

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139747.1 - phase: 0 /pseudo
(441 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
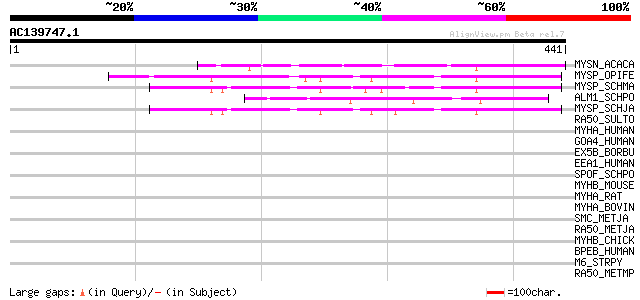
MYSN_ACACA (P05659) Myosin II heavy chain, non muscle 48 6e-05
MYSP_OPIFE (Q9BMQ6) Paramyosin (Fragment) 46 2e-04
MYSP_SCHMA (P06198) Paramyosin 45 4e-04
ALM1_SCHPO (Q9UTK5) Abnormal long morphology protein 1 (Sp8) 45 5e-04
MYSP_SCHJA (Q05870) Paramyosin (Antigen Sj97) 44 6e-04
RA50_SULTO (Q96YR5) DNA double-strand break repair rad50 ATPase 42 0.003
MYHA_HUMAN (P35580) Myosin heavy chain, nonmuscle type B (Cellul... 42 0.003
GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member... 42 0.004
EX5B_BORBU (O51578) Exodeoxyribonuclease V beta chain (EC 3.1.11.5) 41 0.005
EEA1_HUMAN (Q15075) Early endosome antigen 1 (Endosome-associate... 41 0.005
SPOF_SCHPO (Q10411) Sporulation-specific protein 15 41 0.007
MYHB_MOUSE (O08638) Myosin heavy chain, smooth muscle isoform (S... 41 0.007
MYHA_RAT (Q9JLT0) Myosin heavy chain, nonmuscle type B (Cellular... 41 0.007
MYHA_BOVIN (Q27991) Myosin heavy chain, nonmuscle type B (Cellul... 41 0.007
SMC_METJA (Q59037) Chromosome partition protein smc homolog 40 0.012
RA50_METJA (Q58718) DNA double-strand break repair rad50 ATPase 40 0.012
MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle 39 0.035
BPEB_HUMAN (Q8WXK8) Bullous pemphigoid antigen 1, isoform 7 (Bul... 38 0.045
M6_STRPY (P08089) M protein, serotype 6 precursor 38 0.059
RA50_METMP (P62134) DNA double-strand break repair rad50 ATPase 37 0.078
>MYSN_ACACA (P05659) Myosin II heavy chain, non muscle
Length = 1509
Score = 47.8 bits (112), Expect = 6e-05
Identities = 65/298 (21%), Positives = 122/298 (40%), Gaps = 27/298 (9%)
Query: 150 LNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTA----IEEARDISSFKVDELI 205
L ILD+ E D L++K + L + + +A +E+ R + + K EL
Sbjct: 905 LKILDLEGEKADLEE---DNALLQKKVAGLEEELQEETSASNDILEQKRKLEAEK-GELK 960
Query: 206 GSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKKIVKQY 265
SL+ E + + K K E +ELQ ED+ +SL + +++
Sbjct: 961 ASLEEEERNRKALQEAKTK-----VESERNELQDKYEDEAAAHDSLKKKEEDLSRELRE- 1014
Query: 266 DKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*K 325
K + +NI + ++ + D +N E + K++ K +
Sbjct: 1015 TKDALADAENISETLRSKLKNTERGADDVRN---------ELDDVTATKLQLEKTKKSLE 1065
Query: 326 KSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLA--YDELVVSYKRLNDK 383
+ L + + + K E S K A +G+ DA S + L S K D+
Sbjct: 1066 EELAQTRAQLEEEKSGKEAASSK--AKQLGQQLEDARSEVDSLKSKLSAAEKSLKTAKDQ 1123
Query: 384 NTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMAARTD 441
N D+ +QLE+++ + N+++++ K +EL +V L+ Q + Q K + + D
Sbjct: 1124 NRDLDEQLEDERTVRANVDKQKKALEAKLTELEDQVTALDGQKNAAAAQAKTLKTQVD 1181
Score = 31.2 bits (69), Expect = 5.6
Identities = 60/326 (18%), Positives = 123/326 (37%), Gaps = 58/326 (17%)
Query: 113 KIAHEGTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLV 172
K E TKV+S + +L KYE+ D + D++ + ++D + +
Sbjct: 971 KALQEAKTKVESERNEL-QDKYEDEAAAHDSLKKKEE----DLSRELRETKDALADAENI 1025
Query: 173 RKILRSLPKRFDMKVTAIE-EARDISSFKV--DELIGSLQN--FEITVNSKNDKKGKGIA 227
+ LRS K + + E D+++ K+ ++ SL+ + + +K GK A
Sbjct: 1026 SETLRSKLKNTERGADDVRNELDDVTATKLQLEKTKKSLEEELAQTRAQLEEEKSGKEAA 1085
Query: 228 FASSVELDELQVNQEDDEDMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKE 287
+ + +L + + + D +S + K K ++ ++D Q E
Sbjct: 1086 SSKAKQLGQQLEDARSEVDSLKSKLSAAEKSLKTAKDQNR-----------DLDEQLEDE 1134
Query: 288 KMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*KKSLTVSWSD--DDGSKGDGERE 345
+ VR + + K KK+L ++ D + DG++
Sbjct: 1135 RTVRAN---------------------------VDKQKKALEAKLTELEDQVTALDGQKN 1167
Query: 346 SIKHVAALIGRVFSDAESCSEDLAYDELVVSYKRLNDKNTDICKQLEEQKNITNNLEEER 405
+ A + + + E+ + +R N L+E +T +L+ ER
Sbjct: 1168 AAAAQAKTLKTQVDETKRRLEEAEASAARLEKERKN--------ALDEVAQLTADLDAER 1219
Query: 406 VGYLEKNSELNSEVRMLNSQLSNVMK 431
++ +LN+ + L S+L N K
Sbjct: 1220 DSGAQQRRKLNTRISELQSELENAPK 1245
>MYSP_OPIFE (Q9BMQ6) Paramyosin (Fragment)
Length = 638
Score = 46.2 bits (108), Expect = 2e-04
Identities = 82/394 (20%), Positives = 161/394 (40%), Gaps = 54/394 (13%)
Query: 79 GNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLR 138
GNS + L + + KL K A +LE+ A + L + ENL+
Sbjct: 52 GNSSQTHELLKRREMEIGKLRKDLENANASLEM---AETSMRRRHQTALNELAAEVENLQ 108
Query: 139 MLDDESIQDYHLNILDIANSF----------ESAGEKISDEKLVRKILRSLPKRFDMKVT 188
++ +D + I+++ N +SA K+ L+ L ++
Sbjct: 109 KQKGKAEKDKNSLIMEVGNVLGQLDGALKAKQSAESKLEGLDAQLNRLKGLTDDLQRQLN 168
Query: 189 AIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVE--LDELQVNQEDDE- 245
+ A+ + + EL+ + Q +E V + + + SS+E +D+L+ + +D+
Sbjct: 169 DLNAAKARLTSENFELLHANQEYEAQVLNLSKSR-------SSLESAVDDLKRSLDDEAK 221
Query: 246 ---DMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSK--------------EK 288
++ LT L + + +Y++ N+ NQ SK E
