

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
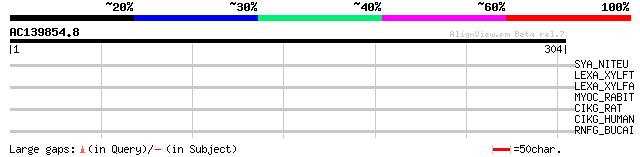
Query= AC139854.8 - phase: 0
(304 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SYA_NITEU (Q82TF8) Alanyl-tRNA synthetase (EC 6.1.1.7) (Alanine-... 32 2.5
LEXA_XYLFT (Q87F45) LexA repressor (EC 3.4.21.88) 32 2.5
LEXA_XYLFA (Q9PH24) LexA repressor (EC 3.4.21.88) 32 2.5
MYOC_RABIT (Q866N2) Myocilin precursor (Trabecular meshwork-indu... 31 3.3
CIKG_RAT (Q63734) Potassium voltage-gated channel subfamily C me... 30 7.4
CIKG_HUMAN (Q03721) Potassium voltage-gated channel subfamily C ... 30 7.4
RNFG_BUCAI (P57217) Electron transport complex protein rnfG homolog 30 9.7
>SYA_NITEU (Q82TF8) Alanyl-tRNA synthetase (EC 6.1.1.7)
(Alanine--tRNA ligase) (AlaRS)
Length = 863
Score = 31.6 bits (70), Expect = 2.5
Identities = 17/48 (35%), Positives = 23/48 (47%)
Query: 35 HEGAAQLMTYLGVSQHEAEKICNQKYGGYISYPRLRDFYTSYLGRANV 82
HE A QLMTY + A + +KYG + + DF T G +V
Sbjct: 612 HEVATQLMTYDDAVKQGAMALFGEKYGDTVRVVAMGDFSTELCGGTHV 659
>LEXA_XYLFT (Q87F45) LexA repressor (EC 3.4.21.88)
Length = 211
Score = 31.6 bits (70), Expect = 2.5
Identities = 21/54 (38%), Positives = 27/54 (49%), Gaps = 10/54 (18%)
Query: 255 GVRRVYRHLPKRVLRQYGYVQTIP--------RHPTDVRDLPTAFHCADVRRLP 300
GVR V HL VL Q G ++ IP +H T+V ++ A DV RLP
Sbjct: 38 GVRAVQHHLD--VLEQQGMIRRIPGQARGIRLKHLTEVDEVALALQSKDVLRLP 89
>LEXA_XYLFA (Q9PH24) LexA repressor (EC 3.4.21.88)
Length = 211
Score = 31.6 bits (70), Expect = 2.5
Identities = 21/54 (38%), Positives = 26/54 (47%), Gaps = 10/54 (18%)
Query: 255 GVRRVYRHLPKRVLRQYGYVQTIPR--------HPTDVRDLPTAFHCADVRRLP 300
GVR V HL VL Q G ++ +PR H T+V + A DV RLP
Sbjct: 38 GVRAVQHHLD--VLEQQGMIRRVPRQARGIRLKHLTEVDETALALQSEDVLRLP 89
>MYOC_RABIT (Q866N2) Myocilin precursor (Trabecular meshwork-induced
glucocorticoid response protein)
Length = 490
Score = 31.2 bits (69), Expect = 3.3
Identities = 30/89 (33%), Positives = 34/89 (37%), Gaps = 18/89 (20%)
Query: 66 YPRLRDFYTSYLGRANVLAGTEDPEELEELARVRTYCVRCYLLYLVGCLLFGDRSNKRIE 125
Y R+ F Y + VL P LE V V LY G G R+ R E
Sbjct: 287 YDRISQFVQGYPSKVYVL-----PRSLESTGAV----VYAGSLYFQGA---GSRTVIRFE 334
Query: 126 LIYLTTMAD------GYAGMRNYSWGGMT 148
L T A+ GY G YSWGG T
Sbjct: 335 LNTETVKAEKEIPGAGYRGQFPYSWGGYT 363
>CIKG_RAT (Q63734) Potassium voltage-gated channel subfamily C
member 4 (Voltage-gated potassium channel subunit Kv3.4)
(Raw3)
Length = 625
Score = 30.0 bits (66), Expect = 7.4
Identities = 17/75 (22%), Positives = 34/75 (44%), Gaps = 12/75 (16%)
Query: 177 WFLAHFPGFFSVDLNTDYLENYPVAARWKLPKG-----HGEGITYWSLLDRIQLDDVCYK 231
+F PG F+ Y+ NY + P E +T+W + D ++ C+
Sbjct: 95 FFFDRHPGVFA------YVLNYYRTGKLHCPADVCGPLFEEELTFWGI-DETDVEPCCWM 147
Query: 232 PYKEHREIQDFEEVF 246
Y++HR+ ++ ++F
Sbjct: 148 TYRQHRDAEEALDIF 162
>CIKG_HUMAN (Q03721) Potassium voltage-gated channel subfamily C
member 4 (Voltage-gated potassium channel subunit Kv3.4)
(KSHIIIC)
Length = 582
Score = 30.0 bits (66), Expect = 7.4
Identities = 17/75 (22%), Positives = 34/75 (44%), Gaps = 12/75 (16%)
Query: 177 WFLAHFPGFFSVDLNTDYLENYPVAARWKLPKG-----HGEGITYWSLLDRIQLDDVCYK 231
+F PG F+ Y+ NY + P E +T+W + D ++ C+
Sbjct: 94 FFFDRHPGVFA------YVLNYYRTGKLHCPADVCGPLFEEELTFWGI-DETDVEPCCWM 146
Query: 232 PYKEHREIQDFEEVF 246
Y++HR+ ++ ++F
Sbjct: 147 TYRQHRDAEEALDIF 161
>RNFG_BUCAI (P57217) Electron transport complex protein rnfG homolog
Length = 164
Score = 29.6 bits (65), Expect = 9.7
Identities = 19/71 (26%), Positives = 33/71 (45%), Gaps = 7/71 (9%)
Query: 119 RSNKRIELIYLTTMADGYAGMRNYSWGGMTLAYLYGELADACRPGHR---ALGGSVTLLT 175
++ K + I TT DGY+G N + AY GE+ +A H+ +G + L
Sbjct: 34 KNKKPVAAIVETTAPDGYSGAINM----LVAAYFNGEIINARVLSHKETPGIGDKIDLSI 89
Query: 176 TWFLAHFPGFF 186
+ ++ F G +
Sbjct: 90 SNWITRFTGMY 100
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.142 0.463
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,255,675
Number of Sequences: 164201
Number of extensions: 1795789
Number of successful extensions: 4023
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 4022
Number of HSP's gapped (non-prelim): 7
length of query: 304
length of database: 59,974,054
effective HSP length: 110
effective length of query: 194
effective length of database: 41,911,944
effective search space: 8130917136
effective search space used: 8130917136
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.5 bits)
S2: 65 (29.6 bits)
Medicago: description of AC139854.8


