

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139601.1 - phase: 0 /pseudo
(2139 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
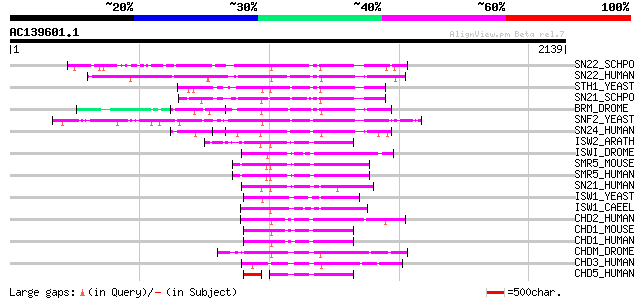
Score E
Sequences producing significant alignments: (bits) Value
SN22_SCHPO (O94421) SNF2-family ATP dependent chromatin remodeli... 435 e-121
SN22_HUMAN (P51531) Possible global transcription activator SNF2... 425 e-118
STH1_YEAST (P32597) Nuclear protein STH1/NPS1 (Chromatin structu... 418 e-116
SN21_SCHPO (Q9UTN6) SNF2-family ATP dependent chromatin remodeli... 416 e-115
BRM_DROME (P25439) Homeotic gene regulator (Brahma protein) 413 e-114
SNF2_YEAST (P22082) Transcription regulatory protein SNF2 (SWI/S... 402 e-111
SN24_HUMAN (P51532) Possible global transcription activator SNF2... 392 e-108
ISW2_ARATH (Q8RWY3) Putative chromatin remodelling complex ATPas... 320 3e-86
ISWI_DROME (Q24368) Chromatin remodelling complex ATPase chain I... 308 1e-82
SMR5_MOUSE (Q91ZW3) SWI/SNF related matrix associated actin depe... 304 2e-81
SMR5_HUMAN (O60264) SWI/SNF related matrix associated actin depe... 303 3e-81
SN21_HUMAN (P28370) Possible global transcription activator SNF2... 303 4e-81
ISW1_YEAST (P38144) Chromatin remodelling complex ATPase chain I... 296 4e-79
ISW1_CAEEL (P41877) Chromatin remodelling complex ATPase chain i... 291 1e-77
CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2 ... 277 3e-73
CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1 ... 265 1e-69
CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1 ... 264 2e-69
CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi... 237 2e-61
CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3 ... 225 9e-58
CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5 ... 212 8e-54
>SN22_SCHPO (O94421) SNF2-family ATP dependent chromatin remodeling
factor snf22
Length = 1680
Score = 435 bits (1118), Expect = e-121
Identities = 407/1422 (28%), Positives = 641/1422 (44%), Gaps = 270/1422 (18%)
Query: 223 QATSPAVSSEGSAHANSSTDVSALV---------GSVKARQT-----APPSH--LGLPIN 266
Q+ +P+V + S+H+ S+ ++ A V GS+ + +PPS L P N
Sbjct: 242 QSRAPSVDTTSSSHSFSARNIPANVSMQQQMGRRGSIPVNPSTFSASSPPSGSMLASPYN 301
Query: 267 AGVAGNSSDTAVQQF--SLHGRDAQGSLKQLIVGVNGMPSMHPQQSSANKSLGADSSLNA 324
G +++ A + S + S + VG G +PQ S+ + ++NA
Sbjct: 302 -GYQNDAASFAHSKLPSSANPNTPFNSTATVDVGAAGSHFPYPQPSNLD-------AINA 353
Query: 325 KASSSRSDPEPAKMQYVRQL-----------SQHASLDGGSTKEVG---------SGNYA 364
K S PA Y L S+ S+D + K S +
Sbjct: 354 KTYFQSSSNSPAPYVYRNNLPPSATSFQPSSSRSPSVDPNTVKSAQHIPRMSPSPSASAL 413
Query: 365 KPQGG-PSQMPQKLNGFTKNQLHVLKAQILAFRRLKKGDGILPQELLEAISPPPLDLHVQ 423
K Q PS + QL +LK+QI+A+ L +G +P + +AI
Sbjct: 414 KTQSHVPSAKVPPTSKLNHAQLAMLKSQIVAYNCLNSPNGQVPPAVQQAIFG-------- 465
Query: 424 QPIHSAGAQNQDKSMGNSVTEQPRQNEPKAKDSQPIVSFDGNSSEQETFVRDQKSTGAEV 483
GA N+ S + +QN P+ +S +++T RD ++
Sbjct: 466 ---RVYGASNEV-----SPSMPFQQNVPQM-----------SSVKKDTPTRDANMRTSKA 506
Query: 484 HMQAMLPVTKVSAGKEDQQSAGFSAKSDKKSEHVINRAPVINDLALDKGKAVASQALVTD 543
+P Q +SA K+E + P ++ + L + T
Sbjct: 507 PYIQNIP--------NQFQRRAYSATIPVKNESLAK--PSVSPMPLQQS---------TG 547
Query: 544 TAQINKPAQSSTVVGLPKDAGPAKKYYGPLFDFPFFTRKQDSFGSSMMANNNNNLSLAYD 603
++ K AQ T V P P+ F + + S S++ S+++D
Sbjct: 548 KTEVAKRAQFPTNVNYSSCVDPRTYVKTPIPFSKFSSSENLSLIPSLLPP-----SISWD 602
Query: 604 VKELLYEEGTEVFNKRRTENLKKIEGLLAVNLERKRIRPDLVLKLQIEEKKLRLLDLQAR 663
L E R + L+K +VN K+I K IE + LRLL+ Q
Sbjct: 603 DVFLSSEIAIACSIANRIDFLEKENRPKSVN---KKILQQDKSKSMIELRCLRLLEKQRS 659
Query: 664 LRGEIDQQQQEIMAMPDRPYRKFVKLCERQRVELARQV------QTSQKALREKQ----- 712
LR I+ ++ R + +RQ ++ A V Q ++ A+R+K+
Sbjct: 660 LRETINSVIPHSDSLAAGNLRLMFRNVKRQTMQEANLVLALAEKQKTEHAMRQKEKLLTH 719
Query: 713 LKSIFQWRKKLLEVHWAIRDARTARNRGVAKYHEKMLKEFSKNKDDDRNKRMEALKNNDV 772
L+SI RK ++ A+T R + + +H + KE K + +R++AL+ +D
Sbjct: 720 LRSIMLHRKSIVTKVDKQNKAKTQRCKDIINFHAHLEKEEKKRIERSARQRLQALRADDE 779