Sbjct: 222 SRFNLQAQLTSLQMDYDNLQAKYEEESEEAS-----NLRNQVSKFNADLAAMKSKFEREL 276
Query: 289 MVRTDE-KNFQYK-GVQCHECEGYGHIKIECASFLKK*KKSLTVSWSDDDGSKGDGERES 346
M +T+E + + K ++ E E + AS L+K K LT+ D E +S
Sbjct: 277 MSKTEEYEELKRKLTLRITELEDTAERERARASNLEKIKAKLTIEIKDLQN-----EVDS 331
Query: 347 IKHVAALIGRVFSDAESCSEDLA--YDELVVSYKRLNDKNTDICKQLEEQKNITNNLEEE 404
+ A + R AE+ + DL DEL + L+ +N+ + + K+ N+L ++
Sbjct: 332 LSAENAELARRAKAAENLANDLQRRVDELTIEINNLHSQNSQLEAENMRLKSQVNDLVDK 391
Query: 405 RVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMAA 438
+N +L+ +V+ L S L + +++ + A
Sbjct: 392 NAALDRENRQLSDQVKDLKSTLRDANRRLTDLEA 425
>MYSP_SCHMA (P06198) Paramyosin
Length = 866
Score = 45.1 bits (105), Expect = 4e-04
Identities = 72/360 (20%), Positives = 151/360 (41%), Gaps = 49/360 (13%)
Query: 112 LKIAHEGTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSF----------ES 161
L++A + L + ENL+ ++ +D I+++ N +S
Sbjct: 101 LELAETSMRRRHQTALNELALEVENLQKQKGKAEKDKSHLIMEVDNVLGQLDGALKAKQS 160
Query: 162 AGEKIS--DEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQNFEITVNSKN 219
A K+ D +L R L+SL ++T + A+ + + EL+ Q++E + + +
Sbjct: 161 AESKLEGLDSQLNR--LKSLTDDLQRQLTELNNAKSRLTSENFELLHINQDYEAQILNYS 218
Query: 220 DKKGKGIAFASSVELDELQVNQEDDE----DMTESLTLLGRQFKKIVKQYDKRPRSIGQN 275
K + ++D+L+ + +D+ ++ LT L + + +YD+ N
Sbjct: 219 KAKS-----SLESQVDDLKRSLDDEAKNRFNLQAQLTSLQMDYDNLQAKYDEESEE-ASN 272
Query: 276 IRPNID----------NQPSKEKMVRTDE-----KNFQYKGVQCHECEGYGHIKIECASF 320
+R + ++ +E M +T+E + F + + + +K A
Sbjct: 273 LRSQVSKFNADIAALKSKFERELMSKTEEFEEMKRKFTMRITELEDTAERERLK---AVS 329
Query: 321 LKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLA--YDELVVSYK 378
L+K K LT+ D E ES+ + + R AES + DL DEL +
Sbjct: 330 LEKLKTKLTLEIKDLQS-----EIESLSLENSELIRRAKAAESLASDLQRRVDELTIEVN 384
Query: 379 RLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMAA 438
L +N+ + + K++ N+L ++ +N ++N +V+ L S L + +++ + A
Sbjct: 385 TLTSQNSQLESENLRLKSLVNDLTDKNNLLERENRQMNDQVKELKSSLRDANRRLTDLEA 444
>ALM1_SCHPO (Q9UTK5) Abnormal long morphology protein 1 (Sp8)
Length = 1727
Score = 44.7 bits (104), Expect = 5e-04
Identities = 49/256 (19%), Positives = 107/256 (41%), Gaps = 23/256 (8%)
Query: 187 VTAIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDED 246
+ + E D+S + L+ S+ F + +++ K A + L E VN E E+
Sbjct: 10 IQLVHEFLDVSFEDIKPLV-SVNGFAVFISAIKTKVKDINALKDQLVLQE--VNHEHKEN 66
Query: 247 -MTESLTLLGRQFKKIVKQYDKRP------RSIGQNIRPNIDNQPSKEKMVRTDEKNFQY 299
+T+ + L +Q + Q ++ R+ ++++ N++NQ + + T+ ++ +
Sbjct: 67 VLTKKINFLEQQLQSSNNQAEESRNLISVLRNENESLKTNLENQNKRFDALTTENQSLRR 126
Query: 300 KGVQCHECEGYGHIKIECAS----FLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIG 355
+ E ++ A L+ L + D E +H+ A
Sbjct: 127 ANSELQEQSKIASEQLSIAKDQIEALQNENSHLGEQVQSAHQALSDIEERKKQHMFA--- 183
Query: 356 RVFSDAESCSEDLAYDE---LVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKN 412
S + E++ E LV L ++ +C++LE +LE++ G ++N
Sbjct: 184 ---SSSSRVKEEILVQEKSALVSDLASLQSDHSKVCEKLEVSSRQVQDLEKKLAGLAQQN 240
Query: 413 SELNSEVRMLNSQLSN 428
+ELN ++++ + SN
Sbjct: 241 TELNEKIQLFEQKRSN 256
>MYSP_SCHJA (Q05870) Paramyosin (Antigen Sj97)
Length = 866
Score = 44.3 bits (103), Expect = 6e-04
Identities = 73/361 (20%), Positives = 147/361 (40%), Gaps = 51/361 (14%)
Query: 112 LKIAHEGTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSF----------ES 161
L++A + L+ + ENL+ ++ +D I+++ N +S
Sbjct: 101 LELAETSMRRRHQTALNELSLEVENLQKQKGKAEKDKSHLIMEVDNVLGQLDGALKAKQS 160
Query: 162 AGEKIS--DEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQNFEITVNSKN 219
A K+ D +L R L++L ++T + A+ + + EL+ Q++E + + +
Sbjct: 161 AESKLEGLDSQLNR--LKTLTDDLQRQLTELNNAKSRLTSENFELLHINQDYEAQILNYS 218
Query: 220 DKKGKGIAFASSVELDELQVNQEDDE----DMTESLTLLGRQFKKIVKQYDKRPRSIGQN 275
K + ++D+L+ + +D+ ++ LT L + + +YD+
Sbjct: 219 KAKS-----SLESQVDDLKRSLDDESRNRFNLQAQLTSLQMDYDNLQAKYDEESEEAS-- 271
Query: 276 IRPNIDNQPSK--------------EKMVRTDEKNFQYKGVQCH--ECEGYGHIKIECAS 319
N+ NQ SK E M +T+E + + E E + A
Sbjct: 272 ---NLRNQVSKFNADIAALKSKFERELMSKTEEFEEMKRKLTMRITELEDVAERERLKAV 328
Query: 320 FLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLA--YDELVVSY 377
L+K K LT+ D E ES+ + R AES + +L DEL +
Sbjct: 329 SLEKLKTKLTLEIKDLQS-----EIESLSLENGELIRRAKSAESLASELQRRVDELTIEV 383
Query: 378 KRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMA 437
L +N + + K++ N+L ++ +N ++N +V+ L S L + +++ +
Sbjct: 384 NTLTSQNNQLESENMRLKSLVNDLTDKNNALERENRQMNDQVKELKSSLRDANRRLTDLE 443
Query: 438 A 438
A
Sbjct: 444 A 444
>RA50_SULTO (Q96YR5) DNA double-strand break repair rad50 ATPase
Length = 879
Score = 42.0 bits (97), Expect = 0.003
Identities = 72/338 (21%), Positives = 139/338 (40%), Gaps = 50/338 (14%)
Query: 130 LTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTA 189
++ K + L LD+E ++ I+ ES E + D + +KIL K+
Sbjct: 116 VSQKIKELLNLDEEVLKS---TIIVGQGKIESVFENLPD--VTKKIL---------KIDK 161
