

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139747.5 + phase: 0 /pseudo
(1608 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
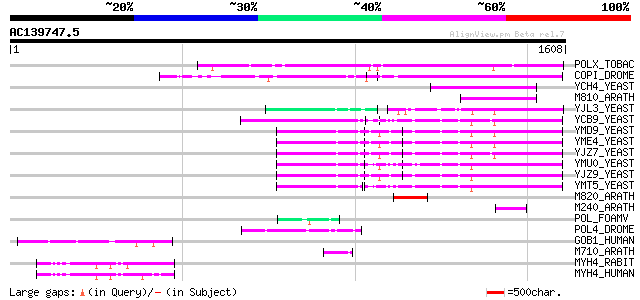
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 535 e-151
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 339 4e-92
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 181 1e-44
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 152 5e-36
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 134 3e-30
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 124 2e-27
YMD9_YEAST (Q03434) Transposon Ty1 protein B 122 8e-27
YME4_YEAST (Q04711) Transposon Ty1 protein B 119 7e-26
YJZ7_YEAST (P47098) Transposon Ty1 protein B 118 1e-25
YMU0_YEAST (Q04670) Transposon Ty1 protein B 116 4e-25
YJZ9_YEAST (P47100) Transposon Ty1 protein B 114 2e-24
YMT5_YEAST (Q04214) Transposon Ty1 protein B 113 4e-24
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 98 2e-19
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 63 6e-09
POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse transcript... 55 2e-06
POL4_DROME (P10394) Retrovirus-related Pol polyprotein from tran... 55 2e-06
GOB1_HUMAN (Q14789) Golgi autoantigen, golgin subfamily B member... 55 2e-06
M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710... 54 3e-06
MYH4_RABIT (Q28641) Myosin heavy chain, skeletal muscle, juvenile 52 1e-05
MYH4_HUMAN (Q9Y623) Myosin heavy chain, skeletal muscle, fetal (... 52 1e-05
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 535 bits (1378), Expect = e-151
Identities = 350/1112 (31%), Positives = 568/1112 (50%), Gaps = 68/1112 (6%)
Query: 544 RSESSQKAEKAKKTCFYCNKSDH-KRQNVTFRKDLLEELTLKDP--------TLHGYLKF 594
R +S +++ + C+ CN+ H KR RK E K+ + F
Sbjct: 218 RGKSKNRSKSRVRNCYNCNQPGHFKRDCPNPRKGKGETSGQKNDDNTAAMVQNNDNVVLF 277
Query: 595 LSCQM*VLPQGARTKPWYLDSGCSRHMTGDRNCFLTFEKKDGGLVTFGNNDKGKIRGKGT 654
++ + + W +D+ S H T R+ F + D G V GN KI G G
Sbjct: 278 INEEEECMHLSGPESEWVVDTAASHHATPVRDLFCRYVAGDFGTVKMGNTSYSKIAGIGD 337
Query: 655 I---GNLNSAKI-ENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEVRQASSNKLFFSG 710
I N+ + ++V++V L+ NL+S L G+E F + R + + G
Sbjct: 338 ICIKTNVGCTLVLKDVRHVPDLRMNLISGIALDRDGYESYFANQ--KWRLTKGSLVIAKG 395
Query: 711 SRRKNLYVLELNDMPAEFCFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFE 770
R LY E E +WHKR GH+S K + L++ L IS+
Sbjct: 396 VARGTLYRTNAEICQGELNAAQDEISVDLWHKRMGHMSEKGLQILAKKSL------ISYA 449
Query: 771 KD---KICEACVKGKQVKSSFKTIEFISTQKPLELLHI---DLFAPVQTASLTGKRYGFV 824
K K C+ C+ GKQ + SF+T S+++ L +L + D+ P++ S+ G +Y
Sbjct: 450 KGTTVKPCDYCLFGKQHRVSFQT----SSERKLNILDLVYSDVCGPMEIESMGGNKYFVT 505
Query: 825 IVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFD 884
+DD SR WV LK KD+ F+ FQ F V+ E G + +RSD+GGE+ + F+ +
Sbjct: 506 FIDDASRKLWVYILKTKDQVFQVFQKFHALVERETGRKLKRLRSDNGGEYTSREFEEYCS 565
Query: 885 ENGIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVS 944
+GI+H + TPQ NGV ER NRT+ E R+ML + + FW EA+ T+CY++NR
Sbjct: 566 SHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLRMAKLPKSFWGEAVQTACYLINRSP 625
Query: 945 IRKVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKG 1004
+ + P +W N + S S+ +FGC + KE+ K D KS IF+GY G
Sbjct: 626 SVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPKEQRTKLDDKSIPCIFIGYGDEEFG 685
Query: 1005 YRVYNLKTQTVEISMHIIFDEYDEHSKPKENEDTE---------APTLQNVPVQNTENTV 1055
YR+++ + V S ++F E + + +E + P+ N P + E+T
Sbjct: 686 YRLWDPVKKKVIRSRDVVFRESEVRTAADMSEKVKNGIIPNFVTIPSTSNNPT-SAESTT 744
Query: 1056 EKEDDQNVQ-----------DQSLQSPPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNN 1104
++ +Q Q D+ ++ + G+ + S V +R
Sbjct: 745 DEVSEQGEQPGEVIEQGEQLDEGVEEVEHPTQ--GEEQHQPLRRSERPRVESRRYPSTEY 802
Query: 1105 MAMISQMEPKSINEAII---DDSWIEVMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFR 1161
+ + EP+S+ E + + ++ M+EE+ ++N + LV + K + +WVF+
Sbjct: 803 VLISDDREPESLKEVLSHPEKNQLMKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVFK 862
Query: 1162 NKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMD 1221
K D + K+VR KARLV +G+ Q++GID+DE F+PV ++ +IR +L+ AA +++ Q+D
Sbjct: 863 LKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQLD 922
Query: 1222 VKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENG 1281
VK+AFL+G L EE+Y+ QP GF K + V KL K+LYGLKQAPR WY + +F+
Sbjct: 923 VKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQT 982
Query: 1282 FSRGKIDTTL-FRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELG 1340
+ + D + F++ + +I+ +YVDD++ + + + F+M +G
Sbjct: 983 YLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPAQ 1042
Query: 1341 FFLGLQI--KQHSNGIFISQEKYIKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISE 1398
LG++I ++ S +++SQEKYI+ +L+++ M AK +STP+ L K ++ E
Sbjct: 1043 QILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVEE 1102
Query: 1399 K------EYRGMIGSLLY-LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTT 1451
K Y +GSL+Y + +RPDI AVG+ +RF + H AVK I RYL GTT
Sbjct: 1103 KGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGTT 1162
Query: 1452 DLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIELSTTE 1511
L + GS L Y DAD AGD RKS++G I W + Q + LSTTE
Sbjct: 1163 GDCLCF-GGSDPILKGYTDADMAGDIDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTTE 1221
Query: 1512 AEYVSAASCCSQILWVRNQLEDYSLRYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHH 1571
AEY++A +++W++ L++ L +YCD+ SAI+LSKN + H+R+KHI++++H
Sbjct: 1222 AEYIAATETGKEMIWLKRFLQELGLHQKEYVVYCDSQSAIDLSKNSMYHARTKHIDVRYH 1281
Query: 1572 FIRDHVQKKNIALSFVDTENQLADIFTKPLVR 1603
+IR+ V +++ + + T AD+ TK + R
Sbjct: 1282 WIREMVDDESLKVLKISTNENPADMLTKVVPR 1313
Score = 40.0 bits (92), Expect = 0.052
Identities = 56/266 (21%), Positives = 104/266 (39%), Gaps = 56/266 (21%)
Query: 47 VPTTKEGAVKAKSAWSTDEKAQVLLNSKARLFLSCALTMEESERVDECTNAKEVWDTLKI 106
V + K +KA+ DE+A S RL LS + + +DE T A+ +W L+
Sbjct: 39 VDSKKPDTMKAEDWADLDERAA----SAIRLHLSDDVV---NNIIDEDT-ARGIWTRLES 90
Query: 107 HHEGTSHVKETRIDIGVRKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRK 166
+ + + + ++ MSE F ++ ++ +LG D+
Sbjct: 91 LYMSKTLTNKLYLK---KQLYALHMSEGTNFLSHLNVFNGLITQLANLGVKIEEEDKAIL 147
Query: 167 ILRCLPSVWRPMVTAITQAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVKTLALKASQQ 226
+L LPS + + T I K ++ L+D+ +L +E
Sbjct: 148 LLNSLPSSYDNLATTILHGKT--TIELKDVTSALLLNE---------------------- 183
Query: 227 TPSVADEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTE-A 285
++ ++ ALI++ R R + + ++ K++
Sbjct: 184 ----------------KMRKKPENQGQALITEGRGRSYQRSSN---NYGRSGARGKSKNR 224
Query: 286 DKSQV-TCYGCNKTGHFKNECPDIKK 310
KS+V CY CN+ GHFK +CP+ +K
Sbjct: 225 SKSRVRNCYNCNQPGHFKRDCPNPRK 250
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 339 bits (869), Expect = 4e-92
Identities = 192/576 (33%), Positives = 320/576 (55%), Gaps = 24/576 (4%)
Query: 1035 NEDTEAPTLQNVPVQNTENTVEKEDDQ--NVQDQSLQSPPRSWRMVGDHPTDQIIGSTTD 1092
NE E+ T +++ +N + + + N + + L++ P+ + + D +
Sbjct: 826 NESRESETAEHLKEIGIDNPTKNDGIEIINRRSERLKTKPQ----ISYNEEDNSLNKVVL 881
Query: 1093 GVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVPNNQDKT 1152
T + N+ I + KS SW E + EL+ + N W + ++K
Sbjct: 882 NAHTIFNDVPNSFDEIQYRDDKS--------SWEEAINTELNAHKINNTWTITKRPENKN 933
Query: 1153 IIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAH 1212
I+ +RWVF K +E G +R KARLVA+G+ Q+ IDY+ETFAPVAR+ + R +L+
Sbjct: 934 IVDSRWVFSVKYNELGNPIRYKARLVARGFTQKYQIDYEETFAPVARISSFRFILSLVIQ 993
Query: 1213 KSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDR 1272
++K+ QMDVK+AFLNG L EE+Y+ P G ++V KL KA+YGLKQA R W++
Sbjct: 994 YNLKVHQMDVKTAFLNGTLKEEIYMRLPQGI--SCNSDNVCKLNKAIYGLKQAARCWFEV 1051
Query: 1273 LSTFLIENGFSRGKIDTTLF--RKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSE 1330