Query: 773 DRYREMLLEQQTSLPGDAAERYNVLSTFLTQTEEYLQKLGSKIT-SAKNQQEVEESAKAA 831
Y ++L D A+ + + L QT++YL+ L + N S K +
Sbjct: 780 AAYLQLL---------DKAKDTRI-THLLKQTDQYLENLTRAVRIQQSNIHSGNTSGKGS 829
Query: 832 AAAARLQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLR 891
+A E+ AP Y+ +AH ++E+V
Sbjct: 830 NSA-----------------------------ELEAPISEEDKNLDYFKVAHRIHEEV-E 859
Query: 892 QPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYG 951
QP + GTL++YQL GL+WMLSLYNN LNGILADEMGLGKT+Q +A I YL+E K G
Sbjct: 860 QPKIFVGGTLKDYQLKGLEWMLSLYNNNLNGILADEMGLGKTIQTIAFITYLIEKKNQQG 919
Query: 952 PHLIIVPNAVLVNWKCAFNPENCIDHAE*IAYMVTIGVMHFL-CWIEGSSVKIIFST--- 1007
P LIIVP + L NW F E + IAY + L I S+ ++ +T
Sbjct: 920 PFLIIVPLSTLTNWIMEF--EKWAPSVKKIAYKGPPQLRKTLQSQIRSSNFNVLLTTFEY 977
Query: 1008 ---------------------QRMKDRESVLARDLDR-YRCHRRLLLTGTPLQNDLKELW 1045
R+K+ +S L L Y RL+LTGTPLQN+L ELW
Sbjct: 978 IIKDRPLLSRIKWVHMIIDEGHRIKNTQSKLTSTLSTYYHSQYRLILTGTPLQNNLPELW 1037
Query: 1046 SLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFML 1105
+LLN +LP++F++ K+F++WF+ PF + L E+ ++II RLH++L PF+
Sbjct: 1038 ALLNFVLPKIFNSIKSFDEWFNTPFAN---TGGQDKIGLNEEEALLIIKRLHKVLRPFLF 1094
Query: 1106 RRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAK 1165
RR ++VE LP KV V++C +S Q +Y +K G L ++ E+ ++ +
Sbjct: 1095 RRLKKDVEKELPDKVEKVIKCPLSGLQLKLYQQMKKHGMLFVDGEKGKTGI--------- 1145
Query: 1166 QYKTLNNRCMELRKTCNHPLLNYPFFSDLSK---------DFMVKCCGKLWMLDRILIKL 1216
K L N M+L+K CNHP + F D+ + D + + GK +LDRIL KL
Sbjct: 1146 --KGLQNTVMQLKKICNHPFI----FEDVERAIDPSGTNVDLLWRAAGKFELLDRILPKL 1199
Query: 1217 QRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIF 1276
TGH+ L+F MT+++ I+E+YL+ + Y R+DG+T +DR S + FN P SD +IF
Sbjct: 1200 FLTGHKTLMFFQMTQIMTIMEDYLRSKNWKYLRLDGSTKSDDRCSLLAQFNDPKSDVYIF 1259
Query: 1277 LLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKIS 1336
+LS RA G GLNLQ+ADTV+I+D D NP + QA RAHRIGQ +EV+++ + + +K
Sbjct: 1260 MLSTRAGGLGLNLQTADTVIIFDTDWNPHQDLQAQDRAHRIGQTKEVRILRL--ITEK-- 1315
Query: 1337 SHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTT 1396
SIE I S QYK+D+ +VI AG+FD ++T
Sbjct: 1316 ----------------------------SIEENILSR-AQYKLDLDGKVIQAGKFDNKST 1346
Query: 1397 HEERRLTLETLL-HDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEE------- 1448
EER L +LL HD + + + E+N +I+R +EE+ LF ++D+E
Sbjct: 1347 PEEREAFLRSLLEHDGDDDHDLTYGELQDDELNELISRTDEELVLFKKLDKERAATDIYG 1406
Query: 1449 --DWLEEMTRYDQVPDWIRASTREVNAAIAASS---------KRPSKKNALSGGNVVLDS 1497
LE + +++PD+ + EV++ SS ++ ++N++S + LD
Sbjct: 1407 KGKPLERLLTVNELPDFYKV---EVDSFAVQSSSELEDQYLERKRRRRNSISYTELTLDE 1463
Query: 1498 TEIGSE-------RRRGRPKGKKNPSYKELEDSSEEISEDRN 1532
+ R+RGRP+ K N S+E S R+
Sbjct: 1464 LNTVDDPSSTLMPRKRGRPRKKTNSGSSLSTPLSQESSLARS 1505
>SN22_HUMAN (P51531) Possible global transcription activator SNF2L2
(SNF2-alpha) (SWI/SNF related matrix associated actin
dependent regulator of chromatin subfamily A member 2)
Length = 1586
Score = 425 bits (1092), Expect = e-118
Identities = 384/1351 (28%), Positives = 587/1351 (43%), Gaps = 212/1351 (15%)
Query: 301 GMPSMHPQQSSANKSLGADSSLNAKASSSRSDPEPAKMQYVRQLSQHASLDGGSTKEVG- 359
GM HP +G S + S P P+ + +S S G + ++
Sbjct: 98 GMRPPHP-------GMGPPQSPMDQHSQGYMSPHPSPLGAPEHVSSPMSGGGPTPPQMPP 150
Query: 360 SGNYAKPQGGPSQMPQKLNG---FTKNQLHVLKAQILAFRRLKKGDGILPQELLEAISPP 416
S A G P M Q G F+ QLH L+AQILA++ L +G LP+ L A+
Sbjct: 151 SQPGALIPGDPQAMSQPNRGPSPFSPVQLHQLRAQILAYKMLARGQP-LPETLQLAVQGK 209
Query: 417 PLDLHVQQPIHSAGAQNQDKSMGNSVTEQPRQNEPKAKDSQPIVSFD----------GNS 466
+QQ Q Q + +QP Q + + + +V+++ G S
Sbjct: 210 RTLPGLQQQQQQQQQQQQQQQQQQQQQQQPPQPQTQQQQQPALVNYNRPSGPGPELSGPS 269
Query: 467 SEQETFVRDQKSTGAEVHMQAMLPVTKVSAGKEDQQSAGFSAKSDKKSEHVINR-APVIN 525
+ Q+ V + A P G Q A + + +R +P+
Sbjct: 270 TPQKLPVPAPGGRPSPAPPAAAQPPAAAVPGPSVPQPAPGQPSPVLQLQQKQSRISPIQK 329