Query: 190 IEEARDISSFKVDELIGSLQNFEITVNS----KNDKKGKGIAFASSVE-----LDELQVN 240
IE+ RD S+ + E++ + N I + S KN+ + + I +E L++L +
Sbjct: 162 IEKLRD-SNGPIKEVMDKINNKIIELQSLEKYKNESENQKIQKEKELENIKRELEDLNIK 220
Query: 241 QEDDEDMTESLTLLGRQFKKIVKQY-------DKRPRSIGQNIRPNIDNQPSKEKMVRTD 293
+E + E + L + +K K+Y +K I + +R + ++ +
Sbjct: 221 EEKERKKYEDIVKLNEEEEKKEKRYVELISLLNKLKDDISE-LREEVKDENRLREEKEKL 279
Query: 294 EKNFQYKGVQCHECEGY--GHIKIECASFLKK*KKSLTVSWSD------------DDGSK 339
EK+ K E E KI+ A +K K++ ++ +D +D K
Sbjct: 280 EKDILEKDKLIEEKEKIIEAQNKIKLAQEKEKSLKTIKINLTDLEEKLKRKRELEEDYKK 339
Query: 340 G---DGERESIKHVAALIGRVFSDAESCSEDLAYDELVVSYKRLNDKNTDICKQLEEQKN 396
GE E ++ + +S L+ E +S ++++ ++ K+L++
Sbjct: 340 YIEIKGELEELEEKERKFNSLSDRLKSLKIKLSEIESKISNRKISINIEELDKELQKLNE 399
Query: 397 ITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVK 434
NN +ER + E+ + LN L N + QVK
Sbjct: 400 DLNNKNQEREKLASQLGEIKGRIEELNKLLGN-LNQVK 436
Score = 35.8 bits (81), Expect = 0.23
Identities = 43/235 (18%), Positives = 104/235 (43%), Gaps = 20/235 (8%)
Query: 212 EITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKKIVKQYDKRPR- 270
E T++ + + IA ++ +++ DE++ +S ++G+ KI ++ P
Sbjct: 95 EDTISELTNDTRRTIARGATTVSQKIKELLNLDEEVLKSTIIVGQG--KIESVFENLPDV 152
Query: 271 -----SIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*K 325
I + + N P KE M + + K + + ++ ++ E KI+ L+ K
Sbjct: 153 TKKILKIDKIEKLRDSNGPIKEVMDKINNKIIELQSLEKYKNESENQ-KIQKEKELENIK 211
Query: 326 KSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLAYDELVVSYKRLNDKNT 385
+ L +D K + ER+ + + L + E ++ Y EL+ +L D +
Sbjct: 212 REL-----EDLNIKEEKERKKYEDIVKL------NEEEEKKEKRYVELISLLNKLKDDIS 260
Query: 386 DICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMAART 440
++ ++++++ + E+ LEK+ + + +++ +Q + Q K + +T
Sbjct: 261 ELREEVKDENRLREEKEKLEKDILEKDKLIEEKEKIIEAQNKIKLAQEKEKSLKT 315
>MYHA_HUMAN (P35580) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 42.0 bits (97), Expect = 0.003
Identities = 67/348 (19%), Positives = 145/348 (41%), Gaps = 36/348 (10%)
Query: 97 KLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRMLD---DESIQDYHLNIL 153
K KQ + LE LK E T +A+ +L T + + + L +E +++ I
Sbjct: 1136 KAEKQKRDLSEELEALKTELEDTLDTTAAQQELRTKREQEVAELKKALEEETKNHEAQIQ 1195
Query: 154 DIANSFESAGEKISDE-----------KLVRKILRSLPKRFDMKVTAIEEARDISSFKVD 202
D+ +A E++S++ + ++ L + K +V +++ + S K
Sbjct: 1196 DMRQRHATALEELSEQLEQAKRFKANLEKNKQGLETDNKELACEVKVLQQVKAESEHKRK 1255
Query: 203 ELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKKIV 262
+L +Q V+ +G + + + +LQ ++ + E G +F K
Sbjct: 1256 KLDAQVQELHAKVS-----EGDRLRVELAEKASKLQNELDNVSTLLEEAEKKGIKFAKDA 1310
Query: 263 KQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLK 322
+ + + + ++ + + +R E+ Q E E E L+
Sbjct: 1311 ASLESQLQDTQELLQEETRQKLNLSSRIRQLEEEKNSLQEQQEEEE-------EARKNLE 1363
Query: 323 K*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDL-----AYDELVVSY 377
K +L +D K D + +I+ + ++ DAE+ S+ L AYD+L +
Sbjct: 1364 KQVLALQSQLADTK-KKVDDDLGTIESLEEAKKKLLKDAEALSQRLEEKALAYDKLEKTK 1422
Query: 378 KRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQ 425
RL + D+ L+ Q+ + +NLE+++ +K +L +E + ++++
Sbjct: 1423 NRLQQELDDLTVDLDHQRQVASNLEKKQ----KKFDQLLAEEKSISAR 1466
Score = 32.3 bits (72), Expect = 2.5
Identities = 83/380 (21%), Positives = 145/380 (37%), Gaps = 61/380 (16%)
Query: 93 KNMFKLI-KQCIMAKDTLEILKIAHEGTTKVKSAKFQL--LTTKYENLRMLDDESIQDYH 149
KN+ K+ KQ +M D E LK + +++ AK +L TT ++ I +
Sbjct: 1023 KNLAKIRNKQEVMISDLEERLKKEEKTRQELEKAKRKLDGETTDLQDQIAELQAQIDELK 1082
Query: 150 LNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQ 209
L + + A + DE L + + + ++ ++E D S K
Sbjct: 1083 LQLAKKEEELQGALARGDDETLHKNNALKVVRELQAQIAELQE--DFESEKASR------ 1134
Query: 210 NFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGR-------QFKKIV 262
+K +K+ + + S EL+ L+ ED D T + L + KK +
Sbjct: 1135 -------NKAEKQKRDL----SEELEALKTELEDTLDTTAAQQELRTKREQEVAELKKAL 1183
Query: 263 KQYDKRPRSIGQNIRPN-------IDNQPSKEKMVRTD-EKNFQYKGVQCHE--CEGYGH 312
++ K + Q++R + Q + K + + EKN Q E CE
Sbjct: 1184 EEETKNHEAQIQDMRQRHATALEELSEQLEQAKRFKANLEKNKQGLETDNKELACEVKVL 1243
Query: 313 IKIECASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAAL-------IGRVFSDAESCS 365
+++ S K+ K V S+GD R + A+ + + +AE
Sbjct: 1244 QQVKAESEHKRKKLDAQVQELHAKVSEGDRLRVELAEKASKLQNELDNVSTLLEEAEKKG 1303
Query: 366 EDLAYD------------ELVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNS 413
A D EL+ R + +QLEE+KN +L+E++ E
Sbjct: 1304 IKFAKDAASLESQLQDTQELLQEETRQKLNLSSRIRQLEEEKN---SLQEQQEEEEEARK 1360
Query: 414 ELNSEVRMLNSQLSNVMKQV 433
L +V L SQL++ K+V
Sbjct: 1361 NLEKQVLALQSQLADTKKKV 1380
Score = 32.3 bits (72), Expect = 2.5
Identities = 17/62 (27%), Positives = 33/62 (52%)
Query: 378 KRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMA 437
K++ D+ +QL+E++ L+ E+V K ++ E+ +L Q S +K+ K+M
