

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
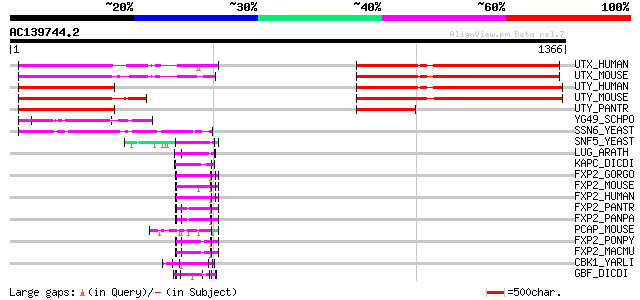
Query= AC139744.2 - phase: 0 /pseudo
(1366 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
UTX_HUMAN (O15550) Ubiquitously transcribed X chromosome tetratr... 694 0.0
UTX_MOUSE (O70546) Ubiquitously transcribed X chromosome tetratr... 693 0.0
UTY_HUMAN (O14607) Ubiquitously transcribed Y chromosome tetratr... 676 0.0
UTY_MOUSE (P79457) Ubiquitously transcribed Y chromosome tetratr... 653 0.0
UTY_PANTR (Q6B4Z3) Ubiquitously transcribed Y chromosome tetratr... 296 3e-79
YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II 172 6e-42
SSN6_YEAST (P14922) Glucose repression mediator protein 146 3e-34
SNF5_YEAST (P18480) Transcription regulatory protein SNF5 (SWI/S... 67 3e-10
LUG_ARATH (Q9FUY2) Transcriptional corepressor LEUNIG 67 3e-10
KAPC_DICDI (P34099) cAMP-dependent protein kinase catalytic subu... 64 4e-09
FXP2_GORGO (Q8MJ99) Forkhead box protein P2 63 6e-09
FXP2_MOUSE (P58463) Forkhead box protein P2 62 1e-08
FXP2_HUMAN (O15409) Forkhead box protein P2 (CAG repeat protein ... 62 1e-08
FXP2_PANTR (Q8MJA0) Forkhead box protein P2 61 2e-08
FXP2_PANPA (Q8HZ00) Forkhead box protein P2 61 2e-08
PCAP_MOUSE (Q924H2) Positive cofactor 2 glutamine/Q-rich-associa... 61 2e-08
FXP2_PONPY (Q8MJ98) Forkhead box protein P2 61 2e-08
FXP2_MACMU (Q8MJ97) Forkhead box protein P2 61 2e-08
CBK1_YARLI (Q6CFS5) Serine/threonine-protein kinase CBK1 (EC 2.7... 59 7e-08
GBF_DICDI (P36417) G-box binding factor (GBF) 58 2e-07
>UTX_HUMAN (O15550) Ubiquitously transcribed X chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the X chromosome)
Length = 1401
Score = 694 bits (1790), Expect = 0.0
Identities = 332/505 (65%), Positives = 391/505 (76%), Gaps = 20/505 (3%)
Query: 853 RGRSLDGISGCSILSTNAPPPQPPDPPPQKLTREQLNPPTPSVYLESKKDAFSPQLQDFC 912
R +G+S SIL PPP+PP P L +++LNPPTPS+YLE+K+DAF P L FC
Sbjct: 901 RNLGKNGLSNSSILLDKCPPPRPPSSPYPPLPKDKLNPPTPSIYLENKRDAFFPPLHQFC 960
Query: 913 LR--NPIAVIRGLAAALKLDLGLFSTKTLVEANPDHGIEVRTQMQQSPDENRDPVLGQRV 970
NP+ VIRGLA ALKLDLGLFSTKTLVEAN +H +EVRTQ+ Q DEN DP +++
Sbjct: 961 TNPNNPVTVIRGLAGALKLDLGLFSTKTLVEANNEHMVEVRTQLLQPADENWDPTGTKKI 1020
Query: 971 WSCISHRSHTSIAKYAQYQASSFQDSLKEEQEKTTGIHCSNLSDSDSKDSTGNVVRRKLK 1030
W C S+RSHT+IAKYAQYQASSFQ+SL+EE EK + H + SDS+S S + RRK
Sbjct: 1021 WHCESNRSHTTIAKYAQYQASSFQESLREENEKRS--HHKDHSDSESTSSDNSGRRRK-- 1076
Query: 1031 NGVHPHGKVAPNGCKMIKFGTNVDLSDDRKWKAQLQELNKLPAFARVVSAGNMLSHVGHV 1090
K IKFGTN+DLSDD+KWK QL EL KLPAF RVVSAGN+LSHVGH
Sbjct: 1077 -----------GPFKTIKFGTNIDLSDDKKWKLQLHELTKLPAFVRVVSAGNLLSHVGHT 1125
Query: 1091 ILGMNTVQLYMKVPGSRTPGHQENNNFCSININIGPGDCEWFAVPDAYWGALNNLCEKNN 1150
ILGMNTVQLYMKVPGSRTPGHQENNNFCS+NINIGPGDCEWF VP+ YWG LN+ CEKNN
Sbjct: 1126 ILGMNTVQLYMKVPGSRTPGHQENNNFCSVNINIGPGDCEWFVVPEGYWGVLNDFCEKNN 1185
Query: 1151 MNYLHGSWWPLLEDLFEANVPVYRFHQRPGDLVWVNSGCVHWVQAVGWCNNIAWNVGPLT 1210
+N+L GSWWP LEDL+EANVPVYRF QRPGDLVW+N+G VHWVQA+GWCNNIAWNVGPLT
Sbjct: 1186 LNFLMGSWWPNLEDLYEANVPVYRFIQRPGDLVWINAGTVHWVQAIGWCNNIAWNVGPLT 1245
Query: 1211 ARQYQLAIERYEWNKLQSYKSIVPMVHLSWNLARNIRIVDPKLFELVKNCLMRTLHHCAL 1270
A QY+LA+ERYEWNKLQS KSIVPMVHLSWN+ARNI++ DPKLFE++K CL+RTL C
Sbjct: 1246 ACQYKLAVERYEWNKLQSVKSIVPMVHLSWNMARNIKVSDPKLFEMIKYCLLRTLKQCQT 1305
Query: 1271 ILEFVKSKGIEVRFHGRRKDEASHYCGQCEVEVFNILFIREQ---EKRHVVHCMDCARKQ 1327
+ E + + G E+ +HGR K+E +HYC CEVEVF++LF+ + K ++VHC DCARK
Sbjct: 1306 LREALIAAGKEIIWHGRTKEEPAHYCSICEVEVFDLLFVTNESNSRKTYIVHCQDCARKT 1365
Query: 1328 SPSLEGFVCLEEYSMSDLCSVYDQF 1352
S +LE FV LE+Y M DL VYDQF
Sbjct: 1366 SGNLENFVVLEQYKMEDLMQVYDQF 1390
Score = 332 bits (852), Expect = 3e-90
Identities = 204/505 (40%), Positives = 279/505 (54%), Gaps = 94/505 (18%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A++AFQ+VLYVDP F RA E+HLR+GLMFKVN D ES+LKH +L+ VD +P T EIQ
Sbjct: 149 AIKAFQEVLYVDPSFCRAKEIHLRVGLMFKVNTDYESSLKHFQLALVDCNPCTLSNAEIQ 208
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTL 141
FHIAHLYE Q+K+ AKE YEQLL+ ++L +KA + +QLGWM+H V+ G K + +
Sbjct: 209 FHIAHLYETQRKYHSAKEAYEQLLQTENLSAQVKATVLQQLGWMHHTVDLLGDKATKESY 268
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
AI+ L+KSLEAD NSGQ+ Y LGRCY+SIGK AF +YR ++K+ +AD WCSIGVLY
Sbjct: 269 AIQYLQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFISYRQSIDKSEASADTWCSIGVLY 328
Query: 202 QQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSVRASKDKAAG 261
QQQ Q +DALQAY+CAVQLD H+A+W++LG LYESC+Q +DA+ CY+++ R+
Sbjct: 329 QQQNQPMDALQAYICAVQLDHGHAAAWMDLGTLYESCNQPQDAIKCYLNATRS------- 381
Query: 262 VGSGTTSSESGPLKPWHISARPQNTDSANSDISQRIAFLQTQLAKTPMPSITSKAE-LPT 320
+ S S ++ RI +LQ QL P S+ +K + LP+
Sbjct: 382 -----------------------KSCSNTSALAARIKYLQAQLCNLPQGSLQNKTKLLPS 418
Query: 321 IEEAWNTPVVSDWNSKHQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPPPPYPGAVPGAG 380
IEEAW+ P+ ++ S+ AQQ N N G AV PP
Sbjct: 419 IEEAWSLPIPAELTSRQGAM-----NTAQQ-----NTSDNWSGGHAVSHPPV-------- 460
Query: 381 PNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNP 440
QQ + L Q+LQ L+ +++ +LNP
Sbjct: 461 -------------------------------QQQAHSWCLTPQKLQHLEQLRANRNNLNP 489
Query: 441 QQRQQMQQFQHHYRLMQQHQQ------QLRMQ-----QQQQQQLHAGVSPVQQPGQVARP 489
Q+ ++Q + + LMQQHQ Q+R L QQP Q+A
Sbjct: 490 AQKLMLEQLESQFVLMQQHQMRPTGVAQVRSTGIPNGPTADSSLPTNSVSGQQP-QLALT 548
Query: 490 PQQSLQQPSVK-SCPPQAV-HIPFS 512
S+ QP V+ +CP Q + + PFS
Sbjct: 549 RVPSVSQPGVRPACPGQPLANGPFS 573
>UTX_MOUSE (O70546) Ubiquitously transcribed X chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the X chromosome) (Fragment)
Length = 1333
Score = 693 bits (1788), Expect = 0.0
Identities = 331/505 (65%), Positives = 391/505 (76%), Gaps = 20/505 (3%)
Query: 853 RGRSLDGISGCSILSTNAPPPQPPDPPPQKLTREQLNPPTPSVYLESKKDAFSPQLQDFC 912
R +G+S SIL PPP+PP P L +++LNPPTPS+YLE+K+DAF P L FC
Sbjct: 833 RNLGKNGLSNSSILLDKCPPPRPPSSPYPPLPKDKLNPPTPSIYLENKRDAFFPPLHQFC 892
Query: 913 LR--NPIAVIRGLAAALKLDLGLFSTKTLVEANPDHGIEVRTQMQQSPDENRDPVLGQRV 970
NP+ VIRGLA ALKLDLGLFSTKTLVEAN +H +EVRTQ+ Q DEN DP +++
Sbjct: 893 TNPNNPVTVIRGLAGALKLDLGLFSTKTLVEANNEHMVEVRTQLLQPADENWDPTGTKKI 952
Query: 971 WSCISHRSHTSIAKYAQYQASSFQDSLKEEQEKTTGIHCSNLSDSDSKDSTGNVVRRKLK 1030
W C S+RSHT+IAKYAQYQASSFQ+SL+EE EK + H + SDS+S S + RRK
Sbjct: 953 WHCESNRSHTTIAKYAQYQASSFQESLREENEKRS--HHKDHSDSESTSSDNSGKRRK-- 1008
Query: 1031 NGVHPHGKVAPNGCKMIKFGTNVDLSDDRKWKAQLQELNKLPAFARVVSAGNMLSHVGHV 1090
K IKFGTN+DLSDD+KWK QL EL KLPAF RVVSAGN+LSHVGH
Sbjct: 1009 -----------GPFKTIKFGTNIDLSDDKKWKLQLHELTKLPAFVRVVSAGNLLSHVGHT 1057
Query: 1091 ILGMNTVQLYMKVPGSRTPGHQENNNFCSININIGPGDCEWFAVPDAYWGALNNLCEKNN 1150
ILGMNTVQLYMKVPGSRTPGHQENNNFCS+NINIGPGDCEWF VP+ YWG LN+ CEKNN
Sbjct: 1058 ILGMNTVQLYMKVPGSRTPGHQENNNFCSVNINIGPGDCEWFVVPEGYWGVLNDFCEKNN 1117
