

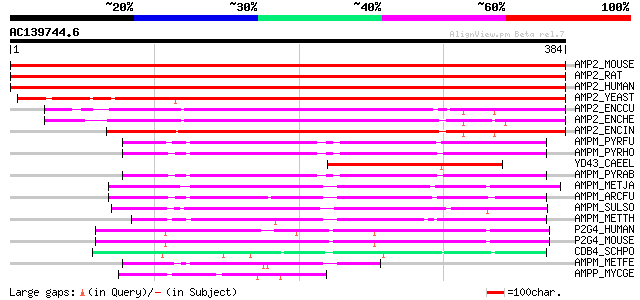
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139744.6 + phase: 1 /partial
(384 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AMP2_MOUSE (O08663) Methionine aminopeptidase 2 (EC 3.4.11.18) (... 612 e-175
AMP2_RAT (P38062) Methionine aminopeptidase 2 (EC 3.4.11.18) (Me... 612 e-175
AMP2_HUMAN (P50579) Methionine aminopeptidase 2 (EC 3.4.11.18) (... 610 e-174
AMP2_YEAST (P38174) Methionine aminopeptidase 2 (EC 3.4.11.18) (... 397 e-110
AMP2_ENCCU (Q8SR45) Methionine aminopeptidase 2 (EC 3.4.11.18) (... 268 1e-71
AMP2_ENCHE (Q6XMH6) Methionine aminopeptidase 2 (EC 3.4.11.18) (... 261 3e-69
AMP2_ENCIN (Q6XMH7) Methionine aminopeptidase 2 (EC 3.4.11.18) (... 258 2e-68
AMPM_PYRFU (P56218) Methionine aminopeptidase (EC 3.4.11.18) (MA... 163 8e-40
AMPM_PYRHO (O58362) Methionine aminopeptidase (EC 3.4.11.18) (MA... 162 2e-39
YD43_CAEEL (P50581) Hypothetical protein T27A8.3 in chromosome X 158 2e-38
AMPM_PYRAB (Q9UYT4) Methionine aminopeptidase (EC 3.4.11.18) (MA... 155 2e-37
AMPM_METJA (Q58725) Methionine aminopeptidase (EC 3.4.11.18) (MA... 147 3e-35
AMPM_ARCFU (O28438) Methionine aminopeptidase (EC 3.4.11.18) (MA... 140 5e-33
AMPM_SULSO (P95963) Methionine aminopeptidase (EC 3.4.11.18) (MA... 137 5e-32
AMPM_METTH (O27355) Methionine aminopeptidase (EC 3.4.11.18) (MA... 119 2e-26
P2G4_HUMAN (Q9UQ80) Proliferation-associated protein 2G4 (Cell c... 93 1e-18
P2G4_MOUSE (P50580) Proliferation-associated protein 2G4 (Prolif... 92 2e-18
CDB4_SCHPO (Q09184) Curved DNA-binding protein (42 kDa protein) 78 4e-14
AMPM_METFE (P22624) Probable methionine aminopeptidase (EC 3.4.1... 75 2e-13
AMPP_MYCGE (P47566) Putative Xaa-Pro aminopeptidase (EC 3.4.11.9... 49 3e-05
>AMP2_MOUSE (O08663) Methionine aminopeptidase 2 (EC 3.4.11.18)
(MetAP 2) (Peptidase M 2) (Initiation factor 2
associated 67 kDa glycoprotein) (p67) (p67eIF2)
Length = 478
Score = 612 bits (1578), Expect = e-175
Identities = 287/384 (74%), Positives = 334/384 (86%)
Query: 1 GGEAQKKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNE 60
G +KKKK KKK+G KVQTDPP++PIC+L+ +GV+P GQ +YP D RTA R T+E
Sbjct: 94 GATGKKKKKKKKKRGPKVQTDPPSVPICDLYPNGVFPKGQECEYPPTQDGRTAAWRTTSE 153
Query: 61 EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGL 120
EK+ALD++ +I+N+ R AAEAHRQ RK++ WIKPGMTMI+ICE+LE+ +RKLIKE+GL
Sbjct: 154 EKKALDQASEEIWNDFREAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGL 213
Query: 121 KAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKY 180
AGLAFPTGCS N+CAAHYTPNAGD TVL+YDD+ KIDFGTHI+GRIIDCAFT++FNPKY
Sbjct: 214 NAGLAFPTGCSLNNCAAHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFTVTFNPKY 273
Query: 181 DKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSI 240
D L+ AV+DATNTGIK AGIDV LC++G AIQEVMESYEVE+DGKTYQVK IRNLNGHSI
Sbjct: 274 DILLTAVKDATNTGIKCAGIDVRLCDVGEAIQEVMESYEVEIDGKTYQVKPIRNLNGHSI 333
Query: 241 SPYRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMP 300
PYRIHAGKTVPIVKGGEAT MEE E YAIETFGSTG+G VHDDM+CSHYMKNFD G++P
Sbjct: 334 GPYRIHAGKTVPIVKGGEATRMEEGEVYAIETFGSTGKGVVHDDMECSHYMKNFDVGHVP 393
Query: 301 LRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGC 360
+RL +K LL+VIN+NF TLAFC+RWLDR G +KY MALK+LCD GIVD YPPLCD+KG
Sbjct: 394 IRLPRTKHLLNVINENFGTLAFCRRWLDRLGESKYLMALKNLCDLGIVDPYPPLCDIKGS 453
Query: 361 YTAQFEHTIMLRPTCKEVVSRGDD 384
YTAQFEHTI+LRPTCKEVVSRGDD
Sbjct: 454 YTAQFEHTILLRPTCKEVVSRGDD 477
>AMP2_RAT (P38062) Methionine aminopeptidase 2 (EC 3.4.11.18) (MetAP
2) (Peptidase M 2) (Initiation factor 2 associated 67