Sbjct: 946 KKMQAHIQDLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEILLLEDQNSKFIKEKKLME 1005
Query: 438 AR 439
R
Sbjct: 1006 DR 1007
>GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member 4
(Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (72.1
protein)
Length = 2230
Score = 41.6 bits (96), Expect = 0.004
Identities = 74/345 (21%), Positives = 152/345 (43%), Gaps = 44/345 (12%)
Query: 110 EILKIAHEGTTKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDE 169
E L++ E +++S + + +TT+ E LR ++S + +FE + +S
Sbjct: 375 ETLEMKEEEIAQLRS-RIKQMTTQGEELREQKEKSERA----------AFEELEKALSTA 423
Query: 170 KLVRKILRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFA 229
+ + R L D ++ IE+ + + + + ++ + V K+ ++ A
Sbjct: 424 QKTEEARRKLKAEMDEQIKTIEKTSEEERISLQQELSRVKQEVVDVMKKSSEE----QIA 479
Query: 230 SSVELDELQVNQEDDEDMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPS---- 285
+L E ++ +++ E +T+ L R+F++ +K ++ +S I + Q S
Sbjct: 480 KLQKLHEKELARKEQE-LTKKLQTREREFQEQMKVALEKSQSEYLKISQEKEQQESLALE 538
Query: 286 ----KEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*KKSLTVSWSDDDGSKGD 341
++K + T+ +N + + +Q E E Y +E S L+K SL + +
Sbjct: 539 ELELQKKAILTESEN-KLRDLQ-QEAETYRTRILELESSLEK---SLQENKNQSKDLAVH 593
Query: 342 GERESIKH---VAALIGRVFSDAESCS--EDLAYDE----LVVSYKRLNDKNTDICKQLE 392
E E KH + ++ + ++ ES +D + E L Y+ +K + C+Q +
Sbjct: 594 LEAEKNKHNKEITVMVEKHKTELESLKHQQDALWTEKLQVLKQQYQTEMEKLREKCEQEK 653
Query: 393 E----QKNIT--NNLEEERVGYLEKNSELNSEVRMLNSQLSNVMK 431
E K I ++EE LEK +E+ L+S+LS V+K
Sbjct: 654 ETLLKDKEIIFQAHIEEMNEKTLEKLDVKQTELESLSSELSEVLK 698
>EX5B_BORBU (O51578) Exodeoxyribonuclease V beta chain (EC 3.1.11.5)
Length = 1169
Score = 41.2 bits (95), Expect = 0.005
Identities = 67/283 (23%), Positives = 117/283 (40%), Gaps = 37/283 (13%)
Query: 64 KP*EEWTAAEDELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEI---LKIAHEGTT 120
KP E+++ DE+ + ++L +D ++L AK T EI +K A+E T
Sbjct: 117 KPKEKFSKEIDEIVYDFLRKSDSLIQALDIKDYELKVFKSDAKKTEEIVLKIKKAYERDT 176
Query: 121 KVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLP 180
+ + T +EN+ + +E I+DY+ I D+ D+ +IL
Sbjct: 177 TQELGDWLKTQTAFENILLKKEELIKDYNKIIEDL------------DKMTKDEILSFYN 224
Query: 181 KRFDMKVTAIEEARDISSFKVDELI------GSLQNFEITVNSK---NDKKGKGIAFASS 231
K IE +++ FK+ E + +L E NSK + K K
Sbjct: 225 KHIQTGKLEIEYSKENDIFKIAETLLKNKFFSTLIEKETKKNSKLSPKELKIKNDLICLG 284
Query: 232 VELDELQVNQEDDEDMT----ESLTLLGRQFK------KIVKQYDKRPRSIGQN-IRPNI 280
+ + + ED+ + + +L ++K K +K+ K +I QN I N+
Sbjct: 285 INIKHEKYKSEDNRNKNRNNLKQYVILKVEYKILKYIEKELKKTIKSTNTIDQNYIISNL 344
Query: 281 DNQ-PSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLK 322
N S++K + KN +YK + E + I+IE LK
Sbjct: 345 KNYLKSEDKKLLNAIKN-RYKIILIDEAQDLSLIQIEIFKILK 386
>EEA1_HUMAN (Q15075) Early endosome antigen 1 (Endosome-associated
protein p162) (Zinc finger FYVE domain containing
protein 2)
Length = 1411
Score = 41.2 bits (95), Expect = 0.005
Identities = 85/380 (22%), Positives = 157/380 (40%), Gaps = 59/380 (15%)
Query: 71 AAEDELALGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLL 130
AA+D + L ++N L + ++++ K+ + I K E+L A T ++ L
Sbjct: 611 AAQDRV-LSLETSVNELNSQLNESKEKVSQLDIQIKAKTELLLSAEAAKTAQRADLQNHL 669
Query: 131 TTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAI 190
T ++QD + I + K+ D++ L S K + K ++
Sbjct: 670 DTA--------QNALQDKQQELNKITTQLDQVTAKLQDKQEHCSQLESHLKEYKEKYLSL 721
Query: 191 EEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTES 250
E+ K +EL G ++ E +S K K A L +LQ ++ + D+
Sbjct: 722 EQ-------KTEELEGQIKKLE--ADSLEVKASKEQA------LQDLQQQRQLNTDLELR 766
Query: 251 LTLLGRQF---KKIVK--QYDKRPRSIG-QNIRPNIDNQPSKEKMVRTD-EKNFQYKGVQ 303
T L +Q K+IV + D + +S ++I+ + Q ++++++ D E Q +Q
Sbjct: 767 ATELSKQLEMEKEIVSSTRLDLQKKSEALESIKQKLTKQEEEKQILKQDFETLSQETKIQ 826
Query: 304 CHECEGYGHIKIECASFLKK*KKSLTVSWSDDD-----------GSKGDGERESIKHVAA 352
E + +K K++L S SK + E+E+ K AA
Sbjct: 827 HEELNNRIQTTVTELQKVKMEKEALMTELSTVKDKLSKVSDSLKNSKSEFEKENQKGKAA 886
Query: 353 LIGRVFSDAESCSEDLAYDELVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKN 412
++ D E ++L + ++ +NT L+EQK + +LE+E+ +
Sbjct: 887 IL-----DLEKTCKELKHQ------LQVQMENT-----LKEQKELKKSLEKEKEASHQLK 930
Query: 413 SELNSEVRMLNSQLSNVMKQ 432
ELNS L Q N +KQ
Sbjct: 931 LELNSMQEQL-IQAQNTLKQ 949
>SPOF_SCHPO (Q10411) Sporulation-specific protein 15
Length = 1957
Score = 40.8 bits (94), Expect = 0.007
Identities = 51/261 (19%), Positives = 109/261 (41%), Gaps = 37/261 (14%)
Query: 210 NFEITVNSKNDKKGKGIAFASSVE--------LDELQVN----QEDDEDMTESLTLLGRQ 257
+ +++ ++K DK + +S E L ++ N Q + D++ L + +
Sbjct: 157 SLQLSSSNKKDKNTSSVTTLTSEEDVSYFQKKLTNMESNFSAKQSEAYDLSRQLLTVTEK 216
Query: 258 FKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHEC-----EGYGH 312
K K Y+K + +I+ ++ + + K +R +++ + V ++ +
Sbjct: 217 LDKKEKDYEKIKEDVS-SIKASLAEEQASNKSLRGEQERLEKLLVSSNKTVSTLRQTENS 275
Query: 313 IKIECASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLA--- 369