Query: 526 DLALDKGKAVASQALVTDTAQINKPAQSSTVVG-LPKDAGPAKKYYGPLFDFPFFTRKQD 584
LD + + + ++ + + G LP D F R+
Sbjct: 330 PQGLDPVEILQEREYRLQARIAHRIQELENLPGSLPPDLRTKATVELKALRLLNFQRQLR 389
Query: 585 SFGSSMMANNNNNLSLAYDVKELLYEEGTEVFNKRRTENLKKIEGLLAVNLERKRIRPDL 644
+ M + L A + K + + R TE L+K + + ERKR +
Sbjct: 390 QEVVACM-RRDTTLETALNSKAYKRSKRQTLREARMTEKLEKQQ---KIEQERKRRQKHQ 445
Query: 645 VLKLQIEEKKLRLLDLQARLRGEIDQQQQEIM---AMPDRPYRKFVKLCERQRVE--LAR 699
I + + + G+I + + + A +R +K + E++R+ +A
Sbjct: 446 EYLNSILQHAKDFKEYHRSVAGKIQKLSKAVATWHANTEREQKKETERIEKERMRRLMAE 505
Query: 700 QVQTSQKALREKQ-------LKSIFQWRKKLLEVHWAIRDARTARNRGVAKYHEKMLKEF 752
+ +K + +K+ L+ ++ L + W + A+ A+ + + +K +E
Sbjct: 506 DEEGYRKLIDQKKDRRLAYLLQQTDEYVANLTNLVWEHKQAQAAKEKKKRRRRKKKAEEN 565
Query: 753 SKNKDD---------DRNKRM-------------------EALKNNDVDRYREMLLEQQT 784
++ + D + +M EA K + +D + EM +
Sbjct: 566 AEGGESALGPDGEPIDESSQMSDLPVKVTHTETGKVLFGPEAPKASQLDAWLEMNPGYEV 625
Query: 785 SLPGDAAERYNVLSTFLTQTEEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQGLSEEE 844
+ D+ E + + E Q+ KI N +EV E Q + +E
Sbjct: 626 APRSDSEESDSDYEEEDEEEESSRQETEEKILLDPNSEEVSEKDAKQIIETAKQDVDDE- 684
Query: 845 VRAAAACAGEEVMIRNRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREY 904
M GS S YY +AHA++E V +Q ++L GTL+ Y
Sbjct: 685 ------------------YSMQYSARGSQS---YYTVAHAISEWVEKQSALLINGTLKHY 723
Query: 905 QLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVN 964
QL GL+WM+SLYNN LNGILADEMGLGKT+Q +ALI YLME K GP+LIIVP + L N
Sbjct: 724 QLQGLEWMVSLYNNNLNGILADEMGLGKTIQTIALITYLMEHKRLNGPYLIIVPLSTLSN 783
Query: 965 WKCAFNPENCIDHAE*IAYMVTIGVMHFLC-WIEGSSVKIIFST---------------- 1007
W F + I+Y T + L + ++ +T
Sbjct: 784 WTYEF--DKWAPSVVKISYKGTPAMRRSLVPQLRSGKFNVLLTTYEYIIKDKHILAKIRW 841
Query: 1008 --------QRMKDRESVLARDLD-RYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDN 1058
RMK+ L + L+ Y RR+LLTGTPLQN L ELW+LLN LLP +F +
Sbjct: 842 KYMIVDEGHRMKNHHCKLTQVLNTHYVAPRRILLTGTPLQNKLPELWALLNFLLPTIFKS 901
Query: 1059 KKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPP 1118
F WF+ PF E L E+ ++II RLH++L PF+LRR +EVE LP
Sbjct: 902 CSTFEQWFNAPFA-----MTGERVDLNEEETILIIRRLHKVLRPFLLRRLKKEVESQLPE 956
Query: 1119 KVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELR 1178
KV V++C MSA Q +Y +++ G L + E+ + + KTL N M+LR
Sbjct: 957 KVEYVIKCDMSALQKILYRHMQAKGILLTDGSEKDKKGKGGA-------KTLMNTIMQLR 1009
Query: 1179 KTCNHPLLNYPFFSDLSKDF---------------MVKCCGKLWMLDRILIKLQRTGHRV 1223
K CNHP + F + + F + + GK +LDRIL KL+ T HRV
Sbjct: 1010 KICNHPYM----FQHIEESFAEHLGYSNGVINGAELYRASGKFELLDRILPKLRATNHRV 1065
Query: 1224 LLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAA 1283
LLF MT L+ I+E+Y +R +Y R+DGTT EDR + + FN P S FIFLLS RA
Sbjct: 1066 LLFCQMTSLMTIMEDYFAFRNFLYLRLDGTTKSEDRAALLKKFNEPGSQYFIFLLSTRAG 1125
Query: 1284 GRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKISSHQKEDE 1343
G GLNLQ+ADTVVI+D D NP + QA RAHRIGQ+ EV+V+ + V
Sbjct: 1126 GLGLNLQAADTVVIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTV------------ 1173
Query: 1344 MRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTTHEERRLT 1403
S+E I + +YK+++ +VI AG FDQ+++ ERR
Sbjct: 1174 --------------------NSVEEKILA-AAKYKLNVDQKVIQAGMFDQKSSSHERRAF 1212
Query: 1404 LETLLHDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMD---EEEDWLE-----EMT 1455
L+ +L EE +E +VP + +N+MIAR EEE +LF +MD ED +
Sbjct: 1213 LQAILEHEEENEEE-DEVPDDETLNQMIARREEEFDLFMRMDMDRRREDARNPKRKPRLM 1271
Query: 1456 RYDQVPDWIRASTREVNAAIAASSK-----RPSKK----------------NALSGGNVV 1494
D++P WI EV + R S++ A+ GN+
Sbjct: 1272 EEDELPSWIIKDDAEVERLTCEEEEEKIFGRGSRQRRDVDYSDALTEKQWLRAIEDGNLE 1331
Query: 1495 LDSTEIGSERRRGRPKGKKNPSYKELEDSSE 1525
E+ ++R+ R K+P+ +++E + +
Sbjct: 1332 EMEEEVRLKKRKRRRNVDKDPAKEDVEKAKK 1362
>STH1_YEAST (P32597) Nuclear protein STH1/NPS1 (Chromatin structure
remodeling complex protein STH1) (SNF2 homolog)
Length = 1359
Score = 418 bits (1074), Expect = e-116