L E F +D ++ K + + + V +YVDD++ + F + +
Sbjct: 1052 FEQALKECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEK 1111
Query: 1331 FEMSMMGELGFFLGLQIKQHSNGIFISQEKYIKDILKKYKMNEAKIMSTPMHPSSSLDKD 1390
F M+ + E+ F+G++I+ + I++SQ Y+K IL K+ M +STP+ + +
Sbjct: 1112 FRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAVSTPLPSKINYELL 1171
Query: 1391 ESGKSISEKEYRGMIGSLLY-LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVG 1449
S + + R +IG L+Y + +RPD+ AV + +R+ + +KR+ RYL G
Sbjct: 1172 NSDEDCN-TPCRSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKG 1230
Query: 1450 TTDLGLWYRKGSSFD--LVAYCDADYAGDKVERKSTSGSC-QFLGQALIGWSCRKQNTIE 1506
T D+ L ++K +F+ ++ Y D+D+AG +++RKST+G + LI W+ ++QN++
Sbjct: 1231 TIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVA 1290
Query: 1507 LSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTS-VPIYCDNTSAINLSKNPIQHSRSKH 1565
S+TEAEY++ + LW++ L +++ + + IY DN I+++ NP H R+KH
Sbjct: 1291 ASSTEAEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKRAKH 1350
Query: 1566 IEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPL 1601
I+IK+HF R+ VQ I L ++ TENQLADIFTKPL
Sbjct: 1351 IDIKYHFAREQVQNNVICLEYIPTENQLADIFTKPL 1386
Score = 190 bits (483), Expect = 2e-47
Identities = 164/669 (24%), Positives = 302/669 (44%), Gaps = 105/669 (15%)
Query: 434 GIMSVPSEVQKENILL----KKEVETLKKVLTGFIKSTETFQNIVGSQNESTKKSGLGFK 489
GI++ + +EN+ L + ++ K+ +++ N + N +T K+ L FK
Sbjct: 154 GIITAIETLSEENLTLAFVKNRLLDQEIKIKNDHNDTSKKVMNAIVHNNNNTYKNNL-FK 212
Query: 490 D----PSKIIGSFVPKAKIRVKCCFCDKYGHNESICHVKKKFIKQNNLYLSSERSHLNRS 545
+ P KI F +K +VKC C + GH + C K+ + N++
Sbjct: 213 NRVTKPKKI---FKGNSKYKVKCHHCGREGHIKKDCFHYKRILN-------------NKN 256
Query: 546 ESSQKAEKAKKTCFYCNKSDHKRQNVTFRKDLLEELTLKDPTLHGYLKFLSCQM*VLPQG 605
+ ++K + + + F + ++ D +C
Sbjct: 257 KENEKQVQTATS-----------HGIAFMVKEVNNTSVMD----------NCG------- 288
Query: 606 ARTKPWYLDSGCSRHMTGDRNCFLTFEKKDGGL----VTFGNNDKGKIRGKGTIGNLNSA 661
+ LDSG S H+ D + + + L G RG + N +
Sbjct: 289 -----FVLDSGASDHLINDESLYTDSVEVVPPLKIAVAKQGEFIYATKRGIVRLRNDHEI 343
Query: 662 KIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEVRQASSNKLFFSGSRRKNLYVLEL 721
+E+V + + NL+S+ +L ++G + F + + + + SG L
Sbjct: 344 TLEDVLFCKEAAGNLMSVKRLQEAGMSIEFDKSGVTISKNGLMVVKNSGM---------L 394
Query: 722 NDMPA----EFCFMSLEKDKW-IWHKRAGHIS------MKTIAKLSQLDLVRGLPKISFE 770
N++P + + K+ + +WH+R GHIS +K S L+ L ++S E
Sbjct: 395 NNVPVINFQAYSINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNL-ELSCE 453
Query: 771 KDKICEACVKGKQVKSSFKTI-EFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDDF 829
ICE C+ GKQ + FK + + ++PL ++H D+ P+ +L K Y + VD F
Sbjct: 454 ---ICEPCLNGKQARLPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQF 510
Query: 830 SRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDENGIK 889
+ + +K+K + F FQ+F + + ++ + D+G E+ + + F + GI
Sbjct: 511 THYCVTYLIKYKSDVFSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGIS 570
Query: 890 HNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIRKVL 949
++ + TPQ NGV ER RT+ E ARTM++ + ++ FW EA+ T+ Y++NR+ R ++
Sbjct: 571 YHLTVPHTPQLNGVSERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALV 630
Query: 950 --NKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRV 1007
+KTPYE+W N KP + + +FG Y+ + K K GKFD KS K+IF+GY G+++
Sbjct: 631 DSSKTPYEMWHNKKPYLKHLRVFGATVYV-HIKNKQGKFDDKSFKSIFVGYE--PNGFKL 687
Query: 1008 YNLKTQTVEISMHIIFDEYDEHSK------------PKENEDTEAPTLQNVPVQNTENTV 1055
++ + ++ ++ DE + + KE+E+ P +Q TE
Sbjct: 688 WDAVNEKFIVARDVVVDETNMVNSRAVKFETVFLKDSKESENKNFPNDSRKIIQ-TEFPN 746
Query: 1056 EKEDDQNVQ 1064
E ++ N+Q
Sbjct: 747 ESKECDNIQ 755
Score = 38.9 bits (89), Expect = 0.12
Identities = 79/446 (17%), Positives = 155/446 (34%), Gaps = 71/446 (15%)
Query: 14 FDGSNYYFWKGKMELFLRSQDNDMWAVITDGDFVPTTKEGAVKAKSAWSTDEKAQVLLNS 73
FDG Y WK ++ L QD + DG +P + + K + + L +S
Sbjct: 11 FDGEKYAIWKFRIRALLAEQDV---LKVVDG-LMPNEVDDSWKKAERCAKSTIIEYLSDS 66
Query: 74 KARLFLSCALTMEESERVDECTNAKEVWDTLKIHHEGTSHVKETRIDIGVRKFEVFEMSE 133
S + E +D K + L + ++ ++S
Sbjct: 67 FLNFATSDITARQILENLDAVYERKSLASQLALR----------------KRLLSLKLSS 110
Query: 134 NETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSVWRPMVTAITQAKDLKSMNL 193
++ + F +++E+ + G D+I +L LPS + ++TAI + +++ L
Sbjct: 111 EMSLLSHFHIFDELISELLAAGAKIEEMDKISHLLITLPSCYDGIITAIETLSE-ENLTL 169
Query: 194 EDLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQEPQELEEVHEEEAEDEL 253
+ L E+ ++ D D + VH +
Sbjct: 170 AFVKNRLLDQEIKIKND---------------------HNDTSKKVMNAIVHNNNNTYKN 208
Query: 254 ALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKV-Q 312
L R+ + KK K N K +V C+ C + GH K +C K++
Sbjct: 209 NLFKNRVTK--------PKKIFKGN-------SKYKVKCHHCGREGHIKKDCFHYKRILN 253
Query: 313 RKPPFKKKAMITWDD-----MEESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKDLEN 367
K +K + T M + + + G + S + +I +SLY D
Sbjct: 254 NKNKENEKQVQTATSHGIAFMVKEVNNTSVMDNCGFVLDSGASDHLI--NDESLYTDSVE 311
Query: 368 KIDSLLY------DSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQDSHFDM 421
+ L + + T R ++ EI E + + + ++K LQ++ +
Sbjct: 312 VVPPLKIAVAKQGEFIYATKRGIVRLRNDHEITLEDVLFCKEAAGNLMSVKRLQEAGMSI 371
Query: 422 SEKQREINRKQKGIMSVPSEVQKENI 447
+ + + G+M V + N+
Sbjct: 372 EFDKSGVTISKNGLMVVKNSGMLNNV 397
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 181 bits (459), Expect = 1e-44
Identities = 105/308 (34%), Positives = 167/308 (54%), Gaps = 3/308 (0%)
Query: 1220 MDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIE 1279
MDV +AFLN ++E +YV QPPGF+N+ P++V++L +YGLKQAP W + ++ L +
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 1280 NGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGEL 1339
GF R + + L+ ++ + + + VYVDD++ A K+ + + + M +G++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 1340 GFFLGLQIKQHSNG-IFISQEKYIKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISE 1398
FLGL I Q +NG I +S + YI + ++N K+ TP+ S L + S
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 1399 KEYRGMIGSLLY-LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWY 1457
Y+ ++G LL+ RPDI + V L +RF + HL + +R+ RYL T + L Y
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 1458 RKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRK-QNTIELSTTEAEYVS 1516
R GS L YCDA + ST G L A + WS +K + I + +TEAEY++
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 1517 AASCCSQI 1524
A+ +I
Sbjct: 301 ASETVMEI 308
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 152 bits (385), Expect = 5e-36
Identities = 80/222 (36%), Positives = 126/222 (56%), Gaps = 1/222 (0%)
Query: 1305 VYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYIKD 1364
+YVDDI+ + + + S F M +G + +FLG+QIK H +G+F+SQ KY +
Sbjct: 5 LYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYAEQ 64
Query: 1365 ILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGL 1424
IL M + K MSTP+ P + K ++R ++G+L YLT +RPDI +AV +
Sbjct: 65 ILNNAGMLDCKPMSTPL-PLKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYAVNI 123
Query: 1425 CARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTS 1484
+ + +KR+ RY+ GT GL+ K S ++ A+CD+D+AG R+ST+
Sbjct: 124 VCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRRSTT 183
Query: 1485 GSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILW 1526
G C FLG +I WS ++Q T+ S+TE EY + A +++ W
Sbjct: 184 GFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 134 bits (336), Expect = 3e-30
Identities = 143/552 (25%), Positives = 244/552 (43%), Gaps = 62/552 (11%)
Query: 1095 RTRLSFQDNNMAMISQMEPKSI--NEAIIDD-------SWIEVMKEELSQFERNKVWNL- 1144
+ R+ +NM +S + ++I NEAI + + + +EL + KV+++
Sbjct: 1250 KNRVKLIPDNMETVSAPKIRAIYYNEAISKNPDLKEKHEYKQAYHKELQNLKDMKVFDVD 1309
Query: 1145 -------VPNNQDKTIIGTRWVFRNKLDEEGKVVRN---KARLVAQGYNQQEGIDYDETF 1194