Query: 1151 MNYLHGSWWPLLEDLFEANVPVYRFHQRPGDLVWVNSGCVHWVQAVGWCNNIAWNVGPLT 1210
+N+L GSWWP LEDL+EANVPVYRF QRPGDLVW+N+G VHWVQA+GWCNNIAWNVGPLT
Sbjct: 1118 LNFLMGSWWPNLEDLYEANVPVYRFIQRPGDLVWINAGTVHWVQAIGWCNNIAWNVGPLT 1177
Query: 1211 ARQYQLAIERYEWNKLQSYKSIVPMVHLSWNLARNIRIVDPKLFELVKNCLMRTLHHCAL 1270
A QY+LA+ERYEWNKLQ+ KSIVPMVHLSWN+ARNI++ DPKLFE++K CL+RTL C
Sbjct: 1178 ACQYKLAVERYEWNKLQNVKSIVPMVHLSWNMARNIKVSDPKLFEMIKYCLLRTLKQCQT 1237
Query: 1271 ILEFVKSKGIEVRFHGRRKDEASHYCGQCEVEVFNILFIREQ---EKRHVVHCMDCARKQ 1327
+ E + + G E+ +HGR K+E +HYC CEVEVF++LF+ + K ++VHC DCARK
Sbjct: 1238 LREALIAAGKEIIWHGRTKEEPAHYCSICEVEVFDLLFVTNESNSRKTYIVHCQDCARKT 1297
Query: 1328 SPSLEGFVCLEEYSMSDLCSVYDQF 1352
S +LE FV LE+Y M DL VYDQF
Sbjct: 1298 SGNLENFVVLEQYKMEDLMQVYDQF 1322
Score = 334 bits (856), Expect = 1e-90
Identities = 199/486 (40%), Positives = 271/486 (54%), Gaps = 94/486 (19%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A++AFQ+VLYVDP F RA E+HLRLGLMFKVN D ES+LKH +L+ VD +P T EIQ
Sbjct: 83 AIKAFQEVLYVDPSFCRAKEIHLRLGLMFKVNTDYESSLKHFQLALVDCNPCTLSNAEIQ 142
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTL 141
FHIAHLYE Q+K+ AKE YEQLL+ ++L +KA I +QLGWM+H V+ G K + +
Sbjct: 143 FHIAHLYETQRKYHSAKEAYEQLLQTENLSAQVKATILQQLGWMHHTVDLLGDKATKESY 202
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
AI+ L+KSLEAD NSGQ+ Y LGRCY+SIGK AF +YR ++K+ +AD WCSIGVLY
Sbjct: 203 AIQYLQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFISYRQSIDKSEASADTWCSIGVLY 262
Query: 202 QQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSVRASKDKAAG 261
QQQ Q +DALQAY+CAVQLD H+A+W++LG LYESC+Q +DA+ CY+++ R
Sbjct: 263 QQQNQPMDALQAYICAVQLDHGHAAAWMDLGTLYESCNQPQDAIKCYLNATR-------- 314
Query: 262 VGSGTTSSESGPLKPWHISARPQNTDSANSDISQRIAFLQTQLAKTPMPSITSKAE-LPT 320
S S+ SG ++ RI +LQ QL P S+ +K + LP+
Sbjct: 315 --SKNCSNTSG--------------------LAARIKYLQAQLCNLPQGSLQNKTKLLPS 352
Query: 321 IEEAWNTPVVSDWNSKHQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPPPPYPGAVPGAG 380
IEEAW+ P+ ++ S+ AQQ + + +GG PPP
Sbjct: 353 IEEAWSLPIPAELTSRQGAM-----NTAQQ------NTSDNWSGGNAPPP---------- 391
Query: 381 PNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNP 440
QQ + L Q+LQ L+ +++ +LNP
Sbjct: 392 ------------------------------VEQQTHSWCLTPQKLQHLEQLRANRNNLNP 421
Query: 441 QQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPP-QQSLQQPSV 499
Q+ ++Q + + LMQQHQ + GV+ V+ G + P SL SV
Sbjct: 422 AQKLMLEQLESQFVLMQQHQMR-----------QTGVAQVRPTGILNGPTVDSSLPTNSV 470
Query: 500 KSCPPQ 505
PQ
Sbjct: 471 SGQQPQ 476
>UTY_HUMAN (O14607) Ubiquitously transcribed Y chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the Y chromosome)
Length = 1347
Score = 676 bits (1745), Expect = 0.0
Identities = 327/513 (63%), Positives = 390/513 (75%), Gaps = 20/513 (3%)
Query: 853 RGRSLDGISGCSILSTNAPPPQPPDPPPQKLTREQLNPPTPSVYLESKKDAFSPQLQDFC 912
R +G+S IL PPP+PP P L +++LNPPTPS+YLE+K+DAF P L FC
Sbjct: 848 RNPGKNGLSNSCILLDKCPPPRPPTSPYPPLPKDKLNPPTPSIYLENKRDAFFPPLHQFC 907
Query: 913 L--RNPIAVIRGLAAALKLDLGLFSTKTLVEANPDHGIEVRTQMQQSPDENRDPVLGQRV 970
+NP+ VIRGLA ALKLDLGLFSTKTLVEAN +H +EVRTQ+ Q DEN DP +++
Sbjct: 908 TNPKNPVTVIRGLAGALKLDLGLFSTKTLVEANNEHMVEVRTQLLQPADENWDPTGTKKI 967
Query: 971 WSCISHRSHTSIAKYAQYQASSFQDSLKEEQEKTTGIHCSNLSDSDSKDSTGNVVRRKLK 1030
W C S+RSHT+IAKYAQYQASSFQ+SL+EE EK T + SD++S S + RRK
Sbjct: 968 WRCESNRSHTTIAKYAQYQASSFQESLREENEKRT--QHKDHSDNESTSSENSGRRRK-- 1023
Query: 1031 NGVHPHGKVAPNGCKMIKFGTNVDLSDDRKWKAQLQELNKLPAFARVVSAGNMLSHVGHV 1090
K IKFGTN+DLSD++KWK QL EL KLPAFARVVSAGN+L+HVGH
Sbjct: 1024 -----------GPFKTIKFGTNIDLSDNKKWKLQLHELTKLPAFARVVSAGNLLTHVGHT 1072
Query: 1091 ILGMNTVQLYMKVPGSRTPGHQENNNFCSININIGPGDCEWFAVPDAYWGALNNLCEKNN 1150
ILGMNTVQLYMKVPGSRTPGHQENNNFCS+NINIGPGDCEWF VP+ YWG LN+ CEKNN
Sbjct: 1073 ILGMNTVQLYMKVPGSRTPGHQENNNFCSVNINIGPGDCEWFVVPEDYWGVLNDFCEKNN 1132
Query: 1151 MNYLHGSWWPLLEDLFEANVPVYRFHQRPGDLVWVNSGCVHWVQAVGWCNNIAWNVGPLT 1210
+N+L SWWP LEDL+EANVPVYRF QRPGDLVW+N+G VHWVQ VGWCNNIAWNVGPLT
Sbjct: 1133 LNFLMSSWWPNLEDLYEANVPVYRFIQRPGDLVWINAGTVHWVQTVGWCNNIAWNVGPLT 1192
Query: 1211 ARQYQLAIERYEWNKLQSYKSIVPMVHLSWNLARNIRIVDPKLFELVKNCLMRTLHHCAL 1270
A QY+LA+ERYEWNKL+S KS VPMVHLSWN+ARNI++ DPKLFE++K CL++ L
Sbjct: 1193 ACQYKLAVERYEWNKLKSVKSPVPMVHLSWNMARNIKVSDPKLFEMIKYCLLKILKQYQT 1252
Query: 1271 ILEFVKSKGIEVRFHGRRKDEASHYCGQCEVEVFNILFIREQ---EKRHVVHCMDCARKQ 1327
+ E + + G EV +HGR DE +HYC CEVEVFN+LF+ + +K ++VHC DCARK
Sbjct: 1253 LREALVAAGKEVIWHGRTNDEPAHYCSICEVEVFNLLFVTNESNTQKTYIVHCHDCARKT 1312
Query: 1328 SPSLEGFVCLEEYSMSDLCSVYDQFKFSSVVSN 1360
S SLE FV LE+Y M DL VYDQF + +S+
Sbjct: 1313 SKSLENFVVLEQYKMEDLIQVYDQFTLALSLSS 1345
Score = 298 bits (762), Expect = 9e-80
Identities = 138/235 (58%), Positives = 183/235 (77%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A++AFQ VLYVDP F RA E+HLRLGLMFKVN D +S+LKH +L+ +D +P T EIQ
Sbjct: 146 AIKAFQDVLYVDPSFCRAKEIHLRLGLMFKVNTDYKSSLKHFQLALIDCNPCTLSNAEIQ 205
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTL 141
FHIAHLYE Q+K+ AKE YEQLL+ ++LP +KA + +QLGWM+HN++ G K + +
Sbjct: 206 FHIAHLYETQRKYHSAKEAYEQLLQTENLPAQVKATVLQQLGWMHHNMDLVGDKATKESY 265
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
AI+ L+KSLEAD NSGQ+ Y LGRCY+SIGK AF +YR ++K+ +AD WCSIGVLY
Sbjct: 266 AIQYLQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFISYRQSIDKSEASADTWCSIGVLY 325
Query: 202 QQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSVRASK 256
QQQ Q +DALQAY+CAVQLD H+A+W++LG LYESC+Q +DA+ CY+++ R+ +
Sbjct: 326 QQQNQPMDALQAYICAVQLDHGHAAAWMDLGTLYESCNQPQDAIKCYLNAARSKR 380
Score = 37.0 bits (84), Expect = 0.37
Identities = 29/99 (29%), Positives = 49/99 (49%), Gaps = 14/99 (14%)
Query: 413 QQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQ 472
QQ L Q+LQ L+ +++ ++LNP Q+ Q++Q + + LMQQ + + + Q +
Sbjct: 413 QQVYSLCLTPQKLQHLEQLRANRDNLNPAQKHQLEQLESQFVLMQQMRHK-EVAQVRTTG 471
Query: 473 LHAG-----------VSPVQQPGQVARPPQQSLQQPSVK 500
+H G VS Q G + R S+ QP V+
Sbjct: 472 IHNGAITDSSLPTNSVSNRQPHGALTR--VSSVSQPGVR 508
>UTY_MOUSE (P79457) Ubiquitously transcribed Y chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein ON the Y chromosome)
(Male-specific histocompatibility antigen H-YDB)
Length = 1212
Score = 653 bits (1685), Expect = 0.0
Identities = 317/513 (61%), Positives = 387/513 (74%), Gaps = 20/513 (3%)
Query: 853 RGRSLDGISGCSILSTNAPPPQPPDPPPQKLTREQLNPPTPSVYLESKKDAFSPQLQDFC 912
R +G+S IL PPP+PP P L +E+LNPPTPS+YLE+K+DAF P L FC
Sbjct: 713 RSLGKNGLSNGHILLDICPPPRPPTSPYPPLPKEKLNPPTPSIYLENKRDAFFPPLHQFC 772
Query: 913 L--RNPIAVIRGLAAALKLDLGLFSTKTLVEANPDHGIEVRTQMQQSPDENRDPVLGQRV 970
+ +NP+ VIRGLA ALKLDLGLFSTKTLVEAN +H +EVRTQ+ Q DEN DP +++
Sbjct: 773 INPKNPVTVIRGLAGALKLDLGLFSTKTLVEANNEHIVEVRTQLLQPADENWDPSGTKKI 832
Query: 971 WSCISHRSHTSIAKYAQYQASSFQDSLKEEQEKTTGIHCSNLSDSDSKDSTGNVVRRKLK 1030
W + SHT+IAKYAQYQA SFQ+SL+EE E+ T + + SD++S S + R+K