Identities = 279/856 (32%), Positives = 427/856 (49%), Gaps = 147/856 (17%)
Query: 646 LKLQIEEKKLRLLDLQARLRGEI-----DQQQQEIMAMPDRPYRKFV--------KLCER 692
+K +E K L+LL Q +R ++ Q I + D P+ K+
Sbjct: 239 IKALVELKSLKLLTKQKSIRQKLINNVASQAHHNIPYLRDSPFTAAAQRSVQIRSKVIVP 298
Query: 693 QRVELARQVQTSQ--------KALREKQLKSIFQWRKKLLEVHWAIRDARTARNRGVAKY 744
Q V LA +++ Q + L +++ SI + K+ W+ ++ R A
Sbjct: 299 QTVRLAEELERQQLLEKRKKERNLHLQKINSIIDFIKERQSEQWSRQERCFQFGRLGASL 358
Query: 745 HEKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQT 804
H +M K+ K + +R+ ALK+ND + Y ++L + + + ++ L QT
Sbjct: 359 HNQMEKDEQKRIERTAKQRLAALKSNDEEAYLKLLDQTKDTR----------ITQLLRQT 408
Query: 805 EEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRNRFME 864
+L L + + +N+ ++ L EEV+
Sbjct: 409 NSFLDSLSEAVRAQQNEAKI---------------LHGEEVQPIT--------------- 438
Query: 865 MNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGIL 924
D + YY +AH + EK+ +QPS+L GTL+EYQL GL+WM+SLYNN LNGIL
Sbjct: 439 -----DEEREKTDYYEVAHRIKEKIDKQPSILVGGTLKEYQLRGLEWMVSLYNNHLNGIL 493
Query: 925 ADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFNP----------ENC 974
ADEMGLGKT+Q ++LI YL E K + GP L+IVP + + NW F +
Sbjct: 494 ADEMGLGKTIQSISLITYLYEVKKDIGPFLVIVPLSTITNWTLEFEKWAPSLNTIIYKGT 553
Query: 975 IDHAE*IAYMVTIGVMHFLCWIEGSSVK-------------IIFSTQRMKDRESVLARDL 1021
+ + + + +G L +K II RMK+ +S L+ +
Sbjct: 554 PNQRHSLQHQIRVGNFDVLLTTYEYIIKDKSLLSKHDWAHMIIDEGHRMKNAQSKLSFTI 613
Query: 1022 DRY-RCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAE 1080
Y R RL+LTGTPLQN+L ELW+LLN +LP++F++ K F DWF+ PF + E
Sbjct: 614 SHYYRTRNRLILTGTPLQNNLPELWALLNFVLPKIFNSAKTFEDWFNTPFANTGTQEKLE 673
Query: 1081 NDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIK 1140
L E+ ++II RLH++L PF+LRR +EVE LP KV V++C++S Q +Y +
Sbjct: 674 ---LTEEETLLIIRRLHKVLRPFLLRRLKKEVEKDLPDKVEKVIKCKLSGLQQQLYQQML 730
Query: 1141 STGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDL------ 1194
L + E + K LNN+ M+LRK CNHP + F ++
Sbjct: 731 KHNALFVGAGTEGATKGG--------IKGLNNKIMQLRKICNHPFV----FDEVEGVVNP 778
Query: 1195 ---SKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRID 1251
+ D + + GK +LDR+L K + +GHRVL+F MT+++DI+E++L+ + L Y R+D
Sbjct: 779 SRGNSDLLFRVAGKFELLDRVLPKFKASGHRVLMFFQMTQVMDIMEDFLRMKDLKYMRLD 838
Query: 1252 GTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAV 1311
G+T E+R + FN+P+SD F FLLS RA G GLNLQ+ADTV+I+D D NP + QA
Sbjct: 839 GSTKTEERTEMLNAFNAPDSDYFCFLLSTRAGGLGLNLQTADTVIIFDTDWNPHQDLQAQ 898
Query: 1312 ARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIR 1371
RAHRIGQK EV+++ + S+E +I
Sbjct: 899 DRAHRIGQKNEVRILRLITT--------------------------------DSVEEVIL 926
Query: 1372 SNIQQYKIDMADEVINAGRFDQRTTHEERRLTLETLLHDEERCQETVHDVPSLQEVNRMI 1431
Q K+D+ +VI AG+FD ++T EE+ L L+ E + E+N +
Sbjct: 927 ERAMQ-KLDIDGKVIQAGKFDNKSTAEEQEAFLRRLIESETNRDDDDKAELDDDELNDTL 985
Query: 1432 ARNEEEVELFDQMDEE 1447
AR+ +E LFD++D+E
Sbjct: 986 ARSADEKILFDKIDKE 1001
>SN21_SCHPO (Q9UTN6) SNF2-family ATP dependent chromatin remodeling
factor snf21
Length = 1199
Score = 416 bits (1068), Expect = e-115
Identities = 288/851 (33%), Positives = 431/851 (49%), Gaps = 155/851 (18%)
Query: 650 IEEKKLRLLDLQARLRGEIDQQQQEIMAMPDRPYRKFVKLCER-----QRVELARQVQTS 704
+E KKLRL+ Q LR ++ Q + + + R C R Q L ++
Sbjct: 199 VELKKLRLIKQQESLRHQVMHCQPHLRTIVNAVERMS---CRRPKLVPQATRLTEVLERQ 255
Query: 705 QKALREKQLKSIFQWRKKLLEVHWAIRDARTAR--------NRGVAKYHEKMLKEFSKNK 756
Q++ RE++LK + + H + RT NR V YH + KE +
Sbjct: 256 QRSDRERRLKQKQCDYLQTVCAHGREINVRTKNAQARAQKANRAVLAYHSHIEKEEQRRA 315
Query: 757 DDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQTEEYLQKLGSKIT 816
+ + +R++ALK ND + Y +++ + + + ++ L QT+ YL L + +
Sbjct: 316 ERNAKQRLQALKENDEEAYLKLIDQAKDTR----------ITHLLRQTDHYLDSLAAAVK 365
Query: 817 SAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKDGSSSVS 876
++Q GE + MN D