+P+N I+ T +F K RN KAR+V +G Q Y
Sbjct: 1310 VKYSRSEIPDN---LIVPTNTIFTKK--------RNGIYKARIVCRGDTQSPDT-YSVIT 1357
Query: 1195 APVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFK 1254
I+I L A ++++ + +D+ AFL L EE+Y+ P V K
Sbjct: 1358 TESLNHNHIKIFLMIANNRNMFMKTLDINHAFLYAKLEEEIYIPHP------HDRRCVVK 1411
Query: 1255 LTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGA 1314
L KALYGLKQ+P+ W D L +L G L+ +T + +L+I VYVDD + A
Sbjct: 1412 LNKALYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY-QTEDKNLMIA-VYVDDCVIAA 1469
Query: 1315 TKIKMCEEFSNLMQSEFEMSMMGEL------GFFLGLQI--KQHSNGIFISQEKYIKDIL 1366
+ + +EF N ++S FE+ + G L LG+ + + I ++ + +I +
Sbjct: 1470 SNEQRLDEFINKLKSNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFINRMD 1529
Query: 1367 KKYKMNEAKI--MSTPMHPSSSLDKDESGKSISEKEYR-------GMIGSLLYLT-ASRP 1416
KKY KI S P + +D + +SE+E+R ++G L Y+ R
Sbjct: 1530 KKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNYVRHKCRY 1589
Query: 1417 DIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD--LVAYCDADYA 1474
DI FAV AR E + +I +YLV D+G+ Y + + D ++A DA
Sbjct: 1590 DIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDAS-V 1648
Query: 1475 GDKVERKSTSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDY 1534
G + + +S G + G + K +S+TEAE + + ++ L++
Sbjct: 1649 GSEYDAQSRIGVILWYGMNIFNVYSNKSTNRCVSSTEAELHAIYEGYADSETLKVTLKEL 1708
Query: 1535 SL-RYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQL 1593
+ + D+ AI Q + K IK I++ +++K+I L + + +
Sbjct: 1709 GEGDNNDIVMITDSKPAIQGLNRSYQQPKEKFTWIKTEIIKEKIKEKSIKLLKITGKGNI 1768
Query: 1594 ADIFTKPLVRID 1605
AD+ TKP+ D
Sbjct: 1769 ADLLTKPVSASD 1780
Score = 79.7 bits (195), Expect = 6e-14
Identities = 76/332 (22%), Positives = 133/332 (39%), Gaps = 19/332 (5%)
Query: 741 HKRAGHISMKTIAK-LSQLDLVRGLPKISFEKDKICEACVKGKQVKSSFKTIEF---IST 796
HKR GH ++ I + L I + C+ C K K + T +
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 797 QKPLELLHIDLFAPVQTASLTGKRYGFVIVDDFSRF--TWVLFLKHKDESFEAFQNFCKR 854
+P +D+F PV +++ KRY ++VD+ +R+ T F K+ + + +
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 855 VQNEKGYNIITVRSDHGGEFENASFKTFFDENGIKHNFSCARTPQQNGVVERKNRTLQEM 914
V+ + + + SD G EF N + +F GI H + + NG ER RT+
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 915 ARTMLNESNVENYFWAEAINTSCYILNRVSIRKVLNKTPYELWKN--IKPSISYFHIFGC 972
A T+L +SN+ FW A+ ++ I N + K K P + + + F FG
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRNYLE-HKSTGKLPLKAISRQPVTVRLMSFLPFGE 800
Query: 973 YCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDEYDEHSKP 1032
I N+ K K P +I L S GY+ + + S D ++ P
Sbjct: 801 KGIIWNHNHK--KLKPSGLPSIILCKDPNSYGYKFFIPSKNKIVTS--------DNYTIP 850
Query: 1033 KENEDTEAPTLQNVPVQNTENTVEKEDDQNVQ 1064
D QN+ + ++ +++ ++
Sbjct: 851 NYTMDGRVRNTQNINKSHQFSSDNDDEEDQIE 882
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 124 bits (312), Expect = 2e-27
Identities = 144/602 (23%), Positives = 266/602 (43%), Gaps = 67/602 (11%)
Query: 1030 SKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIGS 1089
SK + ED E TE V ++ N +SL+ PPRS + + + + S
Sbjct: 1192 SKKRSLEDNE-----------TEIEVSRDTWNNKNMRSLE-PPRSKKRINLIAAIKGVKS 1239
Query: 1090 TTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVW--NLVPN 1147
VRT L + + A+ + K D ++E +E+SQ + W N +
Sbjct: 1240 IKP-VRTTLRYDE---AITYNKDNKE------KDRYVEAYHKEISQLLKMNTWDTNKYYD 1289
Query: 1148 NQD---KTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIR 1204
D K +I + ++F K D +KAR VA+G Q + + A+
Sbjct: 1290 RNDIDPKKVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDSDMQSNTVHHYALM 1344
Query: 1205 ILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQ 1264
L+ A + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLKQ
Sbjct: 1345 TSLSIALDNDYYITQLDISSAYLYADIKEELYIRPPPHLGLNDK---LLRLRKSLYGLKQ 1401
Query: 1265 APRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEF 1323
+ WY+ + ++LI + + +F+ + T + ++VDD+I + + ++
Sbjct: 1402 SGANWYETIKSYLINCCDMQEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKKI 1457
Query: 1324 SNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKYKMN--- 1372
++ +++ ++ E+ + LGL+IK Q S + + EK + + L K +
Sbjct: 1458 ITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRSKYMKLGMEKSLTEKLPKLNVPLNP 1517
Query: 1373 EAKIMSTPMHPSSSLDKDESGKSISEKEYRG-------MIGSLLYLTAS-RPDIVFAVGL 1424
+ K + P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1518 KGKKLRAPGQPGHYIDQDEL--EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINT 1575
Query: 1425 CARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVER 1480
A+ L + +++ T D L + K LVA DA Y G++
Sbjct: 1576 LAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASY-GNQPYY 1634
Query: 1481 KSTSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTS 1540
KS G+ L +IG K + STTEAE + + + + + +++ + +
Sbjct: 1635 KSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPII 1694
Query: 1541 VPIYCDNTSAINLSKNPIQHS-RSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTK 1599
+ D+ S I++ K+ + R++ K +RD V N+ + +++T+ +AD+ TK
Sbjct: 1695 KGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMTK 1754
Query: 1600 PL 1601
PL
Sbjct: 1755 PL 1756
Score = 100 bits (250), Expect = 2e-20
Identities = 101/425 (23%), Positives = 180/425 (41%), Gaps = 28/425 (6%)
Query: 668 YVEGLKHNLLSISQLCDSGFEVIFKPNICEVRQASSNKLF-----FSGSRRKNLYVLELN 722
+ + ++LLS+S+L + F N E + F +K L ++
Sbjct: 514 HTPNIAYDLLSLSELANQNITACFTRNTLERSDGTVLAPIVKHGDFYWLSKKYLIPSHIS 573
Query: 723 DMPAEFCFMSLEKDKWIW---HKRAGHISMKTIAKLSQLDLVRGLPKISFEKDKI----C 775
+ S +K+ + H+ GH + ++I K + + V L + E C
Sbjct: 574 KLTINNVNKSKSVNKYPYPLIHRMLGHANFRSIQKSLKKNAVTYLKESDIEWSNASTYQC 633
Query: 776 EACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDDFSRF 832
C+ GK K +++ + +P + LH D+F PV + Y D+ +RF
Sbjct: 634 PDCLIGKSTKHRHVKGSRLKYQESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRF 693
Query: 833 TWVLFLKHKDES--FEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDENGIKH 890
WV L + E F + ++N+ ++ ++ D G E+ N + FF GI
Sbjct: 694 QWVYPLHDRREESILNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITA 753
Query: 891 NFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIRKVLN 950
++ + +GV ER NRTL RT+L+ S + N+ W A+ S I N + + +
Sbjct: 754 CYTTTADSRAHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSL-VSPKND 812
Query: 951 KTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVY-- 1008
K+ + I+ FG I+NN K P+ L S S GY +Y
Sbjct: 813 KSARQHAGLAGLDITTILPFG-QPVIVNNHNPDSKIHPRGIPGYALHPSRNSYGYIIYLP 871
Query: 1009 NLKTQTVEISMHIIFDEYDEHSKPKE-NEDTEA--PTLQNVPVQNTENTVEKEDDQNVQD 1065
+LK +TV+ + ++I D+ SK + N DT L + N ++ +E+ + + D
Sbjct: 872 SLK-KTVDTTNYVILQ--DKQSKLDQFNYDTLTFDDDLNRLTAHN-QSFIEQNETEQSYD 927
Query: 1066 QSLQS 1070
Q+ +S
Sbjct: 928 QNTES 932
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 122 bits (306), Expect = 8e-27
Identities = 139/603 (23%), Positives = 265/603 (43%), Gaps = 67/603 (11%)
Query: 1029 HSKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIG 1088
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 1089 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVP-- 1146
S +RT L + + + K I E + +IE +E++Q + K W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDEYY 846
Query: 1147 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1203
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 847 DRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 901
Query: 1204 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1263
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 902 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 958
Query: 1264 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1322
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + + ++