Sbjct: 833 WRYENKSSHTTIAKYAQYQACSFQESLREENERRTQV--KDYSDNESTCSDNSGRRQK-- 888
Query: 1031 NGVHPHGKVAPNGCKMIKFGTNVDLSDDRKWKAQLQELNKLPAFARVVSAGNMLSHVGHV 1090
AP K IK G N+DLSD++KWK QL EL KLPAF RVVSAGN+LSHVG+
Sbjct: 889 ---------AP--FKTIKCGINIDLSDNKKWKLQLHELTKLPAFVRVVSAGNLLSHVGYT 937
Query: 1091 ILGMNTVQLYMKVPGSRTPGHQENNNFCSININIGPGDCEWFAVPDAYWGALNNLCEKNN 1150
ILGMN+VQL MKVPGSR PGHQENNNFCS+NINIGPGDCEWF VP+ YWG LN+ CEKNN
Sbjct: 938 ILGMNSVQLCMKVPGSRIPGHQENNNFCSVNINIGPGDCEWFVVPEDYWGVLNDFCEKNN 997
Query: 1151 MNYLHGSWWPLLEDLFEANVPVYRFHQRPGDLVWVNSGCVHWVQAVGWCNNIAWNVGPLT 1210
+N+L SWWP LEDL+EANVPVYRF QRPGDLVW+N+G VHWVQA+GWCNNI WNVGPLT
Sbjct: 998 LNFLMSSWWPNLEDLYEANVPVYRFIQRPGDLVWINAGTVHWVQAIGWCNNITWNVGPLT 1057
Query: 1211 ARQYQLAIERYEWNKLQSYKSIVPMVHLSWNLARNIRIVDPKLFELVKNCLMRTLHHCAL 1270
A QY+LA+ERYEWNKLQS KS+VPMVHLSWN+ARNI++ DPKLFE++K CL++ L HC
Sbjct: 1058 AFQYKLAVERYEWNKLQSVKSVVPMVHLSWNMARNIKVSDPKLFEMIKYCLLKILKHCQT 1117
Query: 1271 ILEFVKSKGIEVRFHGRRKDEASHYCGQCEVEVFNILFIREQ---EKRHVVHCMDCARKQ 1327
+ E + + G EV +HGR DE + YC CEVEVFN+LF+ + +K ++VHC +CARK
Sbjct: 1118 LREALVAAGKEVLWHGRINDEPAPYCSICEVEVFNLLFVTNESNSQKTYIVHCQNCARKT 1177
Query: 1328 SPSLEGFVCLEEYSMSDLCSVYDQFKFSSVVSN 1360
S +LE FV LE+Y M DL VYDQF + +S+
Sbjct: 1178 SGNLENFVVLEQYKMEDLIQVYDQFTLAPSLSS 1210
Score = 310 bits (793), Expect = 2e-83
Identities = 158/316 (50%), Positives = 211/316 (66%), Gaps = 31/316 (9%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A+RAFQ+VLYVDP F RA E+HLRLG MFK+N D ES+LKH +L+ +D + T +EIQ
Sbjct: 144 AIRAFQEVLYVDPNFCRAKEIHLRLGFMFKMNTDYESSLKHFQLALIDCNVCTLSSVEIQ 203
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTL 141
FHIAHLYE Q+K+ AK YEQLL+ + LP +KA + +QLGWM+HN++ G T +
Sbjct: 204 FHIAHLYETQRKYHSAKAAYEQLLQIESLPSQVKATVLQQLGWMHHNMDLIGDNTTKERY 263
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
AI+ L+KSLE D NSGQ+ Y LGRCY+ IGK AF +YR ++K+ +AD WCSIGVLY
Sbjct: 264 AIQYLQKSLEEDPNSGQSWYFLGRCYSCIGKVQDAFVSYRQSIDKSEASADTWCSIGVLY 323
Query: 202 QQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSVRASKDKAAG 261
QQQ Q +DALQAY+CAVQLD H+A+W++LGILYESC+Q +DA+ CY+++ R
Sbjct: 324 QQQNQPMDALQAYICAVQLDHGHAAAWMDLGILYESCNQPQDAIKCYLNAAR-------- 375
Query: 262 VGSGTTSSESGPLKPWHISARPQNTDSANSDISQRIAFLQTQLAKTPMPSITSKAE-LPT 320
S NT S ++ RI FLQ QL P S+ +K + LP+
Sbjct: 376 ------------------SKSCNNT----SALTSRIKFLQAQLCNLPQSSLQNKTKLLPS 413
Query: 321 IEEAWNTPVVSDWNSK 336
IEEAW+ P+ ++ S+
Sbjct: 414 IEEAWSLPIPAELTSR 429
>UTY_PANTR (Q6B4Z3) Ubiquitously transcribed Y chromosome
tetratricopeptide repeat protein (Ubiquitously
transcribed TPR protein on the Y chromosome)
Length = 1079
Score = 296 bits (758), Expect = 3e-79
Identities = 137/235 (58%), Positives = 182/235 (77%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A++AFQ VLYVDP F RA E+HLRLGLMFKVN D +S+LKH +L+ +D +P T EIQ
Sbjct: 146 AIKAFQDVLYVDPSFCRAKEIHLRLGLMFKVNTDYKSSLKHFQLALIDCNPCTLSSAEIQ 205
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTL 141
FHIAHLYE Q+K+ AKE YEQLL+ ++LP +KA + +QLGWM+HN++ G K + +
Sbjct: 206 FHIAHLYETQRKYHSAKEAYEQLLQTENLPAQVKATVLQQLGWMHHNMDLVGDKATKESY 265
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
AI L+KSLEAD NSGQ+ Y LGRCY+SIGK AF +YR ++++ +AD WCSIGVLY
Sbjct: 266 AIPYLQKSLEADPNSGQSWYFLGRCYSSIGKVQDAFVSYRQSIDRSEASADTWCSIGVLY 325
Query: 202 QQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESCSQLKDALACYIHSVRASK 256
QQQ Q +DALQAY+CAVQLD H+A+W++LG LYESC+Q +DA+ CY+++ R+ +
Sbjct: 326 QQQNQPIDALQAYICAVQLDHGHAAAWMDLGTLYESCNQPQDAIKCYLNAARSKR 380
Score = 183 bits (464), Expect = 3e-45
Identities = 90/148 (60%), Positives = 111/148 (74%), Gaps = 2/148 (1%)
Query: 853 RGRSLDGISGCSILSTNAPPPQPPDPPPQKLTREQLNPPTPSVYLESKKDAFSPQLQDFC 912
R +G+S IL PPP+PP P L +++LNPPTPS+YLE+K+DAF P L FC
Sbjct: 848 RNPGKNGLSNSCILLDKCPPPRPPTSPYPPLPKDKLNPPTPSIYLENKRDAFFPPLHQFC 907
Query: 913 L--RNPIAVIRGLAAALKLDLGLFSTKTLVEANPDHGIEVRTQMQQSPDENRDPVLGQRV 970
+NP+ VIRGLA ALKLDLGLFSTKTLVEAN +H +EVRTQ+ Q DEN DP +++
Sbjct: 908 TNPKNPVTVIRGLAGALKLDLGLFSTKTLVEANNEHIVEVRTQLLQPADENWDPTGTKKI 967
Query: 971 WSCISHRSHTSIAKYAQYQASSFQDSLK 998
W C S+RSHT+IAKYAQYQASSFQ+SL+
Sbjct: 968 WRCESNRSHTTIAKYAQYQASSFQESLR 995
Score = 36.6 bits (83), Expect = 0.48
Identities = 27/94 (28%), Positives = 47/94 (49%), Gaps = 4/94 (4%)
Query: 413 QQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQ 472
QQ L Q+LQ L+ +++ ++LNP Q+ Q++Q + + LMQQ + + + Q +
Sbjct: 413 QQVYSLCLTPQKLQHLEQLRANRDNLNPAQKHQLEQLESQFVLMQQMRHK-EVAQVRTTG 471
Query: 473 LHAGV---SPVQQPGQVARPPQQSLQQPSVKSCP 503
+H G S + R P +L + S S P
Sbjct: 472 IHNGAIADSSLPTNSVSNRQPHAALTRVSSVSQP 505
>YG49_SCHPO (O60184) Protein C23E6.09 in chromosome II
Length = 1102
Score = 172 bits (436), Expect = 6e-42
Identities = 118/338 (34%), Positives = 168/338 (48%), Gaps = 42/338 (12%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A AF Q L +DP F++ NE++ RLG+++K + +L+ + +D P L+I
Sbjct: 457 AEEAFMQCLRMDPNFEKVNEIYFRLGIIYKQQHKFAQSLELFR-HILDNPPKPLTVLDIY 515
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTL 141
F I H+YE +K++KLAKE YE++L E P H K + +QLGW+ H S L
Sbjct: 516 FQIGHVYEQRKEYKLAKEAYERVLAET--PNHAK--VLQQLGWLCHQQS---SSFTNQDL 568
Query: 142 AIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGVLY 201
AI+ L KSLEAD Q+ YL+GRCY + K + A++AY+ V + WCSIGVLY
Sbjct: 569 AIQYLTKSLEADDTDAQSWYLIGRCYVAQQKYNKAYEAYQQAVYRDGRNPTFWCSIGVLY 628
Query: 202 QQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESC-SQLKDALACYIHSVRASKDKAA 260
Q QY DAL AY A++L+ S W +LG LYESC +Q+ DAL Y
Sbjct: 629 YQINQYQDALDAYSRAIRLNPYISEVWYDLGTLYESCHNQISDALDAY------------ 676
Query: 261 GVGSGTTSSESGPLKPWHISARPQNTDSANSDISQRIAFLQTQLAKTPMPSITSKAELPT 320
++E P P HI AR Q N++ Q ++ P ++ +
Sbjct: 677 -----QRAAELDPTNP-HIKARLQLLRGPNNE--------QHKIVNAPPSNVPNVQTAKY 722
Query: 321 IEEAW----NTPVV---SDWNSKHQPQQKRHPGMAQQG 351
I + N PV +W H PQ + Q G
Sbjct: 723 INQPGVPYSNVPVAQLSGNWQPPHLPQAQLPSATGQSG 760
Score = 50.4 bits (119), Expect = 3e-05
Identities = 50/202 (24%), Positives = 89/202 (43%), Gaps = 27/202 (13%)
Query: 54 NDPESALKHLKLSYVDASPSTFCKLEIQFHIAHLYEVQKKHKLAKELYEQLLKEKDLPLH 113
N PES ++HL + + T+ ++ +A L++ Q K A YE L++ P
Sbjct: 321 NAPES-VRHL----ISLNEETWIQIG---RLAELFDDQDK---ALSAYESALRQN--PYS 367
Query: 114 LKADIYRQLGWMYHNVEAFGSKTERMTLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKA 173
+ A + Q+ + N E F LAI + L+ D G+ LG CY
Sbjct: 368 IPAML--QIATILRNREQF-------PLAIEYYQTILDCDPKQGEIWSALGHCYLMQDDL 418
Query: 174 DHAFDAYRNVVEKTTPTAD--VWCSIGVLYQQQKQYVDALQAYVCAVQLD---KCHSASW 228
A+ AYR + D +W IG+LY + + A +A++ +++D + + +
Sbjct: 419 SRAYSAYRQALYHLKDPKDPKLWYGIGILYDRYGSHEHAEEAFMQCLRMDPNFEKVNEIY 478
Query: 229 INLGILYESCSQLKDALACYIH 250
LGI+Y+ + +L + H
Sbjct: 479 FRLGIIYKQQHKFAQSLELFRH 500
Score = 43.1 bits (100), Expect = 0.005