Sbjct: 366 VQQSQ------------------------------FGESAYDEDMDRRMNPEDDRKID-- 393
Query: 877 KYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQV 936
YYN+AH + E V QPS+L G L+EYQL GLQWM+SLYNN LNGILADEMGLGKT+Q
Sbjct: 394 -YYNVAHNIREVVTEQPSILVGGKLKEYQLRGLQWMISLYNNHLNGILADEMGLGKTIQT 452
Query: 937 MALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAF----------------------NPENC 974
++LI +L+E K GP L+IVP + L NW F +P+
Sbjct: 453 ISLITHLIEKKRQNGPFLVIVPLSTLTNWTMEFERWAPSIVKIVYKGPPQVRKALHPQ-- 510
Query: 975 IDHAE*IAYMVTIG-------VMHFLCWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCH 1027
+ H+ + T ++ + WI II RMK+ +S L L Y
Sbjct: 511 VRHSNFQVLLTTYEYIIKDRPLLSRIKWI----YMIIDEGHRMKNTQSKLTNTLTTYYSS 566
Query: 1028 R-RLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLET 1086
R RL+LTGTPLQN+L ELW+LLN +LP +F++ K+F++WF+ PF E L
Sbjct: 567 RYRLILTGTPLQNNLPELWALLNFVLPRIFNSIKSFDEWFNTPFANTGGQDKME---LTE 623
Query: 1087 EKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLR 1146
E+ +++I RLH++L PF+LRR ++VE LP KV V+RC+MS Q +Y +K G L
Sbjct: 624 EESLLVIRRLHKVLRPFLLRRLKKDVEAELPDKVEKVIRCQMSGLQQKLYYQMKKHGMLY 683
Query: 1147 LNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDLSK---------D 1197
+ E ++ K+ + K L N M+L+K CNHP + F D+ + D
Sbjct: 684 V----EDAKRGKTGI------KGLQNTVMQLKKICNHPFV----FEDVERSIDPTGFNYD 729
Query: 1198 FMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALE 1257
+ + GK +LDRIL KL R+GHR+L+F MT++++I+E+YL +R+ Y R+DG+T +
Sbjct: 730 MLWRVSGKFELLDRILPKLFRSGHRILMFFQMTQIMNIMEDYLHYRQWRYLRLDGSTKAD 789
Query: 1258 DRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRI 1317
DR + FN P ++ +FLLS RA G GLNLQ+ADTV+I+D D NP + QA RAHRI
Sbjct: 790 DRSKLLGVFNDPTAEVNLFLLSTRAGGLGLNLQTADTVIIFDSDWNPHQDLQAQDRAHRI 849
Query: 1318 GQKREVKVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQY 1377
GQ +EV++ + + +K S+E I + QY
Sbjct: 850 GQTKEVRIYRL--ITEK------------------------------SVEENILAR-AQY 876
Query: 1378 KIDMADEVINAGRFDQRTTHEERRLTLETLLHDEERCQETVHDVP-SLQEVNRMIARNEE 1436
K+D+ +VI AG+FD ++T EER L +LL +E +E E+N ++AR ++
Sbjct: 877 KLDIDGKVIQAGKFDNKSTPEEREAFLRSLLENENGEEENDEKGELDDDELNEILARGDD 936
Query: 1437 EVELFDQMDEE 1447
E+ LF QM E+
Sbjct: 937 ELRLFKQMTED 947
>BRM_DROME (P25439) Homeotic gene regulator (Brahma protein)
Length = 1638
Score = 413 bits (1062), Expect = e-114
Identities = 307/921 (33%), Positives = 446/921 (48%), Gaps = 140/921 (15%)
Query: 619 RRTENLKKIEGLLAVNLERKRIRPDLVLKLQIEEKKLRLLDLQARLRGEIDQQQQEIMAM 678
R TE L+K + L A ERKR + L + + L + + ++ + + +M
Sbjct: 492 RATEKLEKQQKLEA---ERKRRQKHLEFLAAVLQHGKDLREFHRNNKAQLARMNKAVMNH 548
Query: 679 PDRPYRKFVKLCERQRVELARQVQTSQKALREKQLKSIFQWRKKLLEVHWAIRDARTARN 738
R+ K ER E R++ + E K I Q + K L + D +
Sbjct: 549 HANAEREQKKEQERIEKERMRRLMAEDE---EGYRKLIDQKKDKRLAFLLSQTDEYISNL 605
Query: 739 RGVAKYHEKMLKEFSKNKDDDRNKRMEALKN--------------NDVDRYREMLLEQQT 784
+ K H K+ K ++ KR+ K + V R ++EQ T
Sbjct: 606 TQMVKQH----KDDQMKKKEEEGKRLIQFKKELLMSGEYIGIDEGSIVADMRVHVVEQCT 661
Query: 785 S--LPGDAAERYNVLSTFLTQ------TEEYLQKLGSKITSAKNQQEVEESAKAAAAAAR 836
L GD A L +L ++ GS +E + + A A+
Sbjct: 662 GKKLTGDDAPMLKHLHRWLNMHPGWDWIDDEEDSCGSNDDHKPKVEEQPTATEDATDKAQ 721
Query: 837 LQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSML 896
G E+ E+ R + YY++AH ++EKV+ Q S++
Sbjct: 722 ATGNDEDAKDLITKAKVEDDEYR-------------TEEQTYYSIAHTIHEKVVEQASIM 768
Query: 897 RAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLII 956
GTL+EYQ+ GL+W++SLYNN LNGILADEMGLGKT+Q ++L+ YLM+ K GP+LII
Sbjct: 769 VNGTLKEYQIKGLEWLVSLYNNNLNGILADEMGLGKTIQTISLVTYLMDRKKVMGPYLII 828
Query: 957 VPNAVLVNWKCAF--------------NPEN---CIDHAE*IAYMVTIGVMHFLCWIEGS 999
VP + L NW F +P+ + + V + ++ +
Sbjct: 829 VPLSTLPNWVLEFEKWAPAVGVVSYKGSPQGRRLLQNQMRATKFNVLLTTYEYVIKDKAV 888
Query: 1000 SVKI------IFSTQRMKDRESVLARDLD-RYRCHRRLLLTGTPLQNDLKELWSLLNLLL 1052
KI I RMK+ L + L+ Y RLLLTGTPLQN L ELW+LLN LL