Sbjct: 959 QSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKK 1014
Query: 1323 FSNLMQSEFEMSMM------GELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKYKM--- 1371
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1015 IITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1074
Query: 1372 NEAKIMSTPMHPSSSLDKDESGKSISEKEYR-------GMIGSLLYLTAS-RPDIVFAVG 1423
+ + +S P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1075 PKGRKLSAPGQPGLYIDQDE--LEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYIN 1132
Query: 1424 LCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVE 1479
A+ L + +++ T D L + K + LVA DA Y G++
Sbjct: 1133 TLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPY 1191
Query: 1480 RKSTSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSLRYT 1539
KS G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1192 YKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPI 1251
Query: 1540 SVPIYCDNTSAINLSKNPIQHS-RSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFT 1598
+ D+ S I++ K+ + R++ K +RD V N+ + +++T+ +AD+ T
Sbjct: 1252 IKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMT 1311
Query: 1599 KPL 1601
KPL
Sbjct: 1312 KPL 1314
Score = 96.7 bits (239), Expect = 5e-19
Identities = 97/387 (25%), Positives = 167/387 (43%), Gaps = 28/387 (7%)
Query: 772 DKICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDD 828
D C C+ GK K +++ ++ +P + LH D+F PV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 829 FSRFTWVLFL--KHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 886
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI ++ + +GV ER NRTL + RT L S + N+ W AI S + N ++
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
Query: 947 KVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYR 1006
K K+ + IS FG I+N+ K P+ L S S GY
Sbjct: 387 K-SKKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYI 444
Query: 1007 VY--NLKTQTVEISMHIIF----DEYDEHSKPKENEDTEAPTLQNVPVQN--TENTVEKE 1058
+Y +LK +TV+ + ++I D+ + D + L Q+ N +++
Sbjct: 445 IYLPSLK-KTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRL-TASYQSFIASNEIQQS 502
Query: 1059 DDQNVQ-DQSLQS-------PPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQ 1110
DD N++ D QS PR+ PTD ST R+S N+ +
Sbjct: 503 DDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVS--KTNIRAPRE 560
Query: 1111 MEPKSINEAIIDDSWIEVMKEELSQFE 1137
++P +I+E+ I S ++S E
Sbjct: 561 VDP-NISESNILPSKKRSSTPQISNIE 586
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 119 bits (298), Expect = 7e-26
Identities = 137/603 (22%), Positives = 265/603 (43%), Gaps = 67/603 (11%)
Query: 1029 HSKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIG 1088
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 1089 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVP-- 1146
S +RT L + + + K I E + +IE +E++Q + W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMNTWDTDKYY 846
Query: 1147 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1203
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 847 DRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 901
Query: 1204 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1263
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 902 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 958
Query: 1264 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1322
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + + ++
Sbjct: 959 QSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKK 1014
Query: 1323 FSNLMQSEFEMSMM------GELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKYKM--- 1371
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1015 IITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1074
Query: 1372 NEAKIMSTPMHPSSSLDKDESGKSISEKEYR-------GMIGSLLYLTAS-RPDIVFAVG 1423
+ + +S P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1075 PKGRKLSAPGQPGLYIDQDE--LEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYIN 1132
Query: 1424 LCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVE 1479
A+ L + +++ T D L + K + LVA DA Y G++
Sbjct: 1133 TLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPY 1191
Query: 1480 RKSTSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSLRYT 1539
KS G+ L +IG K + STTEAE + + + + + +++ + +
Sbjct: 1192 YKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPI 1251
Query: 1540 SVPIYCDNTSAIN-LSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFT 1598
+ + D+ S I+ + N + R++ K +RD V ++ + +++T+ +AD+ T
Sbjct: 1252 TKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMT 1311
Query: 1599 KPL 1601
KPL
Sbjct: 1312 KPL 1314
Score = 96.7 bits (239), Expect = 5e-19
Identities = 97/387 (25%), Positives = 167/387 (43%), Gaps = 28/387 (7%)
Query: 772 DKICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDD 828
D C C+ GK K +++ ++ +P + LH D+F PV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 829 FSRFTWVLFL--KHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 886
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI ++ + +GV ER NRTL + RT L S + N+ W AI S + N ++
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
Query: 947 KVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYR 1006
K K+ + IS FG I+N+ K P+ L S S GY
Sbjct: 387 K-SKKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYI 444
Query: 1007 VY--NLKTQTVEISMHIIF----DEYDEHSKPKENEDTEAPTLQNVPVQN--TENTVEKE 1058
+Y +LK +TV+ + ++I D+ + D + L Q+ N +++
Sbjct: 445 IYLPSLK-KTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRL-TASYQSFIASNEIQQS 502
Query: 1059 DDQNVQ-DQSLQS-------PPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQ 1110
DD N++ D QS PR+ PTD ST R+S N+ +
Sbjct: 503 DDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVS--KTNIRAPRE 560
Query: 1111 MEPKSINEAIIDDSWIEVMKEELSQFE 1137
++P +I+E+ I S ++S E
Sbjct: 561 VDP-NISESNILPSKKRSSTPQISNIE 586
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 118 bits (296), Expect = 1e-25
Identities = 138/603 (22%), Positives = 264/603 (42%), Gaps = 67/603 (11%)
Query: 1029 HSKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIG 1088
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 1176 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 1223
Query: 1089 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVP-- 1146
S +RT L + + + K I E + +IE +E++Q + K W+
Sbjct: 1224 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDEYY 1273
Query: 1147 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1203
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 1274 DRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDTGMQSNTVHHYAL 1328
Query: 1204 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1263
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+ YGLK
Sbjct: 1329 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSHYGLK 1385
Query: 1264 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1322
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + + ++
Sbjct: 1386 QSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDLNANKK 1441
Query: 1323 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKYKMN-- 1372
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1442 IITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1501
Query: 1373 -EAKIMSTPMHPSSSLDKDESGKSISEKEYRG-------MIGSLLYLTAS-RPDIVFAVG 1423
+ + +S P P +D+DE I E EY+ +IG Y+ R D+++ +
Sbjct: 1502 PKGRKLSAPGQPGLYIDQDEL--EIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYIN 1559
Query: 1424 LCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFD----LVAYCDADYAGDKVE 1479
A+ L + +++ T D L + K + LVA DA Y G++
Sbjct: 1560 TLAQHILFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPY 1618