Identities = 49/165 (29%), Positives = 64/165 (38%), Gaps = 26/165 (15%)
Query: 337 HQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPPPP------YPGAVPGAGPNAKRFKPNG 390
H P+ HP ++QQ P G + PP P P + +P
Sbjct: 56 HLPRSTLHPLLSQQQQPAQQSPSLGPAQQNIQQPPSVSIASQPHYAEAIVPIQQVLQPQQ 115
Query: 391 --EVDNGMVGAAGMGHARPPQAQPQ---QPPPYYLNQ--QQLQLLQYYQSHV-----ESL 438
++ MV A PQ PQ P NQ QQL L ++ +
Sbjct: 116 YRQLPPNMVAATNA-----PQQHPQLQRMMPILSSNQPIQQLPLPNQASPYIPVPLQQQQ 170
Query: 439 NPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQP 483
Q +QQ QQ QH Q QQ QQQQQ+QLH+G +QQP
Sbjct: 171 QSQPQQQPQQQQHQQPQQPQPPQQPLQQQQQQRQLHSG---IQQP 212
Score = 41.6 bits (96), Expect = 0.015
Identities = 45/125 (36%), Positives = 53/125 (42%), Gaps = 42/125 (33%)
Query: 407 PPQAQPQQPP-------PYYLN-----QQQLQLLQYYQ---SHVESLN-PQQRQQMQ--- 447
P Q QQPP P+Y QQ LQ QY Q + V + N PQQ Q+Q
Sbjct: 81 PAQQNIQQPPSVSIASQPHYAEAIVPIQQVLQPQQYRQLPPNMVAATNAPQQHPQLQRMM 140
Query: 448 ----------------QFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQ 491
Q + + Q QQQ + QQQ QQQ H QQP Q +PPQ
Sbjct: 141 PILSSNQPIQQLPLPNQASPYIPVPLQQQQQSQPQQQPQQQQH------QQP-QQPQPPQ 193
Query: 492 QSLQQ 496
Q LQQ
Sbjct: 194 QPLQQ 198
Score = 36.2 bits (82), Expect = 0.63
Identities = 39/113 (34%), Positives = 49/113 (42%), Gaps = 20/113 (17%)
Query: 407 PPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQ 466
P QA P P P QQQ Q Q Q QQ QQ QQ Q + +QQ QQQ ++
Sbjct: 155 PNQASPYIPVPLQ-QQQQSQPQQQPQQ-------QQHQQPQQPQPPQQPLQQQQQQRQLH 206
Query: 467 QQQQQQLHAGVS---------PVQQP--GQ-VARPPQQSLQQPSVKSCPPQAV 507
QQ + VS V P GQ +A P + Q ++ PPQA+
Sbjct: 207 SGIQQPVSTIVSQNGTYYSIPAVNHPMAGQPIAIAPVPAPNQAALPPIPPQAL 259
>SSN6_YEAST (P14922) Glucose repression mediator protein
Length = 966
Score = 146 bits (369), Expect = 3e-34
Identities = 142/486 (29%), Positives = 206/486 (42%), Gaps = 61/486 (12%)
Query: 22 AVRAFQQVLYVDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQ 81
A AF +VL +DP F++ANE++ RLG+++K AL+ + + P+ + +I
Sbjct: 169 AEEAFAKVLELDPHFEKANEIYFRLGIIYKHQGKWSQALECFRY-ILPQPPAPLQEWDIW 227
Query: 82 FHIAHLYEVQKKHKLAKELYEQLLKEKDLPLHLKADIYRQLGWMY--HNVEAFGSKTERM 139
F + + E + + AKE YE +L + A + +QLG +Y NV+ + +
Sbjct: 228 FQLGSVLESMGEWQGAKEAYEHVLAQNQH----HAKVLQQLGCLYGMSNVQFYDPQK--- 280
Query: 140 TLAIRCLEKSLEADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVEKTTPTADVWCSIGV 199
A+ L KSLEAD + T Y LGR + A+DA++ V + + WCSIGV
Sbjct: 281 --ALDYLLKSLEADPSDATTWYHLGRVHMIRTDYTAAYDAFQQAVNRDSRNPIFWCSIGV 338
Query: 200 LYQQQKQYVDALQAYVCAVQLDKCHSASWINLGILYESC-SQLKDALACYIHSVRASKDK 258
LY Q QY DAL AY A++L+ S W +LG LYE+C +QL DAL Y + R +
Sbjct: 339 LYYQISQYRDALDAYTRAIRLNPYISEVWYDLGTLYETCNNQLSDALDAYKQAARLDVNN 398
Query: 259 A---AGVGSGTTSSESGPLKPWHISARPQNTDSANSDISQRIAFLQTQLAKTPMPSITSK 315
+ + T E+ P N + +N T + P P I
Sbjct: 399 VHIRERLEALTKQLEN-----------PGNINKSNG--------APTNASPAPPPVILQP 439
Query: 316 AELPTIE-EAWNTPV-VSDWNSKHQPQQKRHPGMAQQGGPKPNHYVNGVNGGAVPPPPYP 373
P + NT + N+ Q++HP Y NG + P
Sbjct: 440 TLQPNDQGNPLNTRISAQSANATASMVQQQHPAQQTPINSSATMYSNGAS-------PQL 492
Query: 374 GAVPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQS 433
A A A+ A A+ Q + Q Q Q Q+
Sbjct: 493 QAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAQAHAQAQAQAQAKAQAQA 552
Query: 434 HVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQS 493
++ QQ+QQ QQ Q QQ QQQ + QQQQQQQL QP P+Q
Sbjct: 553 QAQAQQQQQQQQQQQQQQ----QQQQQQQQQQQQQQQQQL--------QP-----LPRQQ 595
Query: 494 LQQPSV 499
LQQ V
Sbjct: 596 LQQKGV 601
Score = 43.1 bits (100), Expect = 0.005
Identities = 51/235 (21%), Positives = 99/235 (41%), Gaps = 21/235 (8%)
Query: 32 VDPVFQRANEVHLRLGLMFKVNNDPESALKHLKLSYVDASPSTFCKLEIQFHIAHLYEVQ 91
+DP+ Q E L + + + D + A + + +PS+ L +AHLY +
Sbjct: 38 LDPLTQSTAETWLSIASLAETLGDGDRAAMAYDAT-LQFNPSSAKALT---SLAHLYRSR 93
Query: 92 KKHKLAKELYEQ-LLKEKDLPLHLKADIYRQLGWMYHNVEAFGSKTERMTLAIRCLEKSL 150
+ A ELYE+ LL +L +D++ LG Y ++ A+ L
Sbjct: 94 DMFQRAAELYERALLVNPEL-----SDVWATLGHCYLMLDDLQRAYNAYQQALYHL---- 144
Query: 151 EADRNSGQTLYLLGRCYASIGKADHAFDAYRNVVE---KTTPTADVWCSIGVLYQQQKQY 207
++ N + + +G Y G D+A +A+ V+E +++ +G++Y+ Q ++
Sbjct: 145 -SNPNVPKLWHGIGILYDRYGSLDYAEEAFAKVLELDPHFEKANEIYFRLGIIYKHQGKW 203
Query: 208 VDALQAYVCAVQLDKCHSAS---WINLGILYESCSQLKDALACYIHSVRASKDKA 259
AL+ + + W LG + ES + + A Y H + ++ A
Sbjct: 204 SQALECFRYILPQPPAPLQEWDIWFQLGSVLESMGEWQGAKEAYEHVLAQNQHHA 258
>SNF5_YEAST (P18480) Transcription regulatory protein SNF5 (SWI/SNF
complex component SNF5) (Transcription factor TYE4)
Length = 905
Score = 67.0 bits (162), Expect = 3e-10
Identities = 76/273 (27%), Positives = 104/273 (37%), Gaps = 44/273 (16%)
Query: 282 RPQNTDSANSDISQRIA------FLQTQLAKTPMPSITSKAELPTIEEAWNTPVVSDWNS 335
+PQ T+S + I + F Q+ + S+T + +L I++ + S
Sbjct: 4 QPQGTNSVPNSIGNIFSNIGTPSFNMAQIPQQLYQSLTPQ-QLQMIQQRHQQLLRSRLQQ 62
Query: 336 KHQPQQKRHPGMAQQGGPKP-----NHYVNGVNGGAVPPPPYP-------GAVPGAG--- 380
+ Q QQ+ P P P N PPPP P G VP A
Sbjct: 63 QQQQQQQTSPPPQTHQSPPPPPQQSQPIANQSATSTPPPPPAPHNLHPQIGQVPLAPAPI 122
Query: 381 ---------PNAKR------------FKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYY 419
P A + K N +V N + A + Q + QQ
Sbjct: 123 NLPPQIAQLPLATQQQVLNKLRQQAIAKNNPQVVNAITVAQQQVQRQIEQQKGQQTAQTQ 182
Query: 420 LNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSP 479
L QQ+ QLL Q + N QRQQ QQF+HH ++ QQ Q+Q + QQQ QQQ
Sbjct: 183 LEQQR-QLLVQQQQQQQLRNQIQRQQQQQFRHHVQIQQQQQKQQQQQQQHQQQQQQQQQQ 241
Query: 480 VQQPGQVARPPQQSLQQPSVKSCPPQAVHIPFS 512
QQ Q + QQ QQ + Q IP S
Sbjct: 242 QQQQQQQQQQQQQQQQQQQQQQQQQQQGQIPQS 274
Score = 49.7 bits (117), Expect = 5e-05
Identities = 33/95 (34%), Positives = 45/95 (46%), Gaps = 1/95 (1%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q Q + QQQ + Q Q H + QQ+QQ QQ Q + QQ QQQ + QQQ
Sbjct: 206 QQQQQFRHHVQIQQQQQKQQQQQQQHQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 265
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCP 503
QQQ V Q ++ P ++ QP++ P
Sbjct: 266 QQQGQIPQSQQVPQVRSMSGQPPTNV-QPTIGQLP 299
Score = 41.2 bits (95), Expect = 0.020
Identities = 35/116 (30%), Positives = 49/116 (42%), Gaps = 14/116 (12%)
Query: 404 HARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQL 463
H + Q Q +Q +QQQ Q Q Q + QQ+QQ QQ Q QQ QQQ
Sbjct: 214 HVQIQQQQQKQQQQQQQHQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ------QQQQQQQ 267
Query: 464 RMQQQQQQQLHAGVSPVQQP--------GQVARPPQQSLQQPSVKSCPPQAVHIPF 511
+ Q Q QQ+ S QP GQ+ + P+ +L + P +P+
Sbjct: 268 QGQIPQSQQVPQVRSMSGQPPTNVQPTIGQLPQLPKLNLPKYQTIQYDPPETKLPY 323
>LUG_ARATH (Q9FUY2) Transcriptional corepressor LEUNIG
Length = 931
Score = 67.0 bits (162), Expect = 3e-10
Identities = 43/100 (43%), Positives = 51/100 (51%), Gaps = 3/100 (3%)
Query: 405 ARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQ-HHYRLMQQHQQQL 463
AR Q Q Q P QQQ Q Q + QQ+QQ QQ Q HH++ QQ QQQ