Sbjct: 889 LAKIQWKYMIIDEGHRMKNHHCKLTQVLNTHYIAPYRLLLTGTPLQNKLPELWALLNFLL 948
Query: 1053 PEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEV 1112
P +F + F WF+ PF E L E+ ++II RLH++L PF+LRR +EV
Sbjct: 949 PSIFKSCSTFEQWFNAPFAT-----TGEKVELNEEETILIIRRLHKVLRPFLLRRLKKEV 1003
Query: 1113 EGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNN 1172
E LP KV +++C MSA Q +Y ++S G L L E+ + K K L N
Sbjct: 1004 EHQLPDKVEYIIKCDMSALQRVLYKHMQSKGVL-LTDGSEKGKHGKGGA------KALMN 1056
Query: 1173 RCMELRKTCNHPLLNYPFFSDLSKDF--------------MVKCCGKLWMLDRILIKLQR 1218
++LRK CNHP + F + + + + + GK +LDRIL KL+
Sbjct: 1057 TIVQLRKLCNHPFM----FQHIEEKYCDHTGGHGVVSGPDLYRVSGKFELLDRILPKLKA 1112
Query: 1219 TGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLL 1278
T HRVLLF MT+ + I+E+YL WR+ Y R+DGTT EDR + FN+ SD F+FLL
Sbjct: 1113 TNHRVLLFCQMTQCMTIIEDYLGWRQFGYLRLDGTTKAEDRGELLRKFNAKGSDVFVFLL 1172
Query: 1279 SIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVVDKISSH 1338
S RA G GLNLQ+ADTVVI+D D NP + QA RAHRIGQ+ EV+V+ + V
Sbjct: 1173 STRAGGLGLNLQTADTVVIFDSDWNPHQDLQAQDRAHRIGQRNEVRVLRLMTV------- 1225
Query: 1339 QKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQRTTHE 1398
S+E I + +YK++M ++VI AG FDQ++T
Sbjct: 1226 -------------------------NSVEERILA-AARYKLNMDEKVIQAGMFDQKSTGS 1259
Query: 1399 ERRLTLETLLHDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEE---ED-----W 1450
ER+ L+T+LH ++ +E ++VP + +N MIAR+EEE+E+F +MD E ED
Sbjct: 1260 ERQQFLQTILHQDDNEEEEENEVPDDEMINMMIARSEEEIEIFKRMDAERKKEDEEIHPG 1319
Query: 1451 LEEMTRYDQVPDWIRASTREV 1471
E + ++PDW+ EV
Sbjct: 1320 RERLIDESELPDWLTKDDDEV 1340
Score = 61.2 bits (147), Expect = 3e-08
Identities = 127/608 (20%), Positives = 219/608 (35%), Gaps = 91/608 (14%)
Query: 257 PPSHLGLPINAG----VAGNSSDTAVQQFSLHGRDAQGSLKQLIVGVNGMPS--MHPQQS 310
PP H G+ + G + + + + HG + IV G P P++S
Sbjct: 74 PPHHQGMIFSKGPHMGMQMPPTGPNMSPYQTHGMPPNAPTQPCIVSPGGPPGGPPPPERS 133
Query: 311 SANKSLGADSSLNAKASSS-RSDPEPAKMQYVRQLSQHASLDGGSTKEVGSGNYAKPQGG 369
S ++++ + DP +++ +R S+H L+G
Sbjct: 134 SQENLHALQRAIDSMEEKGLQEDPRYSQLLAMRATSKHQHLNG----------------- 176
Query: 370 PSQMPQKLNGFTKNQLHVLKAQILAFRRLKKGDGI---LPQELLEAISPPPLDLHVQQPI 426
NQ+++L+ QI A+R L + I + Q L A PP + P
Sbjct: 177 -------------NQVNLLRTQITAYRLLARNKPISMQMQQALQAAQQQPPPGPPIGPPG 223
Query: 427 HSAGAQNQDKSMGNS-VTEQPRQNEPKAKDSQPIVSFDGNSSEQETFVRDQKSTGAEV-- 483
G + G V Q +Q P + + P S S+ V QK A
Sbjct: 224 APGGPPPGSQHAGQPPVPPQQQQQPPPSAGTPPQCSTPPASNPYGPPVPGQKMQVAPPPP 283
Query: 484 HMQAMLPVTKVSAGKEDQQSAGFSAKSDKKSEHVINRAPVINDLALDKGKAVAS------ 537
HMQ P+ ++ + P + L G S
Sbjct: 284 HMQQGQPLPPQPPQVGGPPPIQQQQPPQQQQQQSQPPPPEPHQHQLPNGGKPLSMGPSGG 343
Query: 538 QALVTDTAQINKPAQSSTVVGLPKDAGPAKKYYGPLFDFPFFTRKQDSFGSSMMANNNNN 597
Q L+ + +P T+ G+P + + GP ++Q M N
Sbjct: 344 QPLIPSSPM--QPQVRGTLPGMPPGSQVPQPGGGP--------QRQVPPAGMPMPKPNRI 393
Query: 598 LSLAYDV---KELLYEEGTEVFNKRRTENLKKIEGLLAVNLERKRIRPDLVLKLQIEEKK 654
++A V L +E R + +++++ L A E DL L+ IE +
Sbjct: 394 TTVAKPVGLDPITLLQERENRIAARISLRMQELQRLPATMSE------DLRLQAAIELRA 447
Query: 655 LRLLDLQARLRGEIDQQQQEIMAMPDRPYRKFVKLCERQRVELARQVQTSQKAL------ 708
LR+L+ Q +LR E Q + + K K +RQ + AR + +K
Sbjct: 448 LRVLNFQRQLRMEFVQCTRRDTTLETALNIKLYKRTKRQGLREARATEKLEKQQKLEAER 507
Query: 709 --REKQLK---SIFQWRKKLLEVHWAIRDARTARNRGVAKYHEKMLKEFSKNKDDDRNKR 763
R+K L+ ++ Q K L E H + N+ V +H +E K ++ +R
Sbjct: 508 KRRQKHLEFLAAVLQHGKDLREFHRNNKAQLARMNKAVMNHHANAEREQKKEQERIEKER 567
Query: 764 MEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQTEEYLQKLGSKITSAKNQQ- 822
M L D + YR+++ +++ L+ L+QT+EY+ L + K+ Q
Sbjct: 568 MRRLMAEDEEGYRKLIDQKKDKR----------LAFLLSQTDEYISNLTQMVKQHKDDQM 617
Query: 823 -EVEESAK 829
+ EE K
Sbjct: 618 KKKEEEGK 625
>SNF2_YEAST (P22082) Transcription regulatory protein SNF2 (SWI/SNF
complex component SNF2) (Regulatory protein SWI2)
(Regulatory protein GAM1) (Transcription factor TYE3)
Length = 1703
