111
Query: 146 LGNPGKDTVDNWFKIEKYED-DYKFVFCPTVCNFCKVMCRNVGIFRDSNGNQRVAL---T 201
+P + V +FKIEK YK VFCP C ++GI + G + + L
Sbjct: 112 EDHPQGELVRGFFKIEKLGSLAYKLVFCP---KSSSGSCSDIGI--NYEGRRSLVLKSSD 166
Query: 202 DVPYKVRF 209
D P++V F
Sbjct: 167 DSPFRVVF 174
>SPR1_IPOBA (P14715) Sporamin A precursor (Clone PIM0335)
Length = 219
Score = 57.4 bits (137), Expect = 2e-08
Identities = 58/205 (28%), Positives = 90/205 (43%), Gaps = 28/205 (13%)
Query: 18 PLLGEADASPEQVVDTEGKKVRAGVDYYIRPVPTTPCDGRGPCVVGSGFVLIARSPNETC 77
P E +S V+D G +VRAG +YY+ + G G G G L C
Sbjct: 31 PTTHEPASSETPVLDINGDEVRAGGNYYM----VSAIWGAG----GGGLRLAHLDMMSKC 82
Query: 78 PLNVVVVEG--FRGQGVTFTP--VNPKKGVIRVSTDLNIKTSLNTS--CEESTIWTLDDF 131
+V+V G +T TP +P+ V+ ST + ++ T+ C + W +
Sbjct: 83 ATDVIVSPNDLDNGDPITITPATADPESTVVMASTYQTFRFNIATNKLCVNNVNWGI-QH 141
Query: 132 DSSTGQWFVTTGGVLGNPGKDTVDNWFKIEKYE---DDYKFVFCPTVCNFCKVMCRNVGI 188
DS++GQ+F+ G + + N FKIE + + YK T C F C NVG
Sbjct: 142 DSASGQYFLKAGEFVSDN-----SNQFKIELVDANLNSYKL----TYCQFGSDKCYNVGR 192
Query: 189 FRDSN-GNQRVALTDVPYKVRFQPS 212
F D R+AL++ P+ +P+
Sbjct: 193 FHDHMLRTTRLALSNSPFVFVIKPT 217
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.139 0.435
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 27,511,186
Number of Sequences: 164201
Number of extensions: 1245743
Number of successful extensions: 2505
Number of sequences better than 10.0: 66
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 50
Number of HSP's that attempted gapping in prelim test: 2426
Number of HSP's gapped (non-prelim): 74
length of query: 213
length of database: 59,974,054
effective HSP length: 106
effective length of query: 107
effective length of database: 42,568,748
effective search space: 4554856036
effective search space used: 4554856036
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC140022.13