Query: 1480 RKSTSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSLRYT 1539
KS G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1619 YKSQIGNIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPI 1678
Query: 1540 SVPIYCDNTSAINLSKNPIQHS-RSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFT 1598
+ D+ S I++ K+ + R++ K +RD V N+ + +++T+ +AD+ T
Sbjct: 1679 IKGLLTDSRSTISIIKSTNEEKFRNRFFGTKAMRLRDEVSGNNLYVYYIETKKNIADVMT 1738
Query: 1599 KPL 1601
KPL
Sbjct: 1739 KPL 1741
Score = 94.7 bits (234), Expect = 2e-18
Identities = 96/387 (24%), Positives = 167/387 (42%), Gaps = 28/387 (7%)
Query: 772 DKICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDD 828
D C C+ GK K +++ ++ +P + LH D+F PV + Y D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 829 FSRFTWVLFL--KHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 886
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 753
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI ++ + +GV ER NRTL + RT L S + N+ W AI S + N ++
Sbjct: 754 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 813
Query: 947 KVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYR 1006
K K+ + IS FG I+N+ K P+ L S S GY
Sbjct: 814 K-SKKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYI 871
Query: 1007 VY--NLKTQTVEISMHIIF----DEYDEHSKPKENEDTEAPTLQNVPVQN--TENTVEKE 1058
+Y +LK +TV+ + ++I D+ + D + L Q+ N +++
Sbjct: 872 IYLPSLK-KTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRL-TASYQSFIASNEIQES 929
Query: 1059 DDQNVQ-DQSLQS-------PPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQ 1110
+D N++ D QS PR+ PTD ST R+S N+ +
Sbjct: 930 NDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVS--KTNIRAPRE 987
Query: 1111 MEPKSINEAIIDDSWIEVMKEELSQFE 1137
++P +I+E+ I S ++S E
Sbjct: 988 VDP-NISESNILPSKKRSSTPQISNIE 1013
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 116 bits (291), Expect = 4e-25
Identities = 134/606 (22%), Positives = 269/606 (44%), Gaps = 73/606 (12%)
Query: 1029 HSKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIG 1088
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 1089 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLV--- 1145
S +RT L + + + K I E + +I+ +E++Q + K W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIQAYHKEVNQLLKMKTWDTDRYY 846
Query: 1146 --PNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLE-- 1201
K +I + ++F K D +KAR VA+G I + +T+ P +
Sbjct: 847 DRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARG-----DIQHPDTYDPGMQSNTV 896
Query: 1202 ---AIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKA 1258
A+ L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+
Sbjct: 897 HHYALMTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKS 953
Query: 1259 LYGLKQAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKI 1317
LYGLKQ+ WY+ + ++LI+ G + + +F+ + T + ++VDD+I + +
Sbjct: 954 LYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVFKNSQVT----ICLFVDDMILFSKDL 1009
Query: 1318 KMCEEFSNLMQSEFEMSMM------GELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKY 1369
++ ++ +++ ++ E+ + LGL+IK Q + + E + + + K
Sbjct: 1010 NANKKIITTLKKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKL 1069
Query: 1370 KM---NEAKIMSTPMHPSSSLDK-----DESGKSISEKEYRGMIGSLLYLTAS-RPDIVF 1420
+ + + +S P P +D+ +E + E + +IG Y+ R D+++
Sbjct: 1070 NVPLNPKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLY 1129
Query: 1421 AVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGS----SFDLVAYCDADYAGD 1476
+ A+ + L + +++ T D L + K + LV DA Y G+
Sbjct: 1130 YINTLAQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GN 1188
Query: 1477 KVERKSTSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSL 1536
+ KS G+ L +IG K + STTEAE + + + + + +++ +
Sbjct: 1189 QPYYKSQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNK 1248
Query: 1537 RYTSVPIYCDNTSAIN-LSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLAD 1595
+ + + D+ S I+ + N + R++ K +RD V ++ + +++T+ +AD
Sbjct: 1249 KPITKGLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIAD 1308
Query: 1596 IFTKPL 1601
+ TKPL
Sbjct: 1309 VMTKPL 1314
Score = 96.7 bits (239), Expect = 5e-19
Identities = 97/387 (25%), Positives = 167/387 (43%), Gaps = 28/387 (7%)
Query: 772 DKICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDD 828
D C C+ GK K +++ ++ +P + LH D+F PV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 829 FSRFTWVLFL--KHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 886
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI ++ + +GV ER NRTL + RT L S + N+ W AI S + N ++
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
Query: 947 KVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYR 1006
K K+ + IS FG I+N+ K P+ L S S GY
Sbjct: 387 K-SKKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYI 444
Query: 1007 VY--NLKTQTVEISMHIIF----DEYDEHSKPKENEDTEAPTLQNVPVQN--TENTVEKE 1058
+Y +LK +TV+ + ++I D+ + D + L Q+ N +++
Sbjct: 445 IYLPSLK-KTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRL-TASYQSFIASNEIQQS 502
Query: 1059 DDQNVQ-DQSLQS-------PPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQ 1110
DD N++ D QS PR+ PTD ST R+S N+ +
Sbjct: 503 DDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVS--KTNIRAPRE 560
Query: 1111 MEPKSINEAIIDDSWIEVMKEELSQFE 1137
++P +I+E+ I S ++S E
Sbjct: 561 VDP-NISESNILPSKKRSSTPQISNIE 586
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 114 bits (285), Expect = 2e-24
Identities = 132/601 (21%), Positives = 261/601 (42%), Gaps = 63/601 (10%)
Query: 1029 HSKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIG 1088
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 1176 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 1223
Query: 1089 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVP-- 1146
S +RT L + + + K I E + +IE +E++Q + K W+
Sbjct: 1224 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDEYY 1273
Query: 1147 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1203
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 1274 DRKEIDPKRVINSMFIFNKKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 1328
Query: 1204 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1263
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 1329 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 1385
Query: 1264 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1322
Q+ WY+ + ++LI+ G + + +F+ + T + ++VDD++ + + +
Sbjct: 1386 QSGANWYETIKSYLIQQCGMEEVRGWSCVFKNSQVT----ICLFVDDMVLFSKNLNSNKR 1441
Query: 1323 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKYKM--- 1371
++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1442 IIEKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1501
Query: 1372 NEAKIMSTPMHPSSSLDK-----DESGKSISEKEYRGMIGSLLYLTAS-RPDIVFAVGLC 1425
+ + +S P P +D+ +E + E + +IG Y+ R D+++ +
Sbjct: 1502 PKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTL 1561
Query: 1426 ARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGS----SFDLVAYCDADYAGDKVERK 1481
A+ + L + +++ T D L + K + LV DA Y G++ K
Sbjct: 1562 AQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYK 1620
Query: 1482 STSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSV 1541
S G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1621 SQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITK 1680
Query: 1542 PIYCDNTSAIN-LSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKP 1600
+ D+ S I+ + N + R++ K +RD V ++ + +++T+ +AD+ TKP
Sbjct: 1681 GLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKP 1740
Query: 1601 L 1601
L
Sbjct: 1741 L 1741
Score = 96.7 bits (239), Expect = 5e-19
Identities = 97/387 (25%), Positives = 167/387 (43%), Gaps = 28/387 (7%)
Query: 772 DKICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDD 828