++ EC + +K +K +++ SK E +KH A + EDL+
Sbjct: 276 LRAECKTLQEKLEKCAI----NEEDSK---LLEELKHNVANYSDAIVHKDKLIEDLSTRI 328
Query: 370 --YDELVVSYKRLNDKNTDICK-------QLEEQKNITNNLEEERVGYLEKNSELNSEVR 420
+D L L+ KN + K L++ + + LEEE V E N ++S++
Sbjct: 329 SEFDNLKSERDTLSIKNEKLEKLLRNTIGSLKDSRTSNSQLEEEMVELKESNRTIHSQLT 388
Query: 421 MLNSQLSNVMKQVKMMAARTD 441
S+LS+ ++ K + D
Sbjct: 389 DAESKLSSFEQENKSLKGSID 409
Score = 36.6 bits (83), Expect = 0.13
Identities = 72/370 (19%), Positives = 142/370 (37%), Gaps = 62/370 (16%)
Query: 120 TKVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSL 179
T K + QL +E + D +L + S +S +K D + K +++L
Sbjct: 635 TSSKLQQLQLERANFEQKESTLSDENNDLRTKLLKLEESNKSLIKKQEDVDSLEKNIQTL 694
Query: 180 PKRFDMKVTAIE----EARDISSFKVDELIGSLQNFEITVN----SKNDKKGKGIAFAS- 230
+ A+ EA+++ +D L G + E N S +D K +S
Sbjct: 695 KEDLRKSEEALRFSKLEAKNLREV-IDNLKGKHETLEAQRNDLHSSLSDAKNTNAILSSE 753
Query: 231 ----SVELDELQVNQE----DDEDMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDN 282
S ++ L N E D + M +S T L ++ I Y + R N++ +
Sbjct: 754 LTKSSEDVKRLTANVETLTQDSKAMKQSFTSLVNSYQSISNLYHEL-RDDHVNMQSQNNT 812
Query: 283 QPSKEKMVRTDEKNFQYKGV---------------QCHECEGYGHIKIECASFLKK*KKS 327
E ++TD +N + + Q + + + + LK + S
Sbjct: 813 LLESESKLKTDCENLTQQNMTLIDNVQKLMHKHVNQESKVSELKEVNGKLSLDLKNLRSS 872
Query: 328 LTVSWSDDDG-----SKGDGERESIKHVAALIGRVFSDAESCSE---------DLAYDEL 373
L V+ SD+D ++ +S++ +A + E+ + + D+L
Sbjct: 873 LNVAISDNDQILTQLAELSKNYDSLEQESAQLNSGLKSLEAEKQLLHTENEELHIRLDKL 932
Query: 374 VVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGY--------------LEKNSELNSEV 419
K K++D+ K+L ++ +NL+EE + L K+S+L +++
Sbjct: 933 TGKLKIEESKSSDLGKKLTARQEEISNLKEENMSQSQAITSVKSKLDETLSKSSKLEADI 992
Query: 420 RMLNSQLSNV 429
L +++S V
Sbjct: 993 EHLKNKVSEV 1002
Score = 30.8 bits (68), Expect = 7.3
Identities = 48/232 (20%), Positives = 93/232 (39%), Gaps = 19/232 (8%)
Query: 212 EITVNSKNDKKGKGIAFASSVELDE-LQVNQEDDEDMTESLTLLGRQFKKIVKQYDKRPR 270
E+T N + ++GK EL L+ NQ + L L ++ + + + K
Sbjct: 1359 EMTENIHSLEEGKEETKKEIAELSSRLEDNQLATNKLKNQLDHLNQEIR-LKEDVLKEKE 1417
Query: 271 SIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*KKSLTV 330
S+ ++ ++ NQ KE + + ++ KIE + K
Sbjct: 1418 SLIISLEESLSNQRQKESSLLDAKNELEHMLDDTSRKNSSLMEKIESINSSLDDKSFELA 1477
Query: 331 SWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLAYDELVV------------SYK 378
S + G+ ES+ +L+ + S + E + DE + +Y+
Sbjct: 1478 SAVEKLGALQKLHSESL----SLMENIKSQLQEAKEKIQVDESTIQELDHEITASKNNYE 1533
Query: 379 -RLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNV 429
+LNDK++ I E + + N L EE+ ++E SE+ NS+L+++
Sbjct: 1534 GKLNDKDSIIRDLSENIEQLNNLLAEEKSAVKRLSTEKESEILQFNSRLADL 1585
>MYHB_MOUSE (O08638) Myosin heavy chain, smooth muscle isoform (SMMHC)
Length = 1972
Score = 40.8 bits (94), Expect = 0.007
Identities = 74/369 (20%), Positives = 147/369 (39%), Gaps = 53/369 (14%)
Query: 112 LKIAHEGTT---KVKSAKFQLLTTKYENLRMLDDESIQDYHLNILDI-ANSFESAGEKIS 167
LK HE +V+ K + + E L+ + D+H I D+ A E +
Sbjct: 1028 LKSKHESMISELEVRLKKEEKSRQELEKLKRKLEGDASDFHEQIADLQAQIAELKMQLAK 1087
Query: 168 DEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIA 227
E+ ++ L L + K A+++ R++ + +L L + E +K +K+ + +
Sbjct: 1088 KEEELQAALARLDEEIAQKNNALKKIRELEGH-ISDLQEDLDS-ERAARNKAEKQKRDLG 1145
Query: 228 FASSVELDELQVNQEDDEDMT-----------ESLTLLGRQFKKIVKQYDKRPRSIGQNI 276
EL+ L+ ED D T + +T+L + + + ++ + + + Q
Sbjct: 1146 ----EELEALKTELEDTLDSTATQQELRAKREQEVTVLKKALDEETRSHEAQVQEMRQKH 1201
Query: 277 RPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGY---GHIKI--ECASFLKK*KKSLTVS 331
++ + + + + N K Q E E G +++ + ++ KK L V
Sbjct: 1202 TQAVEELTEQLEQFKRAKANLD-KSKQTLEKENADLAGELRVLGQAKQEVEHKKKKLEVQ 1260
Query: 332 WSDDDGSKGDGER-------------ESIKHVAALIGRVFSDAESCSEDLAY-------- 370
D DGER ++ V ++ A ++D+A
Sbjct: 1261 LQDLQSKCSDGERARAELSDKVHKLQNEVESVTGMLNEAEGKAIKLAKDVASLGSQLQDT 1320
Query: 371 DELVVSYKRLNDKNTDICKQLEEQKN-ITNNLEEERVGYLEKNSELNSEVRMLNSQLSNV 429
EL+ R + +QLE+++N + + L+EE +E L V LN QLS+
Sbjct: 1321 QELLQEETRQKLNVSTKLRQLEDERNSLQDQLDEE----MEAKQNLERHVSTLNIQLSDS 1376
Query: 430 MKQVKMMAA 438
K+++ A+
Sbjct: 1377 KKKLQDFAS 1385
Score = 32.3 bits (72), Expect = 2.5
Identities = 17/68 (25%), Positives = 37/68 (54%)
Query: 372 ELVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMK 431
+L K++ + D+ +QLEE++ L+ E+V K +L ++ +++ Q S + K
Sbjct: 940 QLQAERKKMAQQMLDLEEQLEEEEAARQKLQLEKVTAEAKIKKLEDDILVMDDQNSKLSK 999
Query: 432 QVKMMAAR 439
+ K++ R
Sbjct: 1000 ERKLLEER 1007
>MYHA_RAT (Q9JLT0) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 40.8 bits (94), Expect = 0.007
Identities = 67/333 (20%), Positives = 139/333 (41%), Gaps = 42/333 (12%)
Query: 97 KLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENL----RMLDDESIQDYHLNI 152
K KQ + LE LK E T +A+ +L T + + + + L+DE+ +++ I
Sbjct: 1136 KAEKQKRDLSEELEALKTELEDTLDTTAAQQELRTKREQEVAELKKALEDET-KNHEAQI 1194