kDa glycoprotein) (p67) (p67eIF2)
Length = 478
Score = 612 bits (1577), Expect = e-175
Identities = 286/384 (74%), Positives = 335/384 (86%)
Query: 1 GGEAQKKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNE 60
G +KKKK KKK+G +VQTDPP++PIC+L+ +GV+P GQ +YP D RTA R T+E
Sbjct: 94 GAAGKKKKKKKKKRGPRVQTDPPSVPICDLYPNGVFPKGQECEYPPTQDGRTAAWRTTSE 153
Query: 61 EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGL 120
EK+ALD++ +I+N+ R AAEAHRQ RK++ WIKPGMTMI+ICE+LE+ +RKLIKE+GL
Sbjct: 154 EKKALDQASEEIWNDFREAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGL 213
Query: 121 KAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKY 180
AGLAFPTGCS N+CAAHYTPNAGD TVL+YDD+ KIDFGTHI+GRIIDCAFT++FNPKY
Sbjct: 214 NAGLAFPTGCSLNNCAAHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFTVTFNPKY 273
Query: 181 DKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSI 240
D L++AV+DATNTGIK AGIDV LC++G AIQEVMESYEVE+DGKTYQVK IRNLNGHSI
Sbjct: 274 DILLKAVKDATNTGIKCAGIDVRLCDVGEAIQEVMESYEVEIDGKTYQVKPIRNLNGHSI 333
Query: 241 SPYRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMP 300
PYRIHAGKTVPIVKGGEAT MEE E YAIETFGSTG+G VHDDM+CSHYMKNFD G++P
Sbjct: 334 GPYRIHAGKTVPIVKGGEATRMEEGEVYAIETFGSTGKGVVHDDMECSHYMKNFDVGHVP 393
Query: 301 LRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGC 360
+RL +K LL+VIN+NF TLAFC+RWLDR G +KY MALK+LCD GIVD YPPLCD+KG
Sbjct: 394 IRLPRTKHLLNVINENFGTLAFCRRWLDRLGESKYLMALKNLCDLGIVDPYPPLCDIKGS 453
Query: 361 YTAQFEHTIMLRPTCKEVVSRGDD 384
YTAQFEHTI+LRPTCKEVVSRGDD
Sbjct: 454 YTAQFEHTILLRPTCKEVVSRGDD 477
>AMP2_HUMAN (P50579) Methionine aminopeptidase 2 (EC 3.4.11.18)
(MetAP 2) (Peptidase M 2) (Initiation factor 2
associated 67 kDa glycoprotein) (p67) (p67eIF2)
Length = 478
Score = 610 bits (1574), Expect = e-174
Identities = 286/384 (74%), Positives = 334/384 (86%)
Query: 1 GGEAQKKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNE 60
G +KKKK KKK+G KVQTDPP++PIC+L+ +GV+P GQ +YP D RTA R T+E
Sbjct: 94 GATGKKKKKKKKKRGPKVQTDPPSVPICDLYPNGVFPKGQECEYPPTQDGRTAAWRTTSE 153
Query: 61 EKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGL 120
EK+ALD++ +I+N+ R AAEAHRQ RK++ WIKPGMTMI+ICE+LE+ +RKLIKE+GL
Sbjct: 154 EKKALDQASEEIWNDFREAAEAHRQVRKYVMSWIKPGMTMIEICEKLEDCSRKLIKENGL 213
Query: 121 KAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKY 180
AGLAFPTGCS N+CAAHYTPNAGD TVL+YDD+ KIDFGTHI+GRIIDCAFT++FNPKY
Sbjct: 214 NAGLAFPTGCSLNNCAAHYTPNAGDTTVLQYDDICKIDFGTHISGRIIDCAFTVTFNPKY 273
Query: 181 DKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSI 240
D L++AV+DATNTGIK AGIDV LC++G AIQEVMESYEVE+DGKTYQVK IRNLNGHSI
Sbjct: 274 DTLLKAVKDATNTGIKCAGIDVRLCDVGEAIQEVMESYEVEIDGKTYQVKPIRNLNGHSI 333
Query: 241 SPYRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMP 300
YRIHAGKTVPIVKGGEAT MEE E YAIETFGSTG+G VHDDM+CSHYMKNFD G++P
Sbjct: 334 GQYRIHAGKTVPIVKGGEATRMEEGEVYAIETFGSTGKGVVHDDMECSHYMKNFDVGHVP 393
Query: 301 LRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGC 360
+RL +K LL+VIN+NF TLAFC+RWLDR G +KY MALK+LCD GIVD YPPLCD+KG
Sbjct: 394 IRLPRTKHLLNVINENFGTLAFCRRWLDRLGESKYLMALKNLCDLGIVDPYPPLCDIKGS 453
Query: 361 YTAQFEHTIMLRPTCKEVVSRGDD 384
YTAQFEHTI+LRPTCKEVVSRGDD
Sbjct: 454 YTAQFEHTILLRPTCKEVVSRGDD 477
>AMP2_YEAST (P38174) Methionine aminopeptidase 2 (EC 3.4.11.18)
(MetAP 2) (Peptidase M 2)
Length = 421
Score = 397 bits (1021), Expect = e-110
Identities = 214/387 (55%), Positives = 266/387 (68%), Gaps = 15/387 (3%)
Query: 6 KKKKNKKKKGKKVQTDPPTIPICELFSDGVYPIGQIMDY-PTVNDSRTAKDRFTNEEKRA 64
KKKKNKKKK KK + LF DG YP G MDY N RT + + KR