Sbjct: 86 AREQQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHHQQQQQQQQQQ 145
Query: 464 RMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCP 503
+ QQQQQQQ H P QQ Q + P Q QQP+ + P
Sbjct: 146 QQQQQQQQQQHQNQPPSQQQQQQSTPQHQ--QQPTPQQQP 183
Score = 45.8 bits (107), Expect = 8e-04
Identities = 34/84 (40%), Positives = 40/84 (47%), Gaps = 8/84 (9%)
Query: 422 QQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQ 481
+QQLQ Q H + QQ+QQ QQ Q L+Q+ QQQ QQQQQQ H Q
Sbjct: 88 EQQLQQSQ----HPQVSQQQQQQQQQQIQMQQLLLQRAQQQ----QQQQQQQHHHHQQQQ 139
Query: 482 QPGQVARPPQQSLQQPSVKSCPPQ 505
Q Q + QQ QQ P Q
Sbjct: 140 QQQQQQQQQQQQQQQQHQNQPPSQ 163
Score = 42.7 bits (99), Expect = 0.007
Identities = 31/86 (36%), Positives = 40/86 (46%), Gaps = 2/86 (2%)
Query: 419 YLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVS 478
Y+ Q ++ + Q +S +PQ QQ QQ Q MQQ Q QQQQQQQ
Sbjct: 78 YIETQMIKARE--QQLQQSQHPQVSQQQQQQQQQQIQMQQLLLQRAQQQQQQQQQQHHHH 135
Query: 479 PVQQPGQVARPPQQSLQQPSVKSCPP 504
QQ Q + QQ QQ ++ PP
Sbjct: 136 QQQQQQQQQQQQQQQQQQQQHQNQPP 161
Score = 38.5 bits (88), Expect = 0.13
Identities = 23/67 (34%), Positives = 31/67 (45%)
Query: 406 RPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
R Q Q QQ ++ +QQQ Q Q Q + Q + Q Q + QHQQQ
Sbjct: 120 RAQQQQQQQQQQHHHHQQQQQQQQQQQQQQQQQQQQHQNQPPSQQQQQQSTPQHQQQPTP 179
Query: 466 QQQQQQQ 472
QQQ Q++
Sbjct: 180 QQQPQRR 186
>KAPC_DICDI (P34099) cAMP-dependent protein kinase catalytic subunit
(EC 2.7.1.37)
Length = 648
Score = 63.5 bits (153), Expect = 4e-09
Identities = 41/98 (41%), Positives = 52/98 (52%), Gaps = 10/98 (10%)
Query: 406 RPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
+P Q QPQQ P QQQ Q Q Q ++ N QQ+QQ+QQ Q +L QQ QQQ +
Sbjct: 147 QPQQQQPQQQQPQQQQQQQPQQQQQPQQQLQQNNQQQQQQLQQQQLQQQLQQQQQQQQQQ 206
Query: 466 QQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCP 503
QQQQQQ+ QQ Q + QQ L Q + + P
Sbjct: 207 QQQQQQK--------QQKQQ--QQQQQHLHQDGIVNTP 234
Score = 55.1 bits (131), Expect = 1e-06
Identities = 36/90 (40%), Positives = 40/90 (44%)
Query: 407 PPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQ 466
PP+ QQ P QQQ Q Q Q + QQ QQ QQ Q + Q QQQ Q
Sbjct: 130 PPEPNKQQQPQQQPQQQQPQQQQPQQQQPQQQQQQQPQQQQQPQQQLQQNNQQQQQQLQQ 189
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQ QQQL QQ Q + QQ QQ
Sbjct: 190 QQLQQQLQQQQQQQQQQQQQQQQKQQKQQQ 219
Score = 52.8 bits (125), Expect = 6e-06
Identities = 37/90 (41%), Positives = 43/90 (47%), Gaps = 6/90 (6%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQ-MQQFQHHYRLMQQHQQQLRMQ 466
PQ QPQQ P QQ Q Q Q PQQ+QQ QQ Q + + QQ QQ ++Q
Sbjct: 139 PQQQPQQQQPQQQQPQQQQPQQQQQQQ-----PQQQQQPQQQLQQNNQQQQQQLQQQQLQ 193
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQ QQQ QQ Q + QQ QQ
Sbjct: 194 QQLQQQQQQQQQQQQQQQQKQQKQQQQQQQ 223
Score = 38.9 bits (89), Expect = 0.097
Identities = 25/60 (41%), Positives = 30/60 (49%), Gaps = 8/60 (13%)
Query: 437 SLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
SL P + + QQ Q + Q QQQ + QQ QQQQ QQ Q + PQQ LQQ
Sbjct: 127 SLTPPEPNKQQQPQQQPQQQQPQQQQPQQQQPQQQQ--------QQQPQQQQQPQQQLQQ 178
>FXP2_GORGO (Q8MJ99) Forkhead box protein P2
Length = 713
Score = 62.8 bits (151), Expect = 6e-09
Identities = 39/88 (44%), Positives = 47/88 (53%), Gaps = 2/88 (2%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
QA QQ L QQQLQ ++Y+ E L+ Q QQ QQ Q + QQ QQQ + QQQ
Sbjct: 119 QALLQQQQAVMLQQQQLQ--EFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQ 176
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQ Q PG+ A+ QQ QQ
Sbjct: 177 QQQQQQQQQQQQQHPGKQAKEQQQQQQQ 204
Score = 53.1 bits (126), Expect = 5e-06
Identities = 34/86 (39%), Positives = 42/86 (48%), Gaps = 4/86 (4%)
Query: 411 QPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQ 470
Q QQ +Y QQ+ LQ Q + QQ+QQ QQ Q + QQ QQQ QQQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ----QQQQ 186
Query: 471 QQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQ H G +Q Q + Q + QQ
Sbjct: 187 QQQHPGKQAKEQQQQQQQQQQLAAQQ 212
Score = 48.9 bits (115), Expect = 9e-05
Identities = 40/113 (35%), Positives = 51/113 (44%), Gaps = 9/113 (7%)
Query: 411 QPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH----QQQLRMQ 466
Q +Q L QQQ Q Q Q + QQ+QQ QQ Q + QQH ++ + Q
Sbjct: 142 QQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQ 201
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHIPFSFL 514
QQQQQQL A QQ + QQ SLQ+ + S PP +P L
Sbjct: 202 QQQQQQLAAQQLVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAALPVQSL 254
Score = 48.1 bits (113), Expect = 2e-04
Identities = 41/101 (40%), Positives = 46/101 (44%), Gaps = 6/101 (5%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQ-- 157
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQ 505
QQQQQQQ QQ Q + QQ QQ K Q
Sbjct: 158 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQ 198
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/85 (37%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ QQQ Q Q Q H +Q+QQ QQ Q QQQL QQ
Sbjct: 165 QQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQQ 224
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQS 493
QQQ H + +Q+ G ++ PP Q+
Sbjct: 225 LQQQQH--LLSLQRQGLISIPPGQA 247
>FXP2_MOUSE (P58463) Forkhead box protein P2
Length = 714
Score = 61.6 bits (148), Expect = 1e-08
Identities = 40/90 (44%), Positives = 49/90 (54%), Gaps = 4/90 (4%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQ--QRQQMQQFQHHYRLMQQHQQQLRMQ 466
QA QQ L QQQLQ ++Y+ E L+ Q Q+QQ QQ Q + QQ QQQ + Q
Sbjct: 119 QALLQQQQAVMLQQQQLQ--EFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQ 176
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQ Q PG+ A+ QQ QQ
Sbjct: 177 QQQQQQQQQQQQQQQHPGKQAKEQQQQQQQ 206
Score = 52.8 bits (125), Expect = 6e-06
Identities = 41/101 (40%), Positives = 47/101 (45%), Gaps = 4/101 (3%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ +
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQ 159
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQ 505
QQQQQQQ QQ Q + QQ QQ K Q
Sbjct: 160 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQ 200
Score = 52.4 bits (124), Expect = 8e-06
Identities = 45/125 (36%), Positives = 55/125 (44%), Gaps = 21/125 (16%)
Query: 411 QPQQPPPYYLNQQQ---LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQL---- 463
Q QQ +Y QQ+ LQLLQ Q + QQ+QQ QQ Q + QQ QQQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 190
Query: 464 ---------RMQQQQQQQLHAGVSPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHI 509
+ QQQQQQQL A QQ + QQ SLQ+ + S PP +
Sbjct: 191 QHPGKQAKEQQQQQQQQQLAAQQLVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAAL 250
Query: 510 PFSFL 514
P L
Sbjct: 251 PVQSL 255
Score = 46.2 bits (108), Expect = 6e-04