Query: 153 LDIANSFESAGEKISDE-----------KLVRKILRSLPKRFDMKVTAIEEARDISSFKV 201
D+ +A E++S++ + ++ L + K +V +++ + S K
Sbjct: 1195 QDMRQRHATALEELSEQLEQAKRFKANLEKNKQGLETDNKELACEVKVLQQVKAESEHKR 1254
Query: 202 DELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKK- 260
+L +Q V+ +G + + + ++LQ ++ + E G +F K
Sbjct: 1255 KKLDAQVQELHAKVS-----EGDRLRVELAEKANKLQNELDNVSTLLEEAEKKGMKFAKD 1309
Query: 261 ---IVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIEC 317
+ Q + + R + N S+ + + ++ + Q + E E +E
Sbjct: 1310 AAGLESQLQDTQELLQEETRQKL-NLSSRIRQLEEEKNSLQEQ----QEEEEEARKNLEK 1364
Query: 318 ASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDL-----AYDE 372
+ + + T DDD +G E+ K ++ D E+ S+ L AYD+
Sbjct: 1365 QVLALQSQLADTKKKVDDDLGTIEGLEEAKK-------KLLKDVEALSQRLEEKVLAYDK 1417
Query: 373 LVVSYKRLNDKNTDICKQLEEQKNITNNLEEER 405
L + RL + D+ L+ Q+ I +NLE+++
Sbjct: 1418 LEKTKNRLQQELDDLTVDLDHQRQIVSNLEKKQ 1450
Score = 32.7 bits (73), Expect = 1.9
Identities = 18/62 (29%), Positives = 33/62 (53%)
Query: 378 KRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMA 437
K++ D+ +QL+E++ L+ E+V K ++ EV +L Q S +K+ K+M
Sbjct: 946 KKMQAHIQDLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEVLLLEDQNSKFIKEKKLME 1005
Query: 438 AR 439
R
Sbjct: 1006 DR 1007
>MYHA_BOVIN (Q27991) Myosin heavy chain, nonmuscle type B (Cellular
myosin heavy chain, type B) (Nonmuscle myosin heavy
chain-B) (NMMHC-B)
Length = 1976
Score = 40.8 bits (94), Expect = 0.007
Identities = 64/328 (19%), Positives = 134/328 (40%), Gaps = 32/328 (9%)
Query: 97 KLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRMLD---DESIQDYHLNIL 153
K KQ + LE LK E T +A+ +L T + + + L +E + + I
Sbjct: 1136 KAEKQKRDLSEELEALKTELEDTLDTTAAQQELRTKREQEVAELKKALEEETKSHEAQIQ 1195
Query: 154 DIANSFESAGEKISDE-----------KLVRKILRSLPKRFDMKVTAIEEARDISSFKVD 202
D+ +A E++S++ + ++ L + K +V +++ + S K
Sbjct: 1196 DMRQRHATALEELSEQLEQAKRFKANLEKNKQGLETDNKELACEVKVLQQVKAESEHKRK 1255
Query: 203 ELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKKIV 262
+L +Q V+ +G + + + ++LQ ++ + E G +F K
Sbjct: 1256 KLDAQVQELHAKVS-----EGDRLRVELAEKANKLQNELDNVSTLLEEAEKKGIKFAKDA 1310
Query: 263 KQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLK 322
+ + + + ++ + + +R E+ Q E E E L+
Sbjct: 1311 AGLESQLQDTQELLQEETRQKLNLSSRIRQLEEERSSLQEQQEEEE-------EARRSLE 1363
Query: 323 K*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDL-----AYDELVVSY 377
K ++L +D K D + +I+++ ++ D E S+ L AYD+L +
Sbjct: 1364 KQLQALQAQLTDTK-KKVDDDLGTIENLEEAKKKLLKDVEVLSQRLEEKALAYDKLEKTK 1422
Query: 378 KRLNDKNTDICKQLEEQKNITNNLEEER 405
RL + D+ L+ Q+ I +NLE+++
Sbjct: 1423 TRLQQELDDLLVDLDHQRQIVSNLEKKQ 1450
Score = 32.3 bits (72), Expect = 2.5
Identities = 17/62 (27%), Positives = 33/62 (52%)
Query: 378 KRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMA 437
K++ D+ +QL+E++ L+ E+V K ++ E+ +L Q S +K+ K+M
Sbjct: 946 KKMQAHIQDLEEQLDEEEGARQKLQLEKVTAEAKIKKMEEEILLLEDQNSKFIKEKKLME 1005
Query: 438 AR 439
R
Sbjct: 1006 DR 1007
>SMC_METJA (Q59037) Chromosome partition protein smc homolog
Length = 1169
Score = 40.0 bits (92), Expect = 0.012
Identities = 83/400 (20%), Positives = 168/400 (41%), Gaps = 91/400 (22%)
Query: 67 EEWTAAEDELALGNS--KALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKS 124
E+ AE+EL + ++ + V+ N+ KL K+ +D + +K+ E +K+
Sbjct: 176 EKKKKAEEELKKARELIEMIDIRISEVENNLKKLKKE---KEDAEKYIKLNEE----LKA 228
Query: 125 AKFQLLTTKYENLRMLDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFD 184
AK+ L+ K L +L E+IQ+ N+ ++ N F S
Sbjct: 229 AKYALILKKVSYLNVLL-ENIQNDIKNLEELKNEFLS----------------------- 264
Query: 185 MKVTAIEEARDISSFKVDELIGSLQNFEITVNS---KNDKKGKGIAFASSVELDELQVNQ 241
KV E+ ++N ++ +N+ + ++KG + EL+V
Sbjct: 265 ---------------KVREIDVEIENLKLRLNNIINELNEKGNEEVLELHKSIKELEVEI 309
Query: 242 EDDEDMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPS---KEKMVRTDE---K 295
E+D+ + +S + KK+ + + + + I + + I+N+ S KE+ ++ E K
Sbjct: 310 ENDKKVLDSSI---NELKKVEVEIENKKKEIKETQKKIIENRDSIIEKEQQIKEIEEKIK 366
Query: 296 NFQYKGVQCHECEGYGHIKIECASFLKK*KKS-LTVSWSDDDGSKGDGE----RESIKHV 350
N Y+ + E E S +K K+S + ++ D+ +K E ++ + +
Sbjct: 367 NLNYEKERLKEAIA------ESESIIKHLKESEMEIA---DEIAKNQNELYRLKKELNDL 417
Query: 351 AALIGRVFSDAE-------SCSEDLAYDELVVS---YKRLNDKNTDI------CKQLEE- 393
LI R + E E+L E V + Y L + N +I K+LEE
Sbjct: 418 DNLINRKNFEIEKNNEMIKKLKEELETVEDVDTKPLYLELENLNVEIEFSKRGIKELEEK 477
Query: 394 QKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQV 433
+K + L+E Y+++N+ + + M + ++++
Sbjct: 478 KKELQAKLDELHAEYVKENARIKALKEMEELSMDRAIREI 517
>RA50_METJA (Q58718) DNA double-strand break repair rad50 ATPase
Length = 1005
Score = 40.0 bits (92), Expect = 0.012
Identities = 61/307 (19%), Positives = 131/307 (41%), Gaps = 54/307 (17%)
Query: 155 IANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDEL------IGSL 208