Sbjct: 41 KKKKNKKKKKKKSNVKKIEL----LFPDGKYPEGAWMDYHQDFNLQRTTVEE-SRYLKRD 95
Query: 65 LDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARK-------LIKE 117
L+R+E +N+VR AE HR+ R+ ++ I PGM ++ I + +ENT RK L E
Sbjct: 96 LERAEH--WNDVRKGAEIHRRVRRAIKDRIVPGMKLMDIADMIENTTRKYTGAENLLAME 153
Query: 118 DGLKAGLAFPTGCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFN 177
D G+ FPTG S NHCAAH+TPNAGD TVL+Y+DV K+D+G +NG IID AFT+SF+
Sbjct: 154 DPKSQGIGFPTGLSLNHCAAHFTPNAGDKTVLKYEDVMKVDYGVQVNGNIIDSAFTVSFD 213
Query: 178 PKYDKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNG 237
P+YD L+ AV+DAT TGIK AGIDV L +IG AIQEVMESYEVE++G+TYQVK RNL G
Sbjct: 214 PQYDNLLAAVKDATYTGIKEAGIDVRLTDIGEAIQEVMESYEVEINGETYQVKPCRNLCG 273
Query: 238 HSISPYRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAG 297
HSI+PYRIH GK+VPIVK G+ T MEE E++AIETFGSTGRG V + SHY ++ +
Sbjct: 274 HSIAPYRIHGGKSVPIVKNGDTTKMEEGEHFAIETFGSTGRGYVTAGGEVSHYARSAEDH 333
Query: 298 YMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDV 357
+ L S+K+LL I++NF TL FC+R+LDR G KY AL +L G+V YPPL D+
Sbjct: 334 QVMPTLDSAKNLLKTIDRNFGTLPFCRRYLDRLGQEKYLFALNNLVRHGLVQDYPPLNDI 393
Query: 358 KGCYTAQFEHTIMLRPTCKEVVSRGDD 384
G YTAQFEHTI+L KEVVS+GDD
Sbjct: 394 PGSYTAQFEHTILLHAHKKEVVSKGDD 420
>AMP2_ENCCU (Q8SR45) Methionine aminopeptidase 2 (EC 3.4.11.18)
(MetAP 2) (Peptidase M 2) (MetAP2) (MAP-2)
Length = 358
Score = 268 bits (686), Expect = 1e-71
Identities = 154/367 (41%), Positives = 222/367 (59%), Gaps = 28/367 (7%)
Query: 25 IPICELFSDGVYPIGQIMDYPTVNDSRTAK-DRFTNEEKRALDRSESDIYNEVRLAAEAH 83
+PI L DGVY G++ D SR + + FT ESDI + R AAEAH
Sbjct: 12 LPIEFLPKDGVYGKGKLFD------SRNMEIENFT----------ESDILQDARRAAEAH 55
Query: 84 RQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAHYTPNA 143
R+ R +Q ++PG+T+++I +E++ R L+K + G+ FP G S N CAAHYT N
Sbjct: 56 RRARYRVQSIVRPGITLLEIVRSIEDSTRTLLKGER-NNGIGFPAGMSMNSCAAHYTVNP 114
Query: 144 GDPT-VLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVRDATNTGIKAAGIDV 202
G+ VL+ DDV KIDFGTH +GRI+D AFT++F + L+ A R+ T TGIK+ G+DV
Sbjct: 115 GEQDIVLKEDDVLKIDFGTHSDGRIMDSAFTVAFKENLEPLLVAAREGTETGIKSLGVDV 174
Query: 203 PLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVPIVKGGEATVM 262
+C+IG I EV+ SYEVE+ G+ + ++ I +L+GHSIS +RIH G ++P V + T +
Sbjct: 175 RVCDIGRDINEVISSYEVEIGGRMWPIRPISDLHGHSISQFRIHGGISIPAVNNRDTTRI 234
Query: 263 EENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSV---INKNFST 319
+ + +YA+ETF +TG+G + D CSH++ N Y +L +K L+ V + + T
Sbjct: 235 KGDSFYAVETFATTGKGSIDDRPPCSHFVLN---TYKSRKL-FNKDLIKVYEFVKDSLGT 290
Query: 320 LAFCKRWLDRAGCTK--YQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIMLRPTCKE 377
L F R LD G K ++ L G++ YPPL D+ GC AQFEHT+ L KE
Sbjct: 291 LPFSPRHLDYYGLVKGGSLKSVNLLTMMGLLTPYPPLNDIDGCKVAQFEHTVYLSEHGKE 350
Query: 378 VVSRGDD 384
V++RGDD
Sbjct: 351 VLTRGDD 357
>AMP2_ENCHE (Q6XMH6) Methionine aminopeptidase 2 (EC 3.4.11.18)
(MetAP 2) (Peptidase M 2) (MetAP2) (MAP-2)
Length = 358
Score = 261 bits (666), Expect = 3e-69
Identities = 147/367 (40%), Positives = 216/367 (58%), Gaps = 28/367 (7%)
Query: 25 IPICELFSDGVYPIGQIMDYPTVNDSRTAKDRFTNEEKRALDRSESDIYNEVRLAAEAHR 84
+PI L DG Y G+++D T +ESDI + R AAEAHR
Sbjct: 12 LPIEFLPRDGAYRKGRLLDSKNAEVENT---------------TESDILQDARRAAEAHR 56
Query: 85 QTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAHYTPNAG 144
+ R +Q IKPGMT+++I + +E++ R L+ + G+ FP G S N CAAHY+ N G
Sbjct: 57 RVRYKVQSIIKPGMTLLEIVKSIEDSTRILLSGER-NNGIGFPAGMSMNSCAAHYSVNPG 115
Query: 145 DPTV-LEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVRDATNTGIKAAGIDVP 203
+ + L +DV KIDFGTH NGRI+D AFT++F +++ L+ A ++ T TGI++ GID
Sbjct: 116 EKDIILTENDVLKIDFGTHSNGRIMDSAFTIAFKEEFEPLLMAAKEGTETGIRSLGIDAR 175
Query: 204 LCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVPIVKGGEATVME 263
+C+IG I EV+ SYE+E+DGK + ++ + +L+GHSIS ++IH G ++P V + T +
Sbjct: 176 VCDIGRDINEVISSYEMEVDGKKWAIRPVSDLHGHSISQFKIHGGISIPAVNNRDPTRIT 235
Query: 264 ENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSV---INKNFSTL 320
+ +YA+ETF +TG G ++D CSH+M N R +K L+ V + +F TL
Sbjct: 236 GDTFYAVETFATTGEGFINDRSPCSHFMINTHKS----RKLYNKDLIKVYEFVRDSFGTL 291
Query: 321 AFCKRWLDRAGCTKYQMALKD---LCDKGIVDAYPPLCDVKGCYTAQFEHTIMLRPTCKE 377
F R LD + ALK L G+ YPPL D+ G AQFEHT+ L + KE
Sbjct: 292 PFSPRHLDYYNLVE-GSALKSVNLLTMMGLFTPYPPLNDIDGSKVAQFEHTVYLSESGKE 350
Query: 378 VVSRGDD 384
+++RGDD
Sbjct: 351 ILTRGDD 357
>AMP2_ENCIN (Q6XMH7) Methionine aminopeptidase 2 (EC 3.4.11.18)
(MetAP 2) (Peptidase M 2) (MetAP2) (MAP-2)
Length = 358
Score = 258 bits (659), Expect = 2e-68
Identities = 135/323 (41%), Positives = 200/323 (61%), Gaps = 11/323 (3%)
Query: 68 SESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFP 127
++ D+ + R AAEAHR+ R +Q I+PG+T+++I +E++ R L+ E G+ FP
Sbjct: 40 TDCDLLQDARRAAEAHRRARYKVQSIIRPGITLLEIVRSIEDSTRTLL-EGERNNGIGFP 98
Query: 128 TGCSRNHCAAHYTPNAGDPT-VLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEA 186
G S N CAAHYT N G+ VL+ DDV K+DFGTH NGRI+D AFT++F L+ A
Sbjct: 99 AGMSMNSCAAHYTVNPGEEDIVLKEDDVLKVDFGTHSNGRIMDSAFTVAFQENLQPLLMA 158
Query: 187 VRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIH 246
R+ T TGI++ GID +C+IG I EV+ SYEVE++GKT+ ++ + +L+GHSIS ++IH
Sbjct: 159 AREGTETGIRSLGIDARVCDIGRDINEVITSYEVEIEGKTWPIRPVSDLHGHSISQFKIH 218
Query: 247 AGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSS 306
G ++P V + T ++ + +YA+ETF +TG+G ++D CSH+M N R +
Sbjct: 219 GGISIPAVNNRDTTRIKGDTFYAVETFATTGKGFINDRSPCSHFMLNVHKS----RKLFN 274
Query: 307 KSLLSV---INKNFSTLAFCKRWLDRAGCTK--YQMALKDLCDKGIVDAYPPLCDVKGCY 361
K L+ V + +F TL F R LD + ++ L G+ YPPL D+ G
Sbjct: 275 KDLIKVYEFVKSSFGTLPFSPRHLDHYNLVEGGSLKSVNLLTMMGLFTPYPPLNDIDGSK 334
Query: 362 TAQFEHTIMLRPTCKEVVSRGDD 384
AQFEHT+ L KE+++RGDD
Sbjct: 335 VAQFEHTVYLSENGKEILTRGDD 357
>AMPM_PYRFU (P56218) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 295
Score = 163 bits (412), Expect = 8e-40
Identities = 108/295 (36%), Positives = 161/295 (53%), Gaps = 20/295 (6%)
Query: 79 AAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAH 138
A E ++ R+ + +PGM ++++ E +E K+I E G K AFP S N AAH
Sbjct: 9 AGEIAKKVREKAIKLARPGMLLLELAESIE----KMIMELGGKP--AFPVNLSINEIAAH 62
Query: 139 YTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVRDATNTGIKAA 198
YTP GD TVL+ D KID G HI+G I D A T+ + D+L+EA ++A N I A
Sbjct: 63 YTPYKGDTTVLKEGDYLKIDVGVHIDGFIADTAVTVRVGMEEDELMEAAKEALNAAISVA 122
Query: 199 GIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVP-IVKGG 257
V + E+G AI+ E+ G K I NL+GH I Y++HAG ++P I +
Sbjct: 123 RAGVEIKELGKAIEN-----EIRKRG----FKPIVNLSGHKIERYKLHAGISIPNIYRPH 173
Query: 258 EATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNF 317
+ V++E + +AIE F + G GQV + YM D +P+R+ ++ LL+ I + +
Sbjct: 174 DNYVLKEGDVFAIEPFATIGAGQVIEVPPTLIYMYVRD---VPVRVAQARFLLAKIKREY 230
Query: 318 STLAFCKRWLDR-AGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIML 371
TL F RWL + ++ALK L G + YP L +++ AQFEHTI++
Sbjct: 231 GTLPFAYRWLQNDMPEGQLKLALKTLEKAGAIYGYPVLKEIRNGIVAQFEHTIIV 285
>AMPM_PYRHO (O58362) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 295
Score = 162 bits (409), Expect = 2e-39