Identities = 32/85 (37%), Positives = 43/85 (49%), Gaps = 3/85 (3%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ QQQ Q Q Q H +Q+QQ QQ Q + + QQ L+MQQ
Sbjct: 167 QQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQLAAQQLVFQQQLLQMQQL 226
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQS 493
QQQQ + +Q+ G ++ PP Q+
Sbjct: 227 QQQQ---HLLSLQRQGLISIPPGQA 248
>FXP2_HUMAN (O15409) Forkhead box protein P2 (CAG repeat protein 44)
(Trinucleotide repeat-containing gene 10 protein)
Length = 715
Score = 61.6 bits (148), Expect = 1e-08
Identities = 40/90 (44%), Positives = 49/90 (54%), Gaps = 4/90 (4%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQ--QRQQMQQFQHHYRLMQQHQQQLRMQ 466
QA QQ L QQQLQ ++Y+ E L+ Q Q+QQ QQ Q + QQ QQQ + Q
Sbjct: 119 QALLQQQQAVMLQQQQLQ--EFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQ 176
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQ Q PG+ A+ QQ QQ
Sbjct: 177 QQQQQQQQQQQQQQQHPGKQAKEQQQQQQQ 206
Score = 57.4 bits (137), Expect = 3e-07
Identities = 38/89 (42%), Positives = 46/89 (50%), Gaps = 8/89 (8%)
Query: 411 QPQQPPPYYLNQQQ---LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
Q QQ +Y QQ+ LQLLQ Q + QQ+QQ QQ Q QQ QQQ + QQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQ-----QQQQQQQQQQQ 185
Query: 468 QQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQ H G +Q Q + Q + QQ
Sbjct: 186 QQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 214
Score = 52.8 bits (125), Expect = 6e-06
Identities = 41/101 (40%), Positives = 47/101 (45%), Gaps = 4/101 (3%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ +
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQ 159
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQ 505
QQQQQQQ QQ Q + QQ QQ K Q
Sbjct: 160 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQ 200
Score = 48.5 bits (114), Expect = 1e-04
Identities = 41/111 (36%), Positives = 48/111 (42%), Gaps = 8/111 (7%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q QQ QQQ Q Q Q + QQ+QQ QQ Q H + QQQ QQQ
Sbjct: 149 QLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQ---QQQ 205
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHIPFSFL 514
QQQQL A QQ + QQ SLQ+ + S PP +P L
Sbjct: 206 QQQQLAAQQLVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAALPVQSL 256
>FXP2_PANTR (Q8MJA0) Forkhead box protein P2
Length = 716
Score = 61.2 bits (147), Expect = 2e-08
Identities = 40/91 (43%), Positives = 49/91 (52%), Gaps = 5/91 (5%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQ---QRQQMQQFQHHYRLMQQHQQQLRM 465
QA QQ L QQQLQ ++Y+ E L+ Q Q+QQ QQ Q + QQ QQQ +
Sbjct: 119 QALLQQQQAVMLQQQQLQ--EFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQ 176
Query: 466 QQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQQ Q PG+ A+ QQ QQ
Sbjct: 177 QQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQ 207
Score = 58.2 bits (139), Expect = 2e-07
Identities = 38/89 (42%), Positives = 47/89 (52%), Gaps = 7/89 (7%)
Query: 411 QPQQPPPYYLNQQQ---LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
Q QQ +Y QQ+ LQLLQ Q + QQ+QQ QQ Q + QQ QQQ + QQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQ----QQQQQQQQQQQQQQQQQQQQQQQQQQQ 186
Query: 468 QQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQ H G +Q Q + Q + QQ
Sbjct: 187 QQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 215
Score = 52.0 bits (123), Expect = 1e-05
Identities = 39/92 (42%), Positives = 45/92 (48%), Gaps = 4/92 (4%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ +
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQ 159
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQQ QQ Q + QQ QQ
Sbjct: 160 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 191
Score = 47.0 bits (110), Expect = 4e-04
Identities = 37/102 (36%), Positives = 47/102 (45%), Gaps = 9/102 (8%)
Query: 422 QQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH----QQQLRMQQQQQQQLHAGV 477
QQQ Q Q Q + QQ+QQ QQ Q + QQH ++ + QQQQQQQL A
Sbjct: 156 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 215
Query: 478 SPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHIPFSFL 514
QQ + QQ SLQ+ + S PP +P L
Sbjct: 216 LVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAALPVQSL 257
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/85 (37%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ QQQ Q Q Q H +Q+QQ QQ Q QQQL QQ
Sbjct: 168 QQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQQ 227
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQS 493
QQQ H + +Q+ G ++ PP Q+
Sbjct: 228 LQQQQH--LLSLQRQGLISIPPGQA 250
>FXP2_PANPA (Q8HZ00) Forkhead box protein P2
Length = 716
Score = 61.2 bits (147), Expect = 2e-08
Identities = 40/91 (43%), Positives = 49/91 (52%), Gaps = 5/91 (5%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQ---QRQQMQQFQHHYRLMQQHQQQLRM 465
QA QQ L QQQLQ ++Y+ E L+ Q Q+QQ QQ Q + QQ QQQ +
Sbjct: 119 QALLQQQQAVMLQQQQLQ--EFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQ 176
Query: 466 QQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQQ Q PG+ A+ QQ QQ
Sbjct: 177 QQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQ 207
Score = 58.2 bits (139), Expect = 2e-07
Identities = 38/89 (42%), Positives = 47/89 (52%), Gaps = 7/89 (7%)
Query: 411 QPQQPPPYYLNQQQ---LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
Q QQ +Y QQ+ LQLLQ Q + QQ+QQ QQ Q + QQ QQQ + QQ
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQ----QQQQQQQQQQQQQQQQQQQQQQQQQQQ 186
Query: 468 QQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQ H G +Q Q + Q + QQ
Sbjct: 187 QQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 215
Score = 52.0 bits (123), Expect = 1e-05
Identities = 39/92 (42%), Positives = 45/92 (48%), Gaps = 4/92 (4%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ +
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQ 159
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQQ QQ Q + QQ QQ
Sbjct: 160 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQ 191
Score = 47.0 bits (110), Expect = 4e-04
Identities = 37/102 (36%), Positives = 47/102 (45%), Gaps = 9/102 (8%)
Query: 422 QQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH----QQQLRMQQQQQQQLHAGV 477
QQQ Q Q Q + QQ+QQ QQ Q + QQH ++ + QQQQQQQL A
Sbjct: 156 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 215
Query: 478 SPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHIPFSFL 514
QQ + QQ SLQ+ + S PP +P L
Sbjct: 216 LVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAALPVQSL 257
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/85 (37%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ QQQ Q Q Q H +Q+QQ QQ Q QQQL QQ
Sbjct: 168 QQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQQ 227
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQS 493
QQQ H + +Q+ G ++ PP Q+
Sbjct: 228 LQQQQH--LLSLQRQGLISIPPGQA 250