I FE EK+ +E +K+L ++ + +EE + K EL + ++
Sbjct: 220 IKKEFEDI-EKLFNEWENKKLL------YEKFINKLEERKRALELKNQELKILEYDLNTV 272
Query: 209 QNFEITVNSKNDKKGKGIAFASSVE-----LDELQVNQEDDEDMTESLTLLGRQFKKIVK 263
T+N D+ K + + L EL+ + ED +T+ L ++ +K+ +
Sbjct: 273 VEARETLNRHKDEYEKYKSLVDEIRKIESRLRELKSHYEDYLKLTKQLEIIKGDIEKLKE 332
Query: 264 QYDKRPRSIGQNIRPNIDNQPS-----KEKMVRTDE-----KNFQYKGVQCHECEGYGHI 313
+K R +IDN + K+++ R + + + + + E Y I
Sbjct: 333 FINK------SKYRDDIDNLDTLLNKIKDEIERVETIKDLLEELKNLNEEIEKIEKYKRI 386
Query: 314 KIECASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLAYDEL 373
EC + +K + + + + + +++++ L + +S +++ E
Sbjct: 387 CEECKEYYEK--------YLELEEKAVEYNKLTLEYITLL-----QEKKSIEKNINDLET 433
Query: 374 VVSYKRLNDKNTDI------CKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLS 427
++ KN DI K++EE+K + NL++E++ +K E+NSE++ L L
Sbjct: 434 RINKLLEETKNIDIESIENSLKEIEEKKKVLENLQKEKIELNKKLGEINSEIKRLKKILD 493
Query: 428 NVMKQVK 434
+K+V+
Sbjct: 494 E-LKEVE 499
>MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle
Length = 1978
Score = 38.5 bits (88), Expect = 0.035
Identities = 70/341 (20%), Positives = 134/341 (38%), Gaps = 47/341 (13%)
Query: 138 RMLDDESIQDYHLNILDI-ANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDI 196
R L+ ES D H I ++ A E + E+ ++ L L K A+++ R++
Sbjct: 1063 RKLEGES-SDLHEQIAELQAQIAELKAQLAKKEEELQAALARLEDETSQKNNALKKIREL 1121
Query: 197 SSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMT-------- 248
S + +L L++ E +K +K+ + + S EL+ L+ ED D T
Sbjct: 1122 ESH-ISDLQEDLES-EKAARNKAEKQKRDL----SEELEALKTELEDTLDTTATQQELRA 1175
Query: 249 ---ESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCH 305
+ +T+L R ++ + ++ + + + Q ++ + + + + N
Sbjct: 1176 KREQEVTVLKRALEEETRTHEAQVQEMRQKHTQAVEELTEQLEQFKRAKANLDKTKQTLE 1235
Query: 306 ECEGYGHIKIECASFLKK*----KKSLTVSWSDDDGSKGDGERE-------------SIK 348
+ +I S K+ KK L V D DGER ++
Sbjct: 1236 KDNADLANEIRSLSQAKQDVEHKKKKLEVQLQDLQSKYSDGERVRTELNEKVHKLQIEVE 1295
Query: 349 HVAALIGRVFSDAESCSEDLAY--------DELVVSYKRLNDKNTDICKQLEEQKNITNN 400
+V +L+ S ++D+A EL+ R T +QLE+ KN +
Sbjct: 1296 NVTSLLNEAESKNIKLTKDVATLGSQLQDTQELLQEETRQKLNVTTKLRQLEDDKN---S 1352
Query: 401 LEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVKMMAARTD 441
L+E+ +E L + L QLS+ K+++ A +
Sbjct: 1353 LQEQLDEEVEAKQNLERHISTLTIQLSDSKKKLQEFTATVE 1393
Score = 34.7 bits (78), Expect = 0.50
Identities = 65/312 (20%), Positives = 115/312 (36%), Gaps = 35/312 (11%)
Query: 111 ILKIAHEGTTKVKSAKFQLLTTKY--------ENLRMLD------DESIQDYHLNILDIA 156
+LK A E T+ A+ Q + K+ E L D++ Q + D+A
Sbjct: 1183 VLKRALEEETRTHEAQVQEMRQKHTQAVEELTEQLEQFKRAKANLDKTKQTLEKDNADLA 1242
Query: 157 NSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDELIGSLQNFEITVN 216
N S + D + +K L + K + E R + KV +L ++N +N
Sbjct: 1243 NEIRSLSQAKQDVEHKKKKLEVQLQDLQSKYSDGERVRTELNEKVHKLQIEVENVTSLLN 1302
Query: 217 ---SKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKKIVKQYDKRPRSIG 273
SKN K K +A S D ++ QE+ T L +Q + S+
Sbjct: 1303 EAESKNIKLTKDVATLGSQLQDTQELLQEETRQKLNVTTKL--------RQLEDDKNSLQ 1354
Query: 274 QNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIECASFLKK*KKSLTVSWS 333
+ + ++ + + E+ + T + E E L++ +SLT +
Sbjct: 1355 EQLDEEVEAKQNLERHISTLTIQLSDSKKKLQEFTATVETMEEGKKKLQREIESLTQQFE 1414
Query: 334 DDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLAYDELVVSYKRLNDKNTDICKQLEE 393
+ S E+ R+ + + DL +VS K D + L E
Sbjct: 1415 EKAASYDKLEKTK--------NRLQQELDDLVVDLDNQRQLVSNLEKKQKKFD--QMLAE 1464
Query: 394 QKNITNNLEEER 405
+KNI++ +ER
Sbjct: 1465 EKNISSKYADER 1476
Score = 30.4 bits (67), Expect = 9.5
Identities = 16/74 (21%), Positives = 38/74 (50%)
Query: 366 EDLAYDELVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQ 425
E+ +L K++ + D+ +QLEE++ L+ E+V K ++ ++ ++ Q
Sbjct: 939 EEERSQQLQAEKKKMQQQMLDLEEQLEEEEAARQKLQLEKVTADGKIKKMEDDILIMEDQ 998
Query: 426 LSNVMKQVKMMAAR 439
+ + K+ K++ R
Sbjct: 999 NNKLTKERKLLEER 1012
>BPEB_HUMAN (Q8WXK8) Bullous pemphigoid antigen 1, isoform 7 (Bullous
pemphigoid antigen) (BPA) (Hemidesmosomal plaque protein)
(Dystonia musculorum protein) (Dystonin) (Fragment)
Length = 3060
Score = 38.1 bits (87), Expect = 0.045
Identities = 75/358 (20%), Positives = 161/358 (44%), Gaps = 42/358 (11%)
Query: 84 LNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRMLDDE 143
L + NVV +NM ++ M+ ++ LK + G ++A + L YE ++ ++E
Sbjct: 820 LRSELNVVLQNMNQVYS---MSSTYIDKLKTVNLGLKNTQAA--EALVKLYET-KLCEEE 873
Query: 144 SIQDYHLNILDIANSFESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEARDISSFKVDE 203
++ NI ++ ++ + ++ DEK R++ +L +++A+ IS DE
Sbjct: 874 AVIADKNNIENLISTLKQWRSEV-DEK--RQVFHALEDE-------LQKAKAIS----DE 919
Query: 204 LIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGRQFKKI-- 261
+ + + ++ + +K + + +V + ++ D E + +SL + +
Sbjct: 920 MFKTYKERDLDFDWHKEKADQLVERWQNVHV-QIDNRLRDLEGIGKSLKYYRDTYHPLDD 978
Query: 262 -VKQYDKRPRSIGQNIRPN---IDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIEC 317