Identities = 105/295 (35%), Positives = 162/295 (54%), Gaps = 20/295 (6%)
Query: 79 AAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAH 138
A + ++ R+ + KPG++++++ E++E+ I E G K AFP S N AAH
Sbjct: 9 AGKIAKKVREEAVKLAKPGVSLLELAEKIESR----IVELGGKP--AFPANLSLNEVAAH 62
Query: 139 YTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVRDATNTGIKAA 198
YTP GD TVL+ D KID G HI+G I D A T+ +D+L+EA ++A + I A
Sbjct: 63 YTPYKGDQTVLKEGDYLKIDLGVHIDGYIADTAVTVRVGMDFDELMEAAKEALESAISVA 122
Query: 199 GIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVP-IVKGG 257
V + E+G AI+ E+ G I NL+GH I Y++HAG ++P I +
Sbjct: 123 RAGVEVKELGKAIEN-----EIRKRG----FNPIVNLSGHKIERYKLHAGVSIPNIYRPH 173
Query: 258 EATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNF 317
+ V++E + +AIE F +TG GQV + YM DA P+R+ ++ LL+ I + +
Sbjct: 174 DNYVLQEGDVFAIEPFATTGAGQVIEVPPTLIYMYVRDA---PVRMAQARFLLAKIKREY 230
Query: 318 STLAFCKRWLD-RAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIML 371
TL F RWL + ++AL+ L G + YP L +++G QFEHTI++
Sbjct: 231 KTLPFAYRWLQGEMPEGQLKLALRSLERSGALYGYPVLREIRGGMVTQFEHTIIV 285
>YD43_CAEEL (P50581) Hypothetical protein T27A8.3 in chromosome X
Length = 182
Score = 158 bits (400), Expect = 2e-38
Identities = 80/123 (65%), Positives = 93/123 (75%), Gaps = 2/123 (1%)
Query: 221 ELDGKTYQVKSIRNLNGHSISPYRIHAGKTVPIVKGGEATVMEENEYYAIETFGSTGRGQ 280
E G + +VK I +LNG SI+ RIH GKTVPIVKGGE MEENE YAIETFGSTG+G
Sbjct: 29 EFHGNSNEVKPIGSLNGQSIAQCRIHTGKTVPIVKGGEQARMEENEIYAIETFGSTGKGY 88
Query: 281 VHDDMDCSHYMKNFDAG--YMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMA 338
HDDM+ SHYMKNF+ +PLRLQ SK LL +I+KNF+TLAFC+ W+DR TKY MA
Sbjct: 89 FHDDMETSHYMKNFELADEKIPLRLQKSKGLLKLIDKNFATLAFCRCWIDRLEETKYLMA 148
Query: 339 LKD 341
LKD
Sbjct: 149 LKD 151
>AMPM_PYRAB (Q9UYT4) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 295
Score = 155 bits (391), Expect = 2e-37
Identities = 103/295 (34%), Positives = 157/295 (52%), Gaps = 20/295 (6%)
Query: 79 AAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAH 138
A + ++ R+ + KPG++++++ E++E I E G K AFP S N AAH
Sbjct: 9 AGKIAKKVREEAVKLAKPGVSLLELAEKIEGR----IIELGAKP--AFPVNLSLNEIAAH 62
Query: 139 YTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVRDATNTGIKAA 198
YTP GD T L+ D KID G HI+G I D A T+ + D L+EA R+A + I A
Sbjct: 63 YTPYKGDETTLKEGDYLKIDIGVHIDGYIADTAVTVRVGMEEDDLMEAAREALESAISVA 122
Query: 199 GIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVP-IVKGG 257
V + E+G AI++ E+ G I NL+GH I Y++HAG ++P I +
Sbjct: 123 RAGVEIKELGRAIED-----EIRKRG----FNPIVNLSGHKIERYKLHAGISIPNIYRPH 173
Query: 258 EATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNF 317
+ + E + +AIE F +TG GQV + YM DA P+R+ ++ LL+ I + +
Sbjct: 174 DNYKLREGDVFAIEPFATTGAGQVIEVPPTLIYMYVRDA---PVRMVQARFLLAKIKREY 230
Query: 318 STLAFCKRWLD-RAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIML 371
TL F RWL + ++AL+ L G + YP L +++ QFEHTI++
Sbjct: 231 KTLPFAYRWLQGEMPEGQLKLALRTLEKSGALYGYPVLREIRNGLVTQFEHTIIV 285
>AMPM_METJA (Q58725) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 294
Score = 147 bits (372), Expect = 3e-35
Identities = 99/313 (31%), Positives = 153/313 (48%), Gaps = 20/313 (6%)
Query: 69 ESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPT 128
E + Y ++ A + + R+ + I PG+ ++++ E +EN R+L E AFP
Sbjct: 2 EIEGYEKIIEAGKIASKVREEAVKLIXPGVKLLEVAEFVENRIRELGGEP------AFPC 55
Query: 129 GCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVR 188
S N AAHYTP D + DDV K+D G H++G I D A T+ + Y L++A
Sbjct: 56 NISINEIAAHYTPKLNDNLEFKDDDVVKLDLGAHVDGYIADTAITVDLSNSYKDLVKASE 115