D C C+ GK K +++ ++ +P + LH D+F PV + Y D+
Sbjct: 634 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 693
Query: 829 FSRFTWVLFL--KHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 886
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 694 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 753
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI ++ + +GV ER NRTL + RT L S + N+ W AI S + N ++
Sbjct: 754 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 813
Query: 947 KVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYR 1006
K K+ + IS FG I+N+ K P+ L S S GY
Sbjct: 814 K-SKKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYI 871
Query: 1007 VY--NLKTQTVEISMHIIF----DEYDEHSKPKENEDTEAPTLQNVPVQN--TENTVEKE 1058
+Y +LK +TV+ + ++I D+ + D + L Q+ N +++
Sbjct: 872 IYLPSLK-KTVDTTNYVILQGKESRLDQFNYDALTFDEDLNRL-TASYQSFIASNEIQQS 929
Query: 1059 DDQNVQ-DQSLQS-------PPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQ 1110
DD N++ D QS PR+ PTD ST R+S N+ +
Sbjct: 930 DDLNIESDHDFQSDIELHPEQPRNVLSKAVSPTDSTPPSTHTEDSKRVS--KTNIRAPRE 987
Query: 1111 MEPKSINEAIIDDSWIEVMKEELSQFE 1137
++P +I+E+ I S ++S E
Sbjct: 988 VDP-NISESNILPSKKRSSTPQISNIE 1013
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 113 bits (283), Expect = 4e-24
Identities = 132/601 (21%), Positives = 261/601 (42%), Gaps = 63/601 (10%)
Query: 1029 HSKPKENEDTEAPTLQNVPVQNTENTVEKEDDQNVQDQSLQSPPRSWRMVGDHPTDQIIG 1088
+SK + ED E TE V + D N ++ PPRS + + + +
Sbjct: 749 NSKKRSLEDNE-----------TEIKVSR-DTWNTKNMRSLEPPRSKKRIHLIAAVKAVK 796
Query: 1089 STTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVP-- 1146
S +RT L + + + K I E + +IE +E++Q + K W+
Sbjct: 797 SIKP-IRTTLRYDE------AITYNKDIKEK---EKYIEAYHKEVNQLLKMKTWDTDKYY 846
Query: 1147 ---NNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAI 1203
K +I + ++F K D +KAR VA+G Q + A+
Sbjct: 847 DRKEIDPKRVINSMFIFNRKRDGT-----HKARFVARGDIQHPDTYDSGMQSNTVHHYAL 901
Query: 1204 RILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLK 1263
L+ A + + Q+D+ SA+L + EE+Y+ PP +K + +L K+LYGLK
Sbjct: 902 MTSLSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMNDK---LIRLKKSLYGLK 958
Query: 1264 QAPRAWYDRLSTFLIEN-GFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEE 1322
Q+ WY+ + ++LI+ G + + +F + T + ++VDD++ + + +
Sbjct: 959 QSGANWYETIKSYLIKQCGMEEVRGWSCVFENSQVT----ICLFVDDMVLFSKNLNSNKR 1014
Query: 1323 FSNLMQSEFEMSMMG------ELGF-FLGLQIK-QHSNGIFISQEKYIKDILKKYKM--- 1371
+ ++ +++ ++ E+ + LGL+IK Q + + E + + + K +
Sbjct: 1015 IIDKLKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLN 1074
Query: 1372 NEAKIMSTPMHPSSSLDK-----DESGKSISEKEYRGMIGSLLYLTAS-RPDIVFAVGLC 1425
+ + +S P P +D+ +E + E + +IG Y+ R D+++ +
Sbjct: 1075 PKGRKLSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTL 1134
Query: 1426 ARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGS----SFDLVAYCDADYAGDKVERK 1481
A+ + L + +++ T D L + K + LV DA Y G++ K
Sbjct: 1135 AQHILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYK 1193
Query: 1482 STSGSCQFLGQALIGWSCRKQNTIELSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSV 1541
S G+ L +IG K + STTEAE + + + + +++ + +
Sbjct: 1194 SQIGNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITK 1253
Query: 1542 PIYCDNTSAIN-LSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKP 1600
+ D+ S I+ + N + R++ K +RD V ++ + +++T+ +AD+ TKP
Sbjct: 1254 GLLTDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMTKP 1313
Query: 1601 L 1601
L
Sbjct: 1314 L 1314
Score = 94.0 bits (232), Expect = 3e-18
Identities = 69/258 (26%), Positives = 119/258 (45%), Gaps = 10/258 (3%)
Query: 772 DKICEACVKGKQVKSSF---KTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDD 828
D C C+ GK K +++ ++ +P + LH D+F PV + Y D+
Sbjct: 207 DYQCPDCLIGKSTKHRHIKGSRLKYQNSYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDE 266
Query: 829 FSRFTWVLFL--KHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 886
++F WV L + +D + F ++N+ +++ ++ D G E+ N + F ++N
Sbjct: 267 TTKFRWVYPLHDRREDSILDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKN 326
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI ++ + +GV ER NRTL + RT L S + N+ W AI S + N ++
Sbjct: 327 GITPCYTTTADSRAHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
Query: 947 KVLNKTPYELWKNIKPSISYFHIFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYR 1006
K K+ + IS FG I+N+ K P+ L S S GY
Sbjct: 387 K-SKKSARQHAGLAGLDISTLLPFG-QPVIVNDHNPNSKIHPRGIPGYALHPSRNSYGYI 444
Query: 1007 VY--NLKTQTVEISMHII 1022
+Y +LK +TV+ + ++I
Sbjct: 445 IYLPSLK-KTVDTTNYVI 461
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 97.8 bits (242), Expect = 2e-19
Identities = 47/99 (47%), Positives = 66/99 (66%)
Query: 1112 EPKSINEAIIDDSWIEVMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVV 1171
EPKS+ A+ D W + M+EEL RNK W LVP ++ I+G +WVF+ KL +G +
Sbjct: 27 EPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGCKWVFKTKLHSDGTLD 86
Query: 1172 RNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYA 1210
R KARLVA+G++Q+EGI + ET++PV R IR +L A
Sbjct: 87 RLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 63.2 bits (152), Expect = 6e-09
Identities = 29/88 (32%), Positives = 50/88 (55%)
Query: 1409 LYLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAY 1468
+YLT +RPD+ FAV ++F + ++ + + AV ++ Y+ GT GL+Y S L A+
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 1469 CDADYAGDKVERKSTSGSCQFLGQALIG 1496
D+D+A R+S +G C + +G
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLVPLWFLG 88
>POL_FOAMV (P14350) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)]
Length = 886
Score = 54.7 bits (130), Expect = 2e-06
Identities = 49/190 (25%), Positives = 76/190 (39%), Gaps = 25/190 (13%)
Query: 775 CEAC-VKGKQVKSSFKTIEFISTQKPLELLHIDLFAPVQTASLTGKRYGFVIVDDFSRFT 833
C+ C + K+S + QKP + ID P+ + G Y V+VD + FT
Sbjct: 650 CQQCLITNASNKASGPILRPDRPQKPFDKFFIDYIGPLPPSQ--GYLYVLVVVDGMTGFT 707
Query: 834 WVLFLKHKDESFEAFQNFCKRVQNEKGYNIIT-------VRSDHGGEFENASFKTFFDEN 886
W+ K S K N++T + SD G F +++F + E
Sbjct: 708 WLYPTKAPSTSATV-----------KSLNVLTSIAIPKVIHSDQGAAFTSSTFAEWAKER 756
Query: 887 GIKHNFSCARTPQQNGVVERKNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIR 946
GI FS PQ VERKN ++ + +L + W + + LN +
Sbjct: 757 GIHLEFSTPYHPQSGSKVERKNSDIKRLLTKLLVGRPTK---WYDLLPVVQLALNN-TYS 812
Query: 947 KVLNKTPYEL 956
VL TP++L
Sbjct: 813 PVLKYTPHQL 822
>POL4_DROME (P10394) Retrovirus-related Pol polyprotein from
transposon 412 [Contains: Protease (EC 3.4.23.-); Reverse
transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1237
Score = 54.7 bits (130), Expect = 2e-06
Identities = 78/361 (21%), Positives = 147/361 (40%), Gaps = 29/361 (8%)
Query: 672 LKHNLLSISQLCDSGFEVIFKPNICEVRQASSNKLFFSGSRRKNLYVLELNDMPAEFCFM 731
L+ + ISQ+ + ++ IF+ + + NK+ + V ++N+ + +
Sbjct: 838 LQAGIYDISQIKMAPWKKIFEHVSIDKFKNMGNKILKNLKVALLNPVTQINNEKEKEAIL 897
Query: 732 SLEKDKWIWHKRAGHISM-KTIAKLSQLDLVRGLPKISFEKDKICEACVKGKQVKSSFKT 790
S D I + GH + KT+AK+ + + + K E + C+ C K K K +
Sbjct: 898 STLHDDPI---QGGHTGITKTLAKVKRHYYWKNMSKYIKEYVRKCQKCQKAKTTKHTKTP 954
Query: 791 IEFIST-QKPLELLHIDLFAPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEA-- 847
+ T + + + +D P+ + G Y ++ D +++ + + +K A
Sbjct: 955 MTITETPEHAFDRVVVDTIGPLPKSE-NGNEYAVTLICDLTKYLVAIPIANKSAKTVAKA 1013
Query: 848 -FQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDENGIKHNFSCARTPQQNGVVER 906
F++F + K T +D G E++N+ IK+ S A Q GVVER
Sbjct: 1014 IFESFILKYGPMK-----TFITDMGTEYKNSIITDLCKYLKIKNITSTAHHHQTVGVVER 1068
Query: 907 KNRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIRKVLNKTPYEL----WKNIKP 962
+RTL E R+ ++ + W + Y N V N PYEL N+
Sbjct: 1069 SHRTLNEYIRSYISTDKTD---WDVWLQYFVYCFNTTQ-SMVHNYCPYELVFGRTSNLPK 1124