Score = 402 bits (1032), Expect = e-111
Identities = 404/1549 (26%), Positives = 681/1549 (43%), Gaps = 255/1549 (16%)
Query: 163 QIATSDQLRAMQAWAHERNID-LSQPANANFAAQLNLMQ---------TRMVQQSKESGA 212
Q + + R W H RN ++ P+ F ++Q QQ SG+
Sbjct: 7 QFSNEEVNRCYLRWQHLRNEHGMNAPSVPEFIYLTKVLQFAAKQRQELQMQRQQQGISGS 66
Query: 213 QSSSVPVSKQQATSPA-----VSSEGSAHANSSTDVSALVGSVKARQTAPPSHLGLPINA 267
Q + VP S QA P +S+ S H + ++ + +P +P+N+
Sbjct: 67 QQNIVPNSSDQAELPNNASSHISASASPHLAPNMQLNGNETFSTSAHQSPIMQTQMPLNS 126
Query: 268 GVAGNSSDTAVQQFSLHGRDAQGSLKQLIVGVNGMPSMHPQQSSANKSLGADSSLNAKAS 327
G ++ +Q S+ GSL N P+ P + N + D+S + +
Sbjct: 127 N--GGNNMLPQRQSSV------GSLN----ATNFSPT--PANNGENAAEKPDNSNHNNLN 172
Query: 328 SSRSDPEPAKMQYVRQLSQHASLDGGSTKEVGSGNYAKPQGGPSQMPQKLNGFTKNQLHV 387
+ S+ +P Q +++ GS A+ Q Q Q Q
Sbjct: 173 LNNSELQPQNRSLQEHNIQDSNVMPGSQINSPMPQQAQMQQAQFQAQQAQQAQQAQQAQQ 232
Query: 388 LKAQILAFRRLKKGDGILPQ-ELLEA--------ISPPPLDLHVQQPIHSAGAQNQD-KS 437
+A++ RRL Q ELL+A ++ P+ Q I + D K
Sbjct: 233 AQARLQQGRRLPMTMFTAEQSELLKAQITSLKCLVNRKPIPFEFQAVIQKSINHPPDFKR 292
Query: 438 MGNSVTEQPRQNEPKAKDSQPIVSFDGNSSEQETFVRDQKSTGAEVHMQAMLPVTKVSAG 497
M S++E R+ +P +++Q + +G ++ Q+ G H +
Sbjct: 293 MLLSLSEFARRRQPTDQNNQS--NLNGGNNTQQP--------GTNSHYN--------NTN 334
Query: 498 KEDQQSAGFSAKSDKKSEHVINRAPV-------INDLALDKGKAVASQALVTDT------ 544
++ +A D K E+ + +P + LDK S +T T
Sbjct: 335 TDNVSGLTRNAPLDSKDENFASVSPAGPSSVHNAKNGTLDKNSQTVSGTPITQTESKKEE 394
Query: 545 ----AQINKPAQSSTVVGLPKDAGPAKKYYGPL-------------FDFPFFTRKQDSFG 587
+ + K A +S ++ P + PL D DSF
Sbjct: 395 NETISNVAKTAPNSNKTHTEQNNPPKPQKPVPLNVLQDQYKEGIKVVDIDDPDMMVDSFT 454
Query: 588 SSMMANNN---NNLSLAYDVKELLYEEGTEVFNKRRTENLKKIEGLLAVNLERKRIRPDL 644
++++N L D + E G + L+A+NL+ D
Sbjct: 455 MPNISHSNIDYQTLLANSDHAKFTIEPGVLPVGIDTHTATDIYQTLIALNLDTTV--NDC 512
Query: 645 VLKLQIEE-------------KKLRLLDLQARLRGEIDQQQQEIMAMPDRPYRKFVKLCE 691
+ KL +E L+LL LQ +RG + Q + ++ + F+
Sbjct: 513 LDKLLNDECTESTRENALYDYYALQLLPLQKAVRGHVLQFEWHQNSLLTNTHPNFLSKIR 572
Query: 692 RQRVE---LARQVQTSQKALREKQLKS-----IFQWRKKLLEVHWAIRDARTAR----NR 739
V+ L Q+ + + L+ ++ K+ + K + + +D + R +R
Sbjct: 573 NINVQDALLTNQLYKNHELLKLERKKTEAVARLKSMNKSAINQYNRRQDKKNKRLKFGHR 632
Query: 740 GVAKYHEKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLST 799
+A H + ++ K + +R++ALK ND + Y ++L + + + ++
Sbjct: 633 LIAT-HTNLERDEQKRAEKKAKERLQALKANDEEAYIKLLDQTKDTR----------ITH 681
Query: 800 FLTQTEEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIR 859
L QT +L L + K+QQ+ + + + ++ A+ + M+
Sbjct: 682 LLRQTNAFLDSLTRAV---KDQQKYTKE------------MIDSHIKEASEEVDDLSMVP 726
Query: 860 NRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNK 919
E D +S+V YYN+AH + E + +QPS+L GTL++YQ+ GLQWM+SL+NN
Sbjct: 727 KMKDEEYDDDDDNSNVD-YYNVAHRIKEDIKKQPSILVGGTLKDYQIKGLQWMVSLFNNH 785
Query: 920 LNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFNP-------- 971
LNGILADEMGLGKT+Q ++L+ YL E K GP+L+IVP + L NW F
Sbjct: 786 LNGILADEMGLGKTIQTISLLTYLYEMKNIRGPYLVIVPLSTLSNWSSEFAKWAPTLRTI 845
Query: 972 -------ENCIDHAE*IAYMVTIGVMHFLCWIEG----SSVK----IIFSTQRMKDRESV 1016
E A+ A + + F I+ S VK II RMK+ +S
Sbjct: 846 SFKGSPNERKAKQAKIRAGEFDVVLTTFEYIIKERALLSKVKWVHMIIDEGHRMKNAQSK 905
Query: 1017 LARDLD-RYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDP 1075
L+ L+ Y RL+LTGTPLQN+L ELW+LLN +LP++F++ K+F++WF+ PF
Sbjct: 906 LSLTLNTHYHADYRLILTGTPLQNNLPELWALLNFVLPKIFNSVKSFDEWFNTPFANTGG 965
Query: 1076 NQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAI 1135
E L E+ +++I RLH++L PF+LRR ++VE LP KV V++C+MSA Q +
Sbjct: 966 QDKIE---LSEEETLLVIRRLHKVLRPFLLRRLKKDVEKELPDKVEKVVKCKMSALQQIM 1022
Query: 1136 YDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDLS 1195
Y + L + + + + + NN+ M+L+K CNHP + ++
Sbjct: 1023 YQQMLKYRRLFIGDQNNKKMVG---------LRGFNNQIMQLKKICNHPFVFEEVEDQIN 1073
Query: 1196 -----KDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRI 1250