Query: 189 DATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAG 248
DA T IK + + E+G IQEV+ES Y K I NL+GH + Y +H G
Sbjct: 116 DALYTVIKEINPPMNIGEMGKIIQEVIES---------YGYKPISNLSGHVMHRYELHTG 166
Query: 249 KTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKS 308
++P V ++ + AIE F + G G V D + Y P+RL ++
Sbjct: 167 ISIPNVYERTNQYIDVGDLVAIEPFATDGFGMVKDGNLGNIYK---FLAKRPIRLPQARK 223
Query: 309 LLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHT 368
LL VI+KN+ L F +RW+ + ++AL L + YP L + + Q EHT
Sbjct: 224 LLDVISKNYPYLPFAERWVLKN--ESERLALNSLIRASCIYGYPILKERENGIVGQAEHT 281
Query: 369 IMLRPTCKEVVSR 381
I++ E+ ++
Sbjct: 282 ILITENGVEITTK 294
>AMPM_ARCFU (O28438) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 291
Score = 140 bits (353), Expect = 5e-33
Identities = 90/303 (29%), Positives = 151/303 (49%), Gaps = 23/303 (7%)
Query: 69 ESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPT 128
+ ++ ++ A + +Q + I PG+ ++++ E +EN I E G K AFP
Sbjct: 2 DDEVREKLIEAGKILKQAVNEAAEKIAPGVKILEVAEFVENR----IIELGAKP--AFPA 55
Query: 129 GCSRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVR 188
S N AAH+TP D + DV K+D G HI+G I D A T+ +L++A +
Sbjct: 56 NISINSDAAHFTPKKNDERTFKEGDVVKLDVGAHIDGYIADMAVTVDLGDN-TELVKAAK 114
Query: 189 DATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAG 248
+A ++ V + EIG AI++ + + Y K I NL GH + PY HA
Sbjct: 115 EALEAAMEVVRAGVSVSEIGKAIEDAITN---------YGFKPIVNLTGHGLLPYLNHAP 165
Query: 249 KTVPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKS 308
++ + +EE AIE F + G G+V + +C Y P+R++ ++
Sbjct: 166 PSIYNYATEKGVTLEEGMVVAIEPFATNGVGKVGERGECEIYSL---LNPRPVRMKMARE 222
Query: 309 LLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHT 368
+L + +N+ TL F KRWL +A + + L +G++ AYP L +V G +Q+EHT
Sbjct: 223 ILKEVEENYKTLPFAKRWLKKAP----DIIISKLAREGVLRAYPVLTEVSGGLVSQWEHT 278
Query: 369 IML 371
+++
Sbjct: 279 LIV 281
>AMPM_SULSO (P95963) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 301
Score = 137 bits (345), Expect = 5e-32
Identities = 95/305 (31%), Positives = 152/305 (49%), Gaps = 21/305 (6%)
Query: 71 DIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGC 130
D N++ LA + + R + +K ++ ICEE+E+ +I E+ KA +FP
Sbjct: 4 DELNKLLLAGKIAAKARDEVSLDVKASAKVLDICEEVES----IIIEN--KAFPSFPCNI 57
Query: 131 SRNHCAAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDKLIEAVRDA 190
S N AAHY+P D + V K+D G HI+G I D A T+S + +Y +L++A + A
Sbjct: 58 SINSEAAHYSPTINDEKRIPEGAVVKLDLGAHIDGFISDTAITISLDSRYQRLLDASKTA 117
Query: 191 TNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKT 250
I + + EIG I++V+ + K IRNL GH I Y +HAG
Sbjct: 118 LEAAITNFKAGLSIGEIGRVIEKVI---------RAQGYKPIRNLGGHLIRRYELHAGVF 168
Query: 251 VPIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMDCSHY-MKNFDAGYMPLRLQSSKSL 309
+P V V++ + YAIE F + G G+V + + Y +KN + + R L
Sbjct: 169 IPNVYERGLGVIQSDSVYAIEPFATDGGGEVVEGKSITIYSLKNPNIKGLSSR---ENEL 225
Query: 310 LSVINKNFSTLAFCKRWLDR--AGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEH 367
+ I F+ L F +RWL + + +K+L KG + YP L ++K +QFEH
Sbjct: 226 IDFIYTRFNYLPFSERWLKEFSTNVDELRNNIKNLVKKGALRGYPILIEIKKGVVSQFEH 285
Query: 368 TIMLR 372
T++++
Sbjct: 286 TVIVK 290
>AMPM_METTH (O27355) Methionine aminopeptidase (EC 3.4.11.18) (MAP)
(Peptidase M)
Length = 299
Score = 119 bits (297), Expect = 2e-26
Identities = 83/291 (28%), Positives = 140/291 (47%), Gaps = 22/291 (7%)
Query: 85 QTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAHYTPNAG 144
+ R+ I+ G+ +I++ +E+ ++I+E G A FP S N AHYT G
Sbjct: 17 KVRREAAGIIRDGLPIIELVNYVED---RIIEEGGRPA---FPCNVSVNEVTAHYTSPPG 70