Query: 963 SISYFH----IFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEIS 1018
+ H I+ Y +K +L ++ K + + K Y+LK + +E+
Sbjct: 1125 HFNKLHSIEPIYNIDDYAKESKYRLEVAYARARKLL---EAHKEKNKENYDLKVKDIELE 1181
Query: 1019 M 1019
+
Sbjct: 1182 V 1182
>GOB1_HUMAN (Q14789) Golgi autoantigen, golgin subfamily B member 1
(Giantin) (Macrogolgin) (Golgi complex-associated
protein, 372-kDa) (GCP372)
Length = 3259
Score = 54.7 bits (130), Expect = 2e-06
Identities = 92/479 (19%), Positives = 203/479 (42%), Gaps = 47/479 (9%)
Query: 24 GKMELFLRSQDNDMWAVITDGDFVPTTKEGAVKAKSAWSTDEKAQVLLNSKA--RLFLSC 81
G++++ L +D ++ + + D T K+ S+ D++ +V+ +K R F
Sbjct: 2160 GEIQVTLNKKDKEVQQLQENLDSTVTQLAAFTKSMSSLQ-DDRDRVIDEAKKWERKFSDA 2218
Query: 82 ALTMEESERV--DECTNAKEVWDTLKIHHEGTSHVKETRID----IGVRKFEVFEMSENE 135
+ EE R+ D C+ K+ + IH E + +R++ I K + + +
Sbjct: 2219 IQSKEEEIRLKEDNCSVLKDQLRQMSIHMEELK-INISRLEHDKQIWESKAQTEVQLQQK 2277
Query: 136 TIDEMYARFTTIVNEMRSLGKAY-STHDRIRKI---LRCLPSVWRPMVTAITQAKDLKSM 191
D + +++++ Y S+ + + K+ L+ L + ++ + K+ K
Sbjct: 2278 VCDTLQGENKELLSQLEETRHLYHSSQNELAKLESELKSLKDQLTDLSNSLEKCKEQKG- 2336
Query: 192 NLEDLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQEPQE----LEEVHEE 247
NLE G +R E +Q K + L+AS++ S E++ ++ L EE
Sbjct: 2337 NLE---GIIRQQEADIQNSKFSYEQLETDLQASRELTSRLHEEINMKEQKIISLLSGKEE 2393
Query: 248 EAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPD 307
+ +A + ++ + + + + + NI ++ E K+ +KT N+ +
Sbjct: 2394 AIQVAIAELRQQHDKEIKELENLLSQEEEENIVLEEENKKA------VDKT----NQLME 2443
Query: 308 IKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKD--- 364
K +K ++KA + S Q D D +G Q ++ II + D L ++
Sbjct: 2444 TLKTIKKENIQQKAQLDSFVKSMSSLQNDRDRIVGDYQQLEERHLSIILEKDQLIQEAAA 2503
Query: 365 ----LENKIDSLLYDSNFLTNRCHSLIKELSEIKEE-KEILQNKYDESRKTIKI------ 413
L+ +I L + L + L EL + +E+ +++ K + ++ +++
Sbjct: 2504 ENNKLKEEIRGLRSHMDDLNSENAKLDAELIQYREDLNQVITIKDSQQKQLLEVQLQQNK 2563
Query: 414 -LQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKVLTGFIKSTETFQ 471
L++ + + EK +E + + + +Q+E L KE+E+LK ++ + Q
Sbjct: 2564 ELENKYAKLEEKLKESEEANEDLRRSFNALQEEKQDLSKEIESLKVSISQLTRQVTALQ 2622
Score = 43.5 bits (101), Expect = 0.005
Identities = 61/278 (21%), Positives = 124/278 (43%), Gaps = 59/278 (21%)
Query: 194 EDLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQEPQELEEVHEEEAEDEL 253
EDLI +L ++ +Q + +++K L ++ + Q+P+E+ E E A+ +
Sbjct: 1433 EDLIKALHT-QLEMQAKEHDERIKQLQVELCEMK--------QKPEEIGE--ESRAKQQ- 1480
Query: 254 ALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQR 313
I +++Q ++ R + K+ N S++ E ++ T K+ D++
Sbjct: 1481 --IQRKLQAALISRKEALKE----NKSLQEELSLARGTIERLTKS------LADVES--- 1525
Query: 314 KPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLL 373
+ S ++ DT +G +A +E + +I ++D L
Sbjct: 1526 ---------------QVSAQNKEKDTVLGRLALLQEERDKLI-----------TEMDRSL 1559
Query: 374 YDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQK 433
++ L++ C SL L + E+KE L K ES K+ KI + + + EK +E+ ++ +
Sbjct: 1560 LENQSLSSSCESLKLALEGLTEDKEKLV-KEIESLKSSKIAEST--EWQEKHKELQKEYE 1616
Query: 434 GIMSVPSEVQKENILLKKEVETL---KKVLTGFIKSTE 468
++ V E ++ VE + K+ L G ++STE
Sbjct: 1617 ILLQSYENVSNEAERIQHVVEAVRQEKQELYGKLRSTE 1654
Score = 39.3 bits (90), Expect = 0.088
Identities = 34/140 (24%), Positives = 64/140 (45%), Gaps = 17/140 (12%)
Query: 357 KTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEI-------LQNKYDESRK 409
K D L K + K + + Y S L+ + +L K +EI E++++ L+ + E +
Sbjct: 1393 KLDELQKLISKKEEDVSYLSGQLSEKEAALTKIQTEIIEQEDLIKALHTQLEMQAKEHDE 1452
Query: 410 TIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQ----------KENILLKKEVETLKKV 459
IK LQ +M +K EI + + + ++Q KEN L++E+ +
Sbjct: 1453 RIKQLQVELCEMKQKPEEIGEESRAKQQIQRKLQAALISRKEALKENKSLQEELSLARGT 1512
Query: 460 LTGFIKSTETFQNIVGSQNE 479
+ KS ++ V +QN+
Sbjct: 1513 IERLTKSLADVESQVSAQNK 1532
Score = 35.8 bits (81), Expect = 0.98
Identities = 77/398 (19%), Positives = 162/398 (40%), Gaps = 61/398 (15%)
Query: 83 LTMEESERVDECTNAKEVWDTL---KIHHEGTSHVKETRIDIGVRKFEVFEMSENETIDE 139
L E + + E N +E D L K+H E T + E ++ + + EV ++ EN +D
Sbjct: 2127 LEQAEEKHLKEKKNMQEKLDALRREKVHLEET--IGEIQVTLNKKDKEVQQLQEN--LDS 2182
Query: 140 MYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSVWRPMVTAITQAKD----LKSMNLED 195
+ M SL DR R I W + Q+K+ LK N
Sbjct: 2183 TVTQLAAFTKSMSSL-----QDDRDRVIDEA--KKWERKFSDAIQSKEEEIRLKEDNCSV 2235
Query: 196 LIGSLRAHEVVLQGDKPVKKVKTLALKASQQT-PSVADEDVQEPQELEEVHEEEAEDELA 254
L LR + ++ K+ L+ +Q S A +VQ Q++ + + E ++ L+
Sbjct: 2236 LKDQLRQMSIHMEE----LKINISRLEHDKQIWESKAQTEVQLQQKVCDTLQGENKELLS 2291
Query: 255 LISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQRK 314
+ + +N++ K + + ++ K Q+T N K + +++ + R+
Sbjct: 2292 QLEETRHLYHSSQNELAK------LESELKSLKDQLTDLS-NSLEKCKEQKGNLEGIIRQ 2344
Query: 315 PPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLY 374
D S E +TD+ Q+ E T L++++ K ++
Sbjct: 2345 QEA--------DIQNSKFSYEQLETDL----QASRE------LTSRLHEEINMKEQKII- 2385
Query: 375 DSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKG 434
+ L+ + ++ ++E++++ + + E + ++ + + E+ ++ K
Sbjct: 2386 --SLLSGKEEAIQVAIAELRQQHD---KEIKELENLLSQEEEENIVLEEENKKAVDKTNQ 2440
Query: 435 IMSVPSEVQKENILLKKEVETLKKVLTGFIKSTETFQN 472
+M ++KENI K ++++ F+KS + QN
Sbjct: 2441 LMETLKTIKKENIQQKAQLDS-------FVKSMSSLQN 2471
Score = 34.3 bits (77), Expect = 2.8
Identities = 39/166 (23%), Positives = 70/166 (41%), Gaps = 21/166 (12%)
Query: 344 MAQSDDEEEVIIYKTDSLY----KDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEI 399
++Q+ ++E+ I K D L +D+E ++ +T S+ +++ ++ EEK
Sbjct: 864 LSQALSQKELEITKMDQLLLEKKRDVETLQQTIEEKDQQVTEISFSMTEKMVQLNEEKFS 923
Query: 400 LQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGI------MSVPSEVQKENI-----L 448
L + ++ + +L + E+ E N G+ MS ++ KE + L
Sbjct: 924 LGVEIKTLKEQLNLLSRAEEAKKEQVEEDNEVSSGLKQNYDEMSPAGQISKEELQHEFDL 983
Query: 449 LKKEVETLK-KVLTGFIKSTETFQNIVGSQNESTKKSGLGFKDPSK 493
LKKE E K K+ I E Q + + E KD SK
Sbjct: 984 LKKENEQRKRKLQAALINRKELLQRVSRLEEELA-----NLKDESK 1024
Score = 32.7 bits (73), Expect = 8.3
Identities = 25/110 (22%), Positives = 53/110 (47%), Gaps = 7/110 (6%)
Query: 377 NFLTNRCHSLIKELSEIKEEKEILQNKYDESR----KTIKILQDSHFDMSEKQREINRKQ 432
N LT + HSL E + E+LQN+ D+ + + +++ + K+ E+
Sbjct: 792 NLLTEQIHSLSIEAKSKDVKIEVLQNELDDVQLQFSEQSTLIRSLQSQLQNKESEVLEGA 851
Query: 433 KGIMSVPSEVQK-ENILLKKEVETLK--KVLTGFIKSTETFQNIVGSQNE 479
+ + + S+V++ L +KE+E K ++L + ET Q + +++
Sbjct: 852 ERVRHISSKVEELSQALSQKELEITKMDQLLLEKKRDVETLQQTIEEKDQ 901
>M710_ARATH (P92512) Hypothetical mitochondrial protein AtMg00710
(ORF120)
Length = 120
Score = 53.9 bits (128), Expect = 3e-06
Identities = 30/85 (35%), Positives = 45/85 (52%), Gaps = 3/85 (3%)
Query: 908 NRTLQEMARTMLNESNVENYFWAEAINTSCYILNRVSIRKVLNKTPYELWKNIKPSISYF 967
NRT+ E R+ML E + F A+A NT+ +I+N+ + P E+W P+ SY
Sbjct: 2 NRTIIEKVRSMLCECGLPKTFRADAANTAVHIINKYPSTAINFHVPDEVWFQSVPTYSYL 61
Query: 968 HIFGCYCYILNNKEKLGKFDPKSDK 992
FGC YI ++ GK P++ K
Sbjct: 62 RRFGCVAYIHCDE---GKLKPRAKK 83
>MYH4_RABIT (Q28641) Myosin heavy chain, skeletal muscle, juvenile
Length = 1938
Score = 52.0 bits (123), Expect = 1e-05
Identities = 87/428 (20%), Positives = 178/428 (41%), Gaps = 54/428 (12%)
Query: 79 LSCALTMEESERVDECTNAKEVWDTLKI---HHEGTSHVKETRIDIGVRKFEVFEMSENE 135
++ LT ++ + DEC+ K+ D L++ E H E ++ K EM+
Sbjct: 935 INAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV-----KNLTEEMAG-- 987
Query: 136 TIDEMYARFTTIVNEMRSLGKAYS-THDRIRKILRCLPSVWRPMVTAITQAKDLKSMNLE 194
+DE A+ T E ++L +A+ T D ++ V +T+AK ++
Sbjct: 988 -LDETIAKLT---KEKKALQEAHQQTLDDLQ--------AEEDKVNTLTKAKTKLEQQVD 1035
Query: 195 DLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQEPQELEEVHEEE------ 248
DL GSL + + + K+ LK +Q++ + D Q+ E + E E
Sbjct: 1036 DLEGSLEQEKKIRMDLERAKRKLEGDLKLAQESTMDIENDKQQLDEKLKKKEFEMSNLQS 1095