++Q + R I +N N + Q +++KM+ ++ + K + EC+ Y +
Sbjct: 979 WIQQVETTQRKIQENQPENSKTLATQLNQQKMLVSE---IEMKQSKMDECQKYAE---QY 1032
Query: 318 ASFLKK*K-KSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDLAYDELVVS 376
++ +K + +++T D K +R ++ A LI + F D + Y LV
Sbjct: 1033 SATVKDYELQTMTYRAMVDSQQKSPVKRRRMQSSADLIIQEFMDLRT-----RYTALVTL 1087
Query: 377 YKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVK 434
+ D K+LEE++ +LEEE+ ++EK EL V ++ L + K K
Sbjct: 1088 MTQYIKFAGDSLKRLEEEEK---SLEEEKKEHVEKAKELQKWVSNISKTLKDAEKAGK 1142
>M6_STRPY (P08089) M protein, serotype 6 precursor
Length = 483
Score = 37.7 bits (86), Expect = 0.059
Identities = 55/238 (23%), Positives = 98/238 (41%), Gaps = 13/238 (5%)
Query: 72 AEDELA---LGNSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQ 128
AE+E A N +A+ L +D+ + I + +K+T+ LK + T K K AK Q
Sbjct: 147 AEEEAANKERENKEAIGTLKKTLDETVKDKIAKEQESKETIGTLKKTLDETVKDKIAKEQ 206
Query: 129 LLTTKYENLRMLDDESIQDYHLNILDIANSFESAG--EKISDEKLVRKILRSLPKRFDMK 186
L+ DE+++D I S E+ G +KI DE + KI R + D+
Sbjct: 207 ESKETIGTLKKTLDETVKD---KIAKEQESKETIGTLKKILDETVKDKIAREQKSKQDIG 263
Query: 187 VTAIEEARDISSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELD----ELQVNQE 242
E A+ KV E ++ + + K+ + + ELD E Q++
Sbjct: 264 ALKQELAKKDEGNKVSEASRKGLRRDLDASREAKKQVEKDLANLTAELDKVKEEKQISDA 323
Query: 243 DDEDMTESLTLLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYK 300
+ + L K++ K ++ + + N + + SK K+ ++ Q K
Sbjct: 324 SRQGLRRDLDASREAKKQVEKALEEANSKLAALEKLNKELEESK-KLTEKEKAELQAK 380
>RA50_METMP (P62134) DNA double-strand break repair rad50 ATPase
Length = 993
Score = 37.4 bits (85), Expect = 0.078
Identities = 67/359 (18%), Positives = 143/359 (39%), Gaps = 47/359 (13%)
Query: 80 NSKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRM 139
N+K + N N K I++ ++ + L +LK + + K + L E + +
Sbjct: 299 NNKLIGHKSNYESYNKLKTIEESLLKE--LGVLKESLKDNKKNPDELKENLKENDEKILI 356
Query: 140 LDDESIQDYHLNILDIANSFESAGEKISDEKLVRKILRSL---PKRFDMKVTAIEEARDI 196
LD I + E ++I + K+ +K + +L K +D + EE +
Sbjct: 357 LD---------KIKEKIKELEFIEKQIYEIKIHKKTVETLFDSVKIYDDSIKTFEELKTK 407
Query: 197 SSFKVDELIGSLQNFEITVNSKNDKKGKGIAFASSVELDELQVNQEDDEDMTESLTLLGR 256
+ + L+ + E + ++ D+K K I+ + E E ++N E++ L
Sbjct: 408 KN-SYENLLKEKFDLEKKLQNETDEKTKLISELTDFEKIEEKINLENE---------LKE 457
Query: 257 QFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGHIKIE 316
+++ + ++ DK + + + + SK ++ +T + CH C+
Sbjct: 458 KYEDLSEKIDKLNEIVLKKESKISEYKNSKAELEKTKDS--------CHVCQ-------- 501
Query: 317 CASFLKK*KKSLTVSWSDDDGSKGDGERESIKHVAALIGRVFSDAESCSEDL-AYDELVV 375
+ ++ K+ L ++ S+ E+ S + + + + + E L D +
Sbjct: 502 -SKITEEKKQELLEKYN----SEIQNEQLSTESLKKQLEIILNKKEKMKVKLNEIDSFKL 556
Query: 376 SYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLSNVMKQVK 434
Y L +K K E T L E G + + S LN E+ ++ ++L N+ K
Sbjct: 557 KYGELKEKKNYSLKVEESIIETTEKLNE-LTGKINEYSSLNDEISLIENKLKNLENDYK 614
Score = 33.1 bits (74), Expect = 1.5
Identities = 78/359 (21%), Positives = 151/359 (41%), Gaps = 53/359 (14%)
Query: 81 SKALNALFNVVDKNMFKLIKQCIMAKDTLEILKIAHEGTTKVKSAKFQLLTTKYENLRML 140
S+ +N + ++ + KL + + LE L+ ++ + K ++L KYENL L
Sbjct: 164 SEKMNIVKKSYEETLLKLEGELTQEPEILENLEKLKNEVSESEILKEEILK-KYENLEKL 222
Query: 141 DDESIQDYHLNILDIANSF-------ESAGEKISDEKLVRKILRSLPKRFDMKVTAIEEA 193
E + IL + F E+ + IS+ K + +++ ++ EE+
Sbjct: 223 KLEKNSE----ILQMEEKFAENNQLKENLKDIISEIKNINLEIQNFKNSLNL---VAEES 275
Query: 194 RDISSFKVDELIGSLQNFEITVNSKNDKK-GKGIAFASSVELDELQVNQEDDEDMTESLT 252
++IS + +E E+ + N+K G + S +L ++ E + + L
Sbjct: 276 KNIS--ENEENYKKYLELELKIKELNNKLIGHKSNYESYNKLKTIE------ESLLKELG 327
Query: 253 LLGRQFKKIVKQYDKRPRSIGQNIRPNIDNQPSKEKMVRTDEKNFQYKGVQCHECEGYGH 312
+L K+ +K K P + +N++ N EK++ D+ + K ++ E + Y
Sbjct: 328 VL----KESLKDNKKNPDELKENLKEN------DEKILILDKIKEKIKELEFIEKQIY-E 376
Query: 313 IKIECASFLKK*KKSLTVSWSD----DDGSKGDGERESIKHVAALIGRVFSDAESCSEDL 368
IKI KK++ + DD K E ++ K+ + + D E ++
Sbjct: 377 IKIH--------KKTVETLFDSVKIYDDSIKTFEELKTKKNSYENLLKEKFDLEKKLQNE 428
Query: 369 AYDELVVSYKRLNDKNTDICKQLEEQKNITNNLEEERVGYLEKNSELNSEVRMLNSQLS 427
DE L D +++EE+ N+ N L+E+ EK +LN V S++S
Sbjct: 429 T-DEKTKLISELTD-----FEKIEEKINLENELKEKYEDLSEKIDKLNEIVLKKESKIS 481
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,604,490
Number of Sequences: 164201
Number of extensions: 2038224
Number of successful extensions: 7949
Number of sequences better than 10.0: 145
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 120
Number of HSP's that attempted gapping in prelim test: 7726
Number of HSP's gapped (non-prelim): 362
length of query: 441
length of database: 59,974,054
effective HSP length: 113
effective length of query: 328
effective length of database: 41,419,341
effective search space: 13585543848
effective search space used: 13585543848
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC139747.1