Query: 145 DPTVLEYDDVTKIDFGTHINGRIIDCAFTLSFNPKYDK---LIEAVRDATNTGIKAAGID 201
D +V+ D+ K+D G H++G I D A T+ DK +++A R+A I
Sbjct: 71 DDSVIGDGDLVKLDLGAHVDGFIADTAITVPVGDVDDKCHQMMDAAREALENAISTIRAG 130
Query: 202 VPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRIHAGKTVPIVKGGEATV 261
V + EIG I+E + S + + + NL GHS+ + +H+G ++P +
Sbjct: 131 VEVGEIGRVIEETIHS---------HGMNPVSNLTGHSMERWILHSGLSIPNINERNTHQ 181
Query: 262 MEENEYYAIETFGSTGRGQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLA 321
+EE + AIE F + G G V D Y+ F PLRL ++ +L I + + L
Sbjct: 182 LEEGDVLAIEPFATDGVGLVTDMPQT--YIFRF-LRERPLRLVHARRVLGKIREEYHALP 238
Query: 322 FCKRWLDR-AGCTKYQMALKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIML 371
F +RWL+ + +++ L + Y L + G AQ+EHT+++
Sbjct: 239 FAQRWLEEYFDAKRLNASMRMLIQSRAIYPYHVLREKSGAMVAQWEHTVIV 289
>P2G4_HUMAN (Q9UQ80) Proliferation-associated protein 2G4 (Cell
cycle protein p38-2G4 homolog) (hG4-1)
Length = 394
Score = 92.8 bits (229), Expect = 1e-18
Identities = 73/335 (21%), Positives = 149/335 (43%), Gaps = 33/335 (9%)
Query: 60 EEKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEE-----LENTARKL 114
+E++ +E + + ++ + + + + + G++++ +CE+ +E T +
Sbjct: 5 DEQQEQTIAEDLVVTKYKMGGDIANRVLRSLVEASSSGVSVLSLCEKGDAMIMEETGKIF 64
Query: 115 IKEDGLKAGLAFPTGCSRNHCAAHYTPNAGDPT-VLEYDDVTKIDFGTHINGRIIDCAFT 173
KE +K G+AFPT S N+C H++P D +L+ D+ KID G H++G I + A T
Sbjct: 65 KKEKEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVAHT 124
Query: 174 LSFNPKYDKLIEAVRDATNTGIKA---------AGIDVPLCEIGAAIQEVMESYEVELDG 224
+++ + TG KA A + L + G +V E++
Sbjct: 125 F--------VVDVAQGTQVTGRKADVIKAAHLCAEAALRLVKPGNQNTQVTEAWNKV--A 174
Query: 225 KTYQVKSIRNLNGHSISPYRIHAGKTV------PIVKGGEATVMEENEYYAIETFGSTGR 278
++ I + H + + I KT+ K E E +E YA++ S+G
Sbjct: 175 HSFNCTPIEGMLSHQLKQHVIDGEKTIIQNPTDQQKKDHEKAEFEVHEVYAVDVLVSSGE 234
Query: 279 GQVHDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMA 338
G+ D + K + L++++S++ S + + F + F R + K +M
Sbjct: 235 GKAKDAGQRTTIYKRDPSKQYGLKMKTSRAFFSEVERRFDAMPFTLRAFEDE--KKARMG 292
Query: 339 LKDLCDKGIVDAYPPLCDVKGCYTAQFEHTIMLRP 373
+ + ++ + L + +G + AQF+ T++L P
Sbjct: 293 VVECAKHELLQPFNVLYEKEGEFVAQFKFTVLLMP 327
>P2G4_MOUSE (P50580) Proliferation-associated protein 2G4
(Proliferation-associated protein 1) (Protein p38-2G4)
Length = 394
Score = 92.4 bits (228), Expect = 2e-18
Identities = 70/327 (21%), Positives = 144/327 (43%), Gaps = 17/327 (5%)
Query: 60 EEKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQICEE-----LENTARKL 114
+E++ +E + + ++ + + + + + G++++ +CE+ +E T +
Sbjct: 5 DEQQEQTIAEDLVVTKYKMGGDIANRVLRSLVEASSSGVSVLSLCEKGDAMIMEETGKIF 64
Query: 115 IKEDGLKAGLAFPTGCSRNHCAAHYTPNAGDPT-VLEYDDVTKIDFGTHINGRIIDCAFT 173
KE +K G+AFPT S N+C H++P D +L+ D+ KID G H++G I + A T
Sbjct: 65 KKEKEMKKGIAFPTSISVNNCVCHFSPLKSDQDYILKEGDLVKIDLGVHVDGFIANVAHT 124
Query: 174 LSFNPKYDKLIEAVR-DATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSI 232
+ + D A + L + G +V E++ ++ I
Sbjct: 125 FVIGVAQGTQVTGRKADVIKAAHLCAEAALRLVKPGNQNTQVTEAWNKV--AHSFNCTPI 182
Query: 233 RNLNGHSISPYRIHAGKTV------PIVKGGEATVMEENEYYAIETFGSTGRGQVHDDMD 286
+ H + + I KT+ K E E +E YA++ S+G G+ D
Sbjct: 183 EGMLSHQLKQHVIDGEKTIIQNPTDQQKKDHEKAEFEVHEVYAVDVLVSSGEGKAKDAGQ 242
Query: 287 CSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKDLCDKG 346
+ K + L++++S++ S + + F + F R + K +M + +
Sbjct: 243 RTTIYKRDPSKQYGLKMKTSRAFFSEVERRFDAMPFTLRAFEDE--KKARMGVVECAKHE 300
Query: 347 IVDAYPPLCDVKGCYTAQFEHTIMLRP 373
++ + L + +G + AQF+ T++L P
Sbjct: 301 LLQPFNVLYEKEGEFVAQFKFTVLLMP 327