Query: 249 -AEDELAL---ISKRIQRMMLRRNQIRKKFPKTNIS-IKTEADKSQVT------CYGCNK 297
EDE AL + K+I+ + R ++ ++ S K E +S ++ +
Sbjct: 1096 KIEDEQALAMQLQKKIKELQARIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEE 1155
Query: 298 TGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADT---------DMGLMAQSD 348
G + ++ K +R+ F+K D+EE+ Q +A + + +
Sbjct: 1156 AGGATSAQIEMNK-KREAEFQKMRR----DLEEATLQHEATAATLRKKHADSVAELGEQI 1210
Query: 349 DEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESR 408
D + + K + +L+ +ID L + ++ +L K ++++ L+ K +E +
Sbjct: 1211 DNLQRVKQKLEKEKSELKMEIDDLASNMETVSKAKGNLEKMCRTLEDQVSELKTKEEEHQ 1270
Query: 409 KTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKVLTGFIKSTE 468
+ I L + + E +R+ S+ S++ + +++E LK+ L IK+
Sbjct: 1271 RLINDLSAQRARLQTESGEFSRQLDEKDSLVSQLSRGKQAFTQQIEELKRQLEEEIKAKS 1330
Query: 469 TFQNIVGS 476
+ + S
Sbjct: 1331 ALAHALQS 1338
Score = 35.4 bits (80), Expect = 1.3
Identities = 52/321 (16%), Positives = 134/321 (41%), Gaps = 28/321 (8%)
Query: 178 MVTAITQAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQE 237
+V+ +++ K + +E+L + L+ + K AL++++ + E +E
Sbjct: 1300 LVSQLSRGKQAFTQQIEEL-------KRQLEEEIKAKSALAHALQSARHDCDLLREQYEE 1352
Query: 238 PQELEEVHEE---EAEDELALISKRIQRMMLRRNQ----IRKKFPKTNISIKTEADKSQV 290
QE + + +A E+A + + ++R + +KK + + +
Sbjct: 1353 EQEAKAELQRAMSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDAEEHVEAVNA 1412
Query: 291 TCYGCNKTGH-FKNECPD----IKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMA 345
C KT +NE D +++ K +D + + +T L A
Sbjct: 1413 KCASLEKTKQRLQNEVEDLMIDVERTNAACAALDKKQRNFDKILAEWKHKYEETHAELEA 1472
Query: 346 QSDDEEEVI--IYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSE----IKEEKEI 399
+ + ++K + Y++ +++++L ++ L L ++++E I E +++
Sbjct: 1473 SQKESRSLSTEVFKVKNAYEESLDQLETLKRENKNLQQEISDLTEQIAEGGKRIHELEKV 1532
Query: 400 LQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKV 459
+ E + L+++ + ++ +I R Q + V SE+ ++ +E++ LK+
Sbjct: 1533 KKQVEQEKSELQAALEEAEASLEHEEGKILRIQLELNQVKSEIDRKIAEKDEEIDQLKR- 1591
Query: 460 LTGFIKSTETFQNIVGSQNES 480
I+ E+ Q+ + ++ S
Sbjct: 1592 --NHIRVVESMQSTLDAEIRS 1610
>MYH4_HUMAN (Q9Y623) Myosin heavy chain, skeletal muscle, fetal
(Myosin heavy chain IIb) (MyHC-IIb)
Length = 1939
Score = 52.0 bits (123), Expect = 1e-05
Identities = 98/437 (22%), Positives = 184/437 (41%), Gaps = 72/437 (16%)
Query: 79 LSCALTMEESERVDECTNAKEVWDTLKI---HHEGTSHVKETRIDIGVRKFEVFEMSENE 135
++ LT ++ + DEC+ K+ D L++ E H E ++ K EM+
Sbjct: 936 INAELTAKKRKLEDECSELKKDIDDLELTLAKVEKEKHATENKV-----KNLTEEMAG-- 988
Query: 136 TIDEMYARFTTIVNEMRSLGKAYS-THDRIRKILRCLPSVWRPMVTAITQAKDLKSMNLE 194
+DE A+ T E ++L +A+ T D ++ + V +T+AK ++
Sbjct: 989 -LDETIAKLT---KEKKALQEAHQQTLDDLQ--------MEEDKVNTLTKAKTKLEQQVD 1036
Query: 195 DLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQEPQELEEVHEEE------ 248
DL GSL + + + K+ LK +Q++ + D Q+ E + E E
Sbjct: 1037 DLEGSLEQEKKLCMDLERAKRKLEGDLKLAQESTMDTENDKQQLNEKLKKKEFEMSNLQG 1096
Query: 249 -AEDELAL---ISKRIQRMMLRRNQIRKKFPKTNIS-IKTEADKSQVT------CYGCNK 297
EDE AL + K+I+ + R ++ ++ S K E +S ++ +
Sbjct: 1097 KIEDEQALAMQLQKKIKELQARIEELEEEIEAERASRAKAEKQRSDLSRELEEISERLEE 1156
Query: 298 TGHFKNECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVIIYK 357
G + ++ K +R+ F+K D+EES Q +A T L + D + +
Sbjct: 1157 AGGATSAQIELNK-KREAEFQKMRR----DLEESTLQHEA-TAAALRKKHADSVAELGKQ 1210
Query: 358 TDSLYK---DLENKIDSLLYDSNFLTNR--------------CHSLIKELSEIKEEKEIL 400
DSL + LE + L + N L + C +L +LSEIK ++E
Sbjct: 1211 IDSLQRVKQKLEKEKSELKMEINDLASNMETVSKAKANFEKMCRTLEDQLSEIKTKEEEQ 1270
Query: 401 QNKYDE-SRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKV 459
Q +E S + ++ H + E R+++ K ++ S++ + +++E LK+
Sbjct: 1271 QRLINELSAQKARL----HTESGEFSRQLDEKD----AMVSQLSRGKQAFTQQIEELKRQ 1322
Query: 460 LTGFIKSTETFQNIVGS 476
L K+ T + + S
Sbjct: 1323 LEEETKAKSTLAHALQS 1339
Score = 37.0 bits (84), Expect = 0.44
Identities = 44/232 (18%), Positives = 96/232 (40%), Gaps = 21/232 (9%)
Query: 336 DADTDMGLMAQSDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKE 395
+ + +M M + ++ + + KT++ K+LE K+ +L+ + N L + + L++ +E
Sbjct: 848 ETEKEMANMKEEFEKTKEELAKTEAKRKELEEKMVTLMQEKNDLQLQVQAEADALADAEE 907
Query: 396 EKEILQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVET 455
+ L + IK + + D E E+ K++ + SE LKK+++
Sbjct: 908 RCDQLIKTKIQLEAKIKEVTERAEDEEEINAELTAKKRKLEDECSE-------LKKDIDD 960
Query: 456 LKKVLTGFIKSTETFQNIVGSQNESTKKSGLGFKDPSKIIGSFVPKAKIRVKCCFCDKYG 515
L+ L K +N V +N + + +GL + I + K +
Sbjct: 961 LELTLAKVEKEKHATENKV--KNLTEEMAGL-----DETIAKLTKEKKAL-------QEA 1006
Query: 516 HNESICHVKKKFIKQNNLYLSSERSHLNRSESSQKAEKAKKTCFYCNKSDHK 567
H +++ ++ + K N L + + + E+ KK C ++ K
Sbjct: 1007 HQQTLDDLQMEEDKVNTLTKAKTKLEQQVDDLEGSLEQEKKLCMDLERAKRK 1058
Score = 34.3 bits (77), Expect = 2.8
Identities = 56/324 (17%), Positives = 137/324 (42%), Gaps = 26/324 (8%)
Query: 178 MVTAITQAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVKTLALKASQQTPSVADEDVQE 237
MV+ +++ K + +E+L + L+ + K AL++++ + E +E
Sbjct: 1301 MVSQLSRGKQAFTQQIEEL-------KRQLEEETKAKSTLAHALQSARHDCDLLREQYEE 1353
Query: 238 PQELEEVHEE---EAEDELALISKRIQRMMLRRNQ----IRKKFPKTNISIKTEADKSQV 290
QE + + +A E+A + + ++R + +KK + + +
Sbjct: 1354 EQEAKAELQRGMSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDAEEHVEAVNS 1413
Query: 291 TCYGCNKTGH-FKNECPDIK-KVQRKPPF---KKKAMITWDDMEESDSQEDADTDMGLMA 345
C KT +NE D+ V+R K +D + Q+ +T L A
Sbjct: 1414 KCASLEKTKQRLQNEVEDLMIDVERSNAACIALDKKQRNFDKVLAEWKQKYEETQAELEA 1473
Query: 346 QSDDEEEVI--IYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSE----IKEEKEI 399
+ + ++K + Y++ + +++L ++ L L ++++E I E +++
Sbjct: 1474 SQKESRSLSTELFKVKNAYEESLDHLETLKRENKNLQQEISDLTEQIAEGGKHIHELEKV 1533
Query: 400 LQNKYDESRKTIKILQDSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKV 459
+ E + L+++ + ++ +I R Q + V SE+ ++ +E++ LK+
Sbjct: 1534 KKQLDHEKSELQTSLEEAEASLEHEEGKILRIQLELNQVKSEIDRKIAEKDEELDQLKRN 1593
Query: 460 LTGFIKSTE-TFQNIVGSQNESTK 482
++S + T + S+N++ +
Sbjct: 1594 HLRVVESMQSTLDAEIRSRNDALR 1617
Score = 33.5 bits (75), Expect = 4.8
Identities = 39/221 (17%), Positives = 93/221 (41%), Gaps = 11/221 (4%)
Query: 247 EEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECP 306
+E + ++ +S+ Q + +++++ + + T A Q + C+ E
Sbjct: 1296 DEKDAMVSQLSRGKQAFTQQIEELKRQLEEETKAKSTLAHALQSARHDCDLLREQYEEEQ 1355
Query: 307 DIK-KVQRKPPFKKKAMITW------DDMEESDSQEDADTDMGLMAQSDDEE-EVIIYKT 358
+ K ++QR + W D ++ ++ E+A + Q +E E + K
Sbjct: 1356 EAKAELQRGMSKANSEVAQWRTKYETDAIQRTEELEEAKKKLAQRLQDAEEHVEAVNSKC 1415
Query: 359 DSLYKD---LENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDESRKTIKILQ 415
SL K L+N+++ L+ D C +L K+ + + KY+E++ ++ Q
Sbjct: 1416 ASLEKTKQRLQNEVEDLMIDVERSNAACIALDKKQRNFDKVLAEWKQKYEETQAELEASQ 1475
Query: 416 DSHFDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETL 456
+S + ++ + + +++EN L++E+ L
Sbjct: 1476 KESRSLSTELFKVKNAYEESLDHLETLKRENKNLQQEISDL 1516
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.134 0.389
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 185,793,454
Number of Sequences: 164201
Number of extensions: 8062053
Number of successful extensions: 29210
Number of sequences better than 10.0: 449
Number of HSP's better than 10.0 without gapping: 104
Number of HSP's successfully gapped in prelim test: 355
Number of HSP's that attempted gapping in prelim test: 27774
Number of HSP's gapped (non-prelim): 1470
length of query: 1608
length of database: 59,974,054
effective HSP length: 124
effective length of query: 1484
effective length of database: 39,613,130
effective search space: 58785884920
effective search space used: 58785884920
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 73 (32.7 bits)
Medicago: description of AC139747.5