>PCAP_MOUSE (Q924H2) Positive cofactor 2 glutamine/Q-rich-associated
protein (PC2 glutamine/Q-rich-associated protein)
(mPcqap)
Length = 792
Score = 60.8 bits (146), Expect = 2e-08
Identities = 60/170 (35%), Positives = 72/170 (42%), Gaps = 38/170 (22%)
Query: 344 HPGMAQQGGPKPNHYVNGVNGGAVPPPPYPGA----VPGAGPNAKRFKPNGEVDNGMVGA 399
H +Q P + + + GG P PGA +P GP G+ GM G
Sbjct: 70 HNKKSQASVSDPMNALQSLTGG-----PTPGAAGIGMPPRGP--------GQSLGGMGGL 116
Query: 400 AGMGHARPPQAQPQQPP------PYYL-------NQQQLQLLQYYQSHVESLNPQQRQQM 446
MG P QP PP P+ + Q QLQL Q V QQRQQ
Sbjct: 117 GAMGQPLPLSGQP--PPGTSGMAPHGMAVVSTTTPQTQLQLQQ-----VALQQQQQRQQQ 169
Query: 447 QQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQFQ +QQ QQQ + QQQQQQQ A + +QQ Q QQ QQ
Sbjct: 170 QQFQQQQAALQQQQQQ-QQQQQQQQQFQAQQNAMQQQFQAVVQQQQLQQQ 218
Score = 54.3 bits (129), Expect = 2e-06
Identities = 67/216 (31%), Positives = 81/216 (37%), Gaps = 49/216 (22%)
Query: 345 PGMAQQGGPK--PNHYVNGVNG-GAV---------PPPPYPGAVPGAGPNAKRFKPNGEV 392
PG A G P P + G+ G GA+ PPP G P P ++
Sbjct: 94 PGAAGIGMPPRGPGQSLGGMGGLGAMGQPLPLSGQPPPGTSGMAPHGMAVVSTTTPQTQL 153
Query: 393 DNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVES------------LNP 440
V A R Q Q QQ QQQ Q Q Q ++ +
Sbjct: 154 QLQQV-ALQQQQQRQQQQQFQQQQAALQQQQQQQQQQQQQQQFQAQQNAMQQQFQAVVQQ 212
Query: 441 QQRQQMQQFQH----HYRLMQQ--HQQQL---------RMQQQQQQQLHAGVSPVQQPG- 484
QQ QQ QQ QH H++ QQ QQQL + QQQQQQQ P+QQP
Sbjct: 213 QQLQQQQQQQHLIKLHHQSQQQQIQQQQLQRMAQLQLQQQQQQQQQQALQAQPPMQQPSM 272
Query: 485 -QVARPPQQSLQQ-------PSVKSCPPQAVHIPFS 512
Q PP Q+L Q P PPQA P +
Sbjct: 273 QQPQPPPSQALPQQLSQLHHPQHHQPPPQAQQSPIA 308
>FXP2_PONPY (Q8MJ98) Forkhead box protein P2
Length = 714
Score = 60.8 bits (146), Expect = 2e-08
Identities = 41/91 (45%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query: 409 QAQPQQPPPYYLNQQQLQLL---QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
QA QQ L QQQLQ Q Q H++ L QQ+QQ QQ Q + QQ QQQ +
Sbjct: 119 QALLQQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQ 178
Query: 466 QQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQQ Q PG+ A+ QQ QQ
Sbjct: 179 QQQQQQQ----QQQQQHPGKQAKEQQQQQQQ 205
Score = 52.8 bits (125), Expect = 6e-06
Identities = 39/93 (41%), Positives = 45/93 (47%), Gaps = 4/93 (4%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ +
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQ 159
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQP 497
QQQQQQQ QQ Q + QQ Q P
Sbjct: 160 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHP 192
Score = 51.2 bits (121), Expect = 2e-05
Identities = 37/89 (41%), Positives = 45/89 (49%), Gaps = 9/89 (10%)
Query: 411 QPQQPPPYYLNQQQ---LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
Q QQ +Y QQ+ LQLLQ Q + QQ+QQ QQ Q + QQ QQQ Q
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQ--QQQQQQ----Q 184
Query: 468 QQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQ H G +Q Q + Q + QQ
Sbjct: 185 QQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 213
Score = 47.0 bits (110), Expect = 4e-04
Identities = 37/102 (36%), Positives = 47/102 (45%), Gaps = 9/102 (8%)
Query: 422 QQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH----QQQLRMQQQQQQQLHAGV 477
QQQ Q Q Q + QQ+QQ QQ Q + QQH ++ + QQQQQQQL A
Sbjct: 154 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 213
Query: 478 SPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHIPFSFL 514
QQ + QQ SLQ+ + S PP +P L
Sbjct: 214 LVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAALPVQSL 255
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/85 (37%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ QQQ Q Q Q H +Q+QQ QQ Q QQQL QQ
Sbjct: 166 QQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQQ 225
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQS 493
QQQ H + +Q+ G ++ PP Q+
Sbjct: 226 LQQQQH--LLSLQRQGLISIPPGQA 248
Score = 39.3 bits (90), Expect = 0.074
Identities = 34/109 (31%), Positives = 43/109 (39%), Gaps = 13/109 (11%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH-------- 459
P + P QQ Q+LQ + L+PQQ Q + Q Q L QQ
Sbjct: 88 PVSVAMMTPQVITPQQMQQILQQ-----QVLSPQQLQALLQQQQAVMLQQQQLQEFYKKQ 142
Query: 460 QQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
Q+QL +Q QQQQ QQ Q + QQ QQ + Q H
Sbjct: 143 QEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQH 191
>FXP2_MACMU (Q8MJ97) Forkhead box protein P2
Length = 714
Score = 60.8 bits (146), Expect = 2e-08
Identities = 41/91 (45%), Positives = 48/91 (52%), Gaps = 7/91 (7%)
Query: 409 QAQPQQPPPYYLNQQQLQLL---QYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRM 465
QA QQ L QQQLQ Q Q H++ L QQ+QQ QQ Q + QQ QQQ +
Sbjct: 119 QALLQQQQAVMLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQ 178
Query: 466 QQQQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQQQ Q PG+ A+ QQ QQ
Sbjct: 179 QQQQQQQ----QQQQQHPGKQAKEQQQQQQQ 205
Score = 52.8 bits (125), Expect = 6e-06
Identities = 39/93 (41%), Positives = 45/93 (47%), Gaps = 4/93 (4%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQM---QQFQHHYRLMQQHQQQLR 464
PQ Q L+ QQLQ L Q V L QQ Q+ QQ Q H +L+QQ QQQ +
Sbjct: 101 PQQMQQILQQQVLSPQQLQALLQQQQAV-MLQQQQLQEFYKKQQEQLHLQLLQQQQQQQQ 159
Query: 465 MQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQP 497
QQQQQQQ QQ Q + QQ Q P
Sbjct: 160 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHP 192
Score = 51.2 bits (121), Expect = 2e-05
Identities = 37/89 (41%), Positives = 45/89 (49%), Gaps = 9/89 (10%)
Query: 411 QPQQPPPYYLNQQQ---LQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQ 467
Q QQ +Y QQ+ LQLLQ Q + QQ+QQ QQ Q + QQ QQQ Q
Sbjct: 131 QQQQLQEFYKKQQEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQ--QQQQQQ----Q 184
Query: 468 QQQQQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQQQQ H G +Q Q + Q + QQ
Sbjct: 185 QQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 213
Score = 47.0 bits (110), Expect = 4e-04
Identities = 37/102 (36%), Positives = 47/102 (45%), Gaps = 9/102 (8%)
Query: 422 QQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH----QQQLRMQQQQQQQLHAGV 477
QQQ Q Q Q + QQ+QQ QQ Q + QQH ++ + QQQQQQQL A
Sbjct: 154 QQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQ 213
Query: 478 SPVQQPGQVARPPQQ-----SLQQPSVKSCPPQAVHIPFSFL 514
QQ + QQ SLQ+ + S PP +P L
Sbjct: 214 LVFQQQLLQMQQLQQQQHLLSLQRQGLISIPPGQAALPVQSL 255
Score = 46.6 bits (109), Expect = 5e-04
Identities = 32/85 (37%), Positives = 40/85 (46%), Gaps = 2/85 (2%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQ QQQ Q Q Q H +Q+QQ QQ Q QQQL QQ
Sbjct: 166 QQQQQQQQQQQQQQQQQQQQQQQQQHPGKQAKEQQQQQQQQQQLAAQQLVFQQQLLQMQQ 225
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQS 493
QQQ H + +Q+ G ++ PP Q+
Sbjct: 226 LQQQQH--LLSLQRQGLISIPPGQA 248
Score = 39.3 bits (90), Expect = 0.074