>CDB4_SCHPO (Q09184) Curved DNA-binding protein (42 kDa protein)
Length = 381
Score = 77.8 bits (190), Expect = 4e-14
Identities = 83/330 (25%), Positives = 132/330 (39%), Gaps = 25/330 (7%)
Query: 58 TNEEKRALDRSESDIYNEVRLAAEAHRQTRKHMQQWIKPGMTMIQIC---EELENTA-RK 113
T+E S + N+ ++A E + K + + +PG + IC +EL N A +K
Sbjct: 7 TSETAVDYSLSNPETVNKYKIAGEVSQNVIKKVVELCQPGAKIYDICVRGDELLNEAIKK 66
Query: 114 LIKEDGLKAGLAFPTGCSRNHCAAHYTPNAGDP---TVLEYDDVTKIDFGTHING--RII 168
+ + G+AFPT S N AAH +P DP L+ DV KI G HI+G ++
Sbjct: 67 VYRTKDAYKGIAFPTAVSPNDMAAHLSPLKSDPEANLALKSGDVVKILLGAHIDGFASLV 126
Query: 169 DCAFTLSFNPKYDKLIEAVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQ 228
+S P + + A + +KAA + + ++++ +Y
Sbjct: 127 ATTTVVSEEPVTGPAADVIA-AASAALKAAQRTIKPGNTNWQVTDIVDKI-----ATSYG 180
Query: 229 VKSIRNLNGHSISPYRIHAGKTVPIVKGG------EATVMEENEYYAIETFGSTG-RGQV 281
K + + H I K V + + EE E Y ++ ST G+V
Sbjct: 181 CKPVAGMLSHQQEREVIDGKKQVILNPSDSQRSEMDTFTFEEGEVYGVDILVSTSPSGKV 240
Query: 282 HDDMDCSHYMKNFDAGYMPLRLQSSKSLLSVINKNFSTLAFCKRWLDRAGCTKYQMALKD 341
+ K D YM L+LQ+S+ + S I F F R + T M L +
Sbjct: 241 KRSDIATRIYKKTDTTYM-LKLQASRKVYSEIQTKFGPFPFSTRNISFDSRT--NMGLNE 297
Query: 342 LCDKGIVDAYPPLCDVKGCYTAQFEHTIML 371
++ Y L D G A+F TI L
Sbjct: 298 CTSHKLLFPYEVLLDKDGGIVAEFYSTIAL 327
>AMPM_METFE (P22624) Probable methionine aminopeptidase (EC
3.4.11.18) (MAP) (Peptidase M) (Fragment)
Length = 188
Score = 75.5 bits (184), Expect = 2e-13
Identities = 57/191 (29%), Positives = 86/191 (44%), Gaps = 28/191 (14%)
Query: 79 AAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHCAAH 138
A + + RK + +K M ++ + E +EN K+ G K AFP S N AH
Sbjct: 7 AGKIASKVRKKAIKAVKGEMKILDLAEFIENEIEKM----GAKP--AFPCNISVNEITAH 60
Query: 139 YTPNAGDPTVLEYDDVTKIDFGTHINGRIIDCAFTL---------SFN----PKYDKLIE 185
Y+P D + D+ KID G H++G I D A T+ ++N K K+IE
Sbjct: 61 YSPPCNDDRKILPGDLVKIDIGVHVDGFIGDTATTVLVEGYEDLKNYNDELAEKNKKMIE 120
Query: 186 AVRDATNTGIKAAGIDVPLCEIGAAIQEVMESYEVELDGKTYQVKSIRNLNGHSISPYRI 245
A A I V + +IG I+ + + K I NL GH+I + +
Sbjct: 121 AAESALENAINTIRDGVEIGKIGEVIENTINK---------FGFKPISNLTGHTIDRWVL 171
Query: 246 HAGKTVPIVKG 256
H+G ++P VKG
Sbjct: 172 HSGLSIPNVKG 182
>AMPP_MYCGE (P47566) Putative Xaa-Pro aminopeptidase (EC 3.4.11.9)
(X-Pro aminopeptidase) (Aminopeptidase P) (APP)
(Aminoacylproline aminopeptidase)
Length = 354
Score = 48.5 bits (114), Expect = 3e-05
Identities = 38/149 (25%), Positives = 67/149 (44%), Gaps = 11/149 (7%)
Query: 76 VRLAAEAHRQTRKHMQQWIKPGMTMIQICEELENTARKLIKEDGLKAGLAFPTGCSRNHC 135
+ A + R+ ++++IKP MT + I + + N +L+K+ G K +N
Sbjct: 137 IEKAVDITRKVAVKLKRFIKPKMTELFISQWITN---ELVKQGGAKNSFDPIVATGKNGA 193
Query: 136 AAHYTPNAGDPTVLEYDDVTKIDFGTHINGRIIDC--AFTLSFNPKYDKLIEA---VRDA 190
H+ P T+++ D DFGT NG D F + PK KL+ A V +A
Sbjct: 194 NPHHKPTK---TIVKEGDFITCDFGTIYNGYCSDITRTFLVGKKPKSAKLLSAYKKVEEA 250
Query: 191 TNTGIKAAGIDVPLCEIGAAIQEVMESYE 219
GI A + ++ ++++E+ E
Sbjct: 251 NLAGINAVNTTLTGSQVDKVCRDIIENSE 279
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.135 0.405
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,122,882
Number of Sequences: 164201
Number of extensions: 1995557
Number of successful extensions: 5713
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 5621
Number of HSP's gapped (non-prelim): 64
length of query: 384
length of database: 59,974,054
effective HSP length: 112
effective length of query: 272
effective length of database: 41,583,542
effective search space: 11310723424
effective search space used: 11310723424
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC139744.6