Identities = 34/109 (31%), Positives = 43/109 (39%), Gaps = 13/109 (11%)
Query: 408 PQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQH-------- 459
P + P QQ Q+LQ + L+PQQ Q + Q Q L QQ
Sbjct: 88 PVSVAMMTPQVITPQQMQQILQQ-----QVLSPQQLQALLQQQQAVMLQQQQLQEFYKKQ 142
Query: 460 QQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
Q+QL +Q QQQQ QQ Q + QQ QQ + Q H
Sbjct: 143 QEQLHLQLLQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQQH 191
>CBK1_YARLI (Q6CFS5) Serine/threonine-protein kinase CBK1 (EC
2.7.1.37)
Length = 588
Score = 59.3 bits (142), Expect = 7e-08
Identities = 42/111 (37%), Positives = 49/111 (43%), Gaps = 7/111 (6%)
Query: 401 GMGHARPPQAQPQQ--PPPYYL-----NQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHY 453
G G QP PPP L NQ L Q Q + + N QQ+ Q QQ +
Sbjct: 8 GQGQPMYGSQQPHNLMPPPSGLYQQNSNQSNTSLDQQQQFNQQQYNSQQQYQQQQHSQYQ 67
Query: 454 RLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPP 504
+ QQHQQQ + QQQQQQQ QQ Q + QQ QQ SV P
Sbjct: 68 QQQQQHQQQQQQQQQQQQQQQQQQQQQQQQHQQHQQQQQQPQQQSVAQQQP 118
Score = 55.8 bits (133), Expect = 8e-07
Identities = 46/125 (36%), Positives = 57/125 (44%), Gaps = 13/125 (10%)
Query: 376 VPGAGPNAKRFKPNGEVDNGMVGAAGMGHARPPQAQPQQPPPYYLNQQQLQLLQYYQSHV 435
VPG G + + + N M +G+ Q+ NQQQ Q YQ
Sbjct: 6 VPGQG---QPMYGSQQPHNLMPPPSGLYQQNSNQSNTSLDQQQQFNQQQYNSQQQYQQQQ 62
Query: 436 ESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSL- 494
S QQ+QQ QQ Q + QQ QQQ + QQQQQ Q H QQ Q +P QQS+
Sbjct: 63 HSQYQQQQQQHQQQQQQQQQQQQQQQQQQQQQQQQHQQH------QQ--QQQQPQQQSVA 114
Query: 495 -QQPS 498
QQPS
Sbjct: 115 QQQPS 119
Score = 50.8 bits (120), Expect = 2e-05
Identities = 29/71 (40%), Positives = 33/71 (45%)
Query: 419 YLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVS 478
Y QQ Q Q Q H + QQ+QQ QQ Q + QQHQQ + QQQ QQQ A
Sbjct: 58 YQQQQHSQYQQQQQQHQQQQQQQQQQQQQQQQQQQQQQQQHQQHQQQQQQPQQQSVAQQQ 117
Query: 479 PVQQPGQVARP 489
P RP
Sbjct: 118 PSDYVNFTPRP 128
Score = 45.4 bits (106), Expect = 0.001
Identities = 28/72 (38%), Positives = 34/72 (46%)
Query: 423 QQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQQQLHAGVSPVQQ 482
QQ Q QY Q + QQ+QQ QQ Q + QQ QQ + QQQQQQ V+ Q
Sbjct: 59 QQQQHSQYQQQQQQHQQQQQQQQQQQQQQQQQQQQQQQQHQQHQQQQQQPQQQSVAQQQP 118
Query: 483 PGQVARPPQQSL 494
V P+ L
Sbjct: 119 SDYVNFTPRPDL 130
>GBF_DICDI (P36417) G-box binding factor (GBF)
Length = 708
Score = 58.2 bits (139), Expect = 2e-07
Identities = 39/102 (38%), Positives = 49/102 (47%), Gaps = 10/102 (9%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRL--MQQHQQQLRMQ 466
QAQ QPP QQ Q Q++Q + PQ QQMQQ QHH ++ QQH QQ++ Q
Sbjct: 124 QAQQNQPP------QQNQQQQHHQQ--QQQQPQHHQQMQQQQHHQQMQQQQQHHQQMQQQ 175
Query: 467 QQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
Q QQ H + Q Q + QQ QQ + Q H
Sbjct: 176 QHHQQMQHHQLQQHQHQHQQQQQQQQHQQQHHQQQQQQQQQH 217
Score = 54.7 bits (130), Expect = 2e-06
Identities = 37/100 (37%), Positives = 41/100 (41%), Gaps = 7/100 (7%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQ 468
Q Q QQP + QQQ Q Q QQ+Q QQ QHH QHQ Q + QQQ
Sbjct: 141 QQQQQQPQHHQQMQQQQHHQQMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQQQ 200
Query: 469 QQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
Q QQ H QQ Q QQ + PQ H
Sbjct: 201 QHQQQHHQQQQQQQ-------QQHHQQQQHHQHSQPQQQH 233
Score = 54.3 bits (129), Expect = 2e-06
Identities = 38/97 (39%), Positives = 45/97 (46%), Gaps = 11/97 (11%)
Query: 404 HARPPQAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQ-----QFQHHYRLMQQ 458
H + Q QPQ Q Q+ Q Q H + Q QQMQ Q QH ++ QQ
Sbjct: 139 HHQQQQQQPQHHQQMQQQQHHQQMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQ 198
Query: 459 ---HQQQLRMQQQQQQQLHAGVSPVQQPGQVARPPQQ 492
HQQQ QQQQQQQ H QQ Q ++P QQ
Sbjct: 199 QQQHQQQHHQQQQQQQQQH---HQQQQHHQHSQPQQQ 232
Score = 50.4 bits (119), Expect = 3e-05
Identities = 32/86 (37%), Positives = 38/86 (43%)
Query: 411 QPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQQLRMQQQQQ 470
Q QQ ++ QQ Q Q Q H + Q QQ QQ Q H + Q QQQ + Q QQ
Sbjct: 161 QMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQQQQHQQQHHQQQQQQQQQHHQQ 220
Query: 471 QQLHAGVSPVQQPGQVARPPQQSLQQ 496
QQ H P QQ + Q QQ
Sbjct: 221 QQHHQHSQPQQQHQHNQQQQHQHNQQ 246
Score = 49.7 bits (117), Expect = 5e-05
Identities = 30/74 (40%), Positives = 37/74 (49%), Gaps = 7/74 (9%)
Query: 421 NQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHY--RLMQQHQQQLRMQQQQQQQLHAGVS 478
+QQQ Q Q+ Q H + QQ+Q QQ QHH + QQHQ + Q Q QQ H
Sbjct: 193 HQQQQQQQQHQQQHHQQQQQQQQQHHQQQQHHQHSQPQQQHQHNQQQQHQHNQQQHQ--- 249
Query: 479 PVQQPGQVARPPQQ 492
QQ Q+ PQQ
Sbjct: 250 --QQQNQIQMVPQQ 261
Score = 47.0 bits (110), Expect = 4e-04
Identities = 30/75 (40%), Positives = 37/75 (49%), Gaps = 5/75 (6%)
Query: 404 HARPPQAQPQQPPPYYLNQQQLQLLQYYQS--HVESLNPQQRQQMQQFQHHYRLMQQHQQ 461
H Q Q QQ + QQQ Q Q++Q H + PQQ+ Q Q Q H QQHQQ
Sbjct: 191 HQHQQQQQQQQHQQQHHQQQQQQQQQHHQQQQHHQHSQPQQQHQHNQQQQHQHNQQQHQQ 250
Query: 462 ---QLRMQQQQQQQL 473
Q++M QQ Q L
Sbjct: 251 QQNQIQMVPQQPQSL 265
Score = 46.2 bits (108), Expect = 6e-04
Identities = 37/104 (35%), Positives = 45/104 (42%), Gaps = 10/104 (9%)
Query: 409 QAQPQQPPPYYLNQQQL-QLLQYYQ--SHVESLNPQQRQQMQQFQHHYRLMQQ----HQQ 461
Q Q QQ + QQQ Q +Q++Q H QQ+QQ Q QHH + QQ HQQ
Sbjct: 161 QMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQQQQHQQQHHQQQQQQQQQHHQQ 220
Query: 462 QLRMQQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQ 505
Q Q Q QQ H QQ Q QQ ++ P Q
Sbjct: 221 QQHHQHSQPQQQH---QHNQQQQHQHNQQQHQQQQNQIQMVPQQ 261
Score = 43.5 bits (101), Expect = 0.004
Identities = 35/103 (33%), Positives = 41/103 (38%), Gaps = 6/103 (5%)
Query: 409 QAQPQQPPPYYLNQQQLQLLQYYQSHVESLNPQQRQQMQQFQHHYRLMQQHQQ---QLRM 465
Q P Q P Y Q QL+ Q ++ PQQ QQ Q Q + Q HQQ Q
Sbjct: 103 QIHPNQNPHY---NYQYQLMFMQQQAQQNQPPQQNQQQQHHQQQQQQPQHHQQMQQQQHH 159
Query: 466 QQQQQQQLHAGVSPVQQPGQVARPPQQSLQQPSVKSCPPQAVH 508
QQ QQQQ H QQ Q + Q Q + Q H
Sbjct: 160 QQMQQQQQHHQQMQQQQHHQQMQHHQLQQHQHQHQQQQQQQQH 202
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.136 0.428
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 164,209,040
Number of Sequences: 164201
Number of extensions: 7407268
Number of successful extensions: 55497
Number of sequences better than 10.0: 592
Number of HSP's better than 10.0 without gapping: 273
Number of HSP's successfully gapped in prelim test: 336
Number of HSP's that attempted gapping in prelim test: 38345
Number of HSP's gapped (non-prelim): 5653
length of query: 1366
length of database: 59,974,054
effective HSP length: 123
effective length of query: 1243
effective length of database: 39,777,331
effective search space: 49443222433
effective search space used: 49443222433
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 72 (32.3 bits)
Medicago: description of AC139744.2