D + + GK +LDRIL KL+ TGHRVL+F MT+++DI+E++L++ + Y R+
Sbjct: 1074 PTRETNDDIWRVAGKFELLDRILPKLKATGHRVLIFFQMTQIMDIMEDFLRYINIKYLRL 1133
Query: 1251 DGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQA 1310
DG T ++R + FN+P+S+ F+LS RA G GLNLQ+ADTV+I+D D NP + QA
Sbjct: 1134 DGHTKSDERSELLRLFNAPDSEYLCFILSTRAGGLGLNLQTADTVIIFDTDWNPHQDLQA 1193
Query: 1311 VARAHRIGQKREVKVIYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLI 1370
RAHRIGQK EV+++ + S+E +I
Sbjct: 1194 QDRAHRIGQKNEVRILRLITT--------------------------------NSVEEVI 1221
Query: 1371 RSNIQQYKIDMADEVINAGRFDQRTTHEERRLTLETLLHDEE----RCQETVHDVPSLQ- 1425
+ K+D+ +VI AG+FD ++T EE+ L +LL EE + + V + L+
Sbjct: 1222 LERAYK-KLDIDGKVIQAGKFDNKSTSEEQEALLRSLLDAEEERRKKRESGVEEEEELKD 1280
Query: 1426 -EVNRMIARNEEEVELFDQMDEEEDWLEE-------MTRYDQVPDWIRASTREVNAAIAA 1477
E+N ++ARN+EE+ + +MDE+ EE + ++PD +R++ A +
Sbjct: 1281 SEINEILARNDEEMAVLTRMDEDRSKKEEELGVKSRLLEKSELPD---IYSRDIGAEL-- 1335
Query: 1478 SSKRPSKKNALSGGNVVLDSTEIGSERRRGRPKGKKNPSYKELEDSSEEISEDRNEDSAH 1537
+ S+ A+ G R R N S ++ E +++N+ A
Sbjct: 1336 -KREESESAAVYNGR---------GARERKTATYNDNMSEEQWLRQFEVSDDEKNDKQAR 1385
Query: 1538 DEGEIGEFEDDGYSGAGIAQPVDKDKLDDVTPSDAEYECPRSSSESARN 1586
+ E + + G G + K +++ DA+ + PR ++ SA +
Sbjct: 1386 KQRTKKEDKSEAIDGNG------EIKGENI---DADNDGPRINNISAED 1425
>SN24_HUMAN (P51532) Possible global transcription activator SNF2L4
(SNF2-beta) (BRG-1 protein) (Mitotic growth and
transcription activator) (Brahma protein homolog 1)
(SWI/SNF related matrix associated actin dependent
regulator of chromatin subfamily A m
Length = 1647
Score = 392 bits (1007), Expect = e-108
Identities = 277/780 (35%), Positives = 383/780 (48%), Gaps = 135/780 (17%)
Query: 780 LEQQTSLPGDAAERYNVLSTFLTQTEEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQG 839
+E L G A + L +L Y S + +++E EE + AA+
Sbjct: 622 VESGKILTGTDAPKAGQLEAWLEMNPGYEVAPRSDSEESGSEEEEEEEEEEQPQAAQPPT 681
Query: 840 LSEEEVRAAAACAGEEVM-IRNRFMEMNAPKDGSSS----------VSKYYNLAHAVNEK 888
L EE + ++V + R + NA +D + YY +AHAV E+
Sbjct: 682 LPVEEKKKIPDPDSDDVSEVDARHIIENAKQDVDDEYGVSQALARGLQSYYAVAHAVTER 741
Query: 889 VLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKG 948
V +Q +++ G L++YQ+ GL+W++SLYNN LNGILADEMGLGKT+Q +ALI YLME K
Sbjct: 742 VDKQSALMVNGVLKQYQIKGLEWLVSLYNNNLNGILADEMGLGKTIQTIALITYLMEHKR 801
Query: 949 NYGPHLIIVPNAVLVNW----------------------KCAFNPENCIDHAE*I--AYM 984
GP LIIVP + L NW + AF P+ + Y
Sbjct: 802 INGPFLIIVPLSTLSNWAYEFDKWAPSVVKVSYKGSPAARRAFVPQLRSGKFNVLLTTYE 861
Query: 985 VTIGVMHFLCWIEGSSVKIIFSTQRMKDRESVLARDLD-RYRCHRRLLLTGTPLQNDLKE 1043
I H L I + I+ RMK+ L + L+ Y RRLLLTGTPLQN L E
Sbjct: 862 YIIKDKHILAKIRWKYM-IVDEGHRMKNHHCKLTQVLNTHYVAPRRLLLTGTPLQNKLPE 920
Query: 1044 LWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPF 1103
LW+LLN LLP +F + F WF+ PF E L E+ ++II RLH++L PF
Sbjct: 921 LWALLNFLLPTIFKSCSTFEQWFNAPFA-----MTGEKVDLNEEETILIIRRLHKVLRPF 975
Query: 1104 MLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQ 1163
+LRR +EVE LP KV V++C MSA Q +Y +++ G L + E+ + +
Sbjct: 976 LLRRLKKEVEAQLPEKVEYVIKCDMSALQRVLYRHMQAKGVLLTDGSEKDKKGKGGT--- 1032
Query: 1164 AKQYKTLNNRCMELRKTCNHPLLNYPFFSDLSKDF-----------MVKCCGKLWMLDRI 1212
KTL N M+LRK CNHP + S+ + + GK +LDRI
Sbjct: 1033 ----KTLMNTIMQLRKICNHPYMFQHIEESFSEHLGFTGGIVQGLDLYRASGKFELLDRI 1088
Query: 1213 LIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSD 1272
L KL+ T H+VLLF MT L+ I+E+Y +R Y R+DGTT EDR + FN P S+
Sbjct: 1089 LPKLRATNHKVLLFCQMTSLMTIMEDYFAYRGFKYLRLDGTTKAEDRGMLLKTFNEPGSE 1148
Query: 1273 CFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYMEAVV 1332
FIFLLS RA G GLNLQSADTV+I+D D NP + QA RAHRIGQ+ EV+V+ + V
Sbjct: 1149 YFIFLLSTRAGGLGLNLQSADTVIIFDSDWNPHQDLQAQDRAHRIGQQNEVRVLRLCTV- 1207
Query: 1333 DKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFD 1392
S+E I + +YK+++ +VI AG FD
Sbjct: 1208 -------------------------------NSVEEKILA-AAKYKLNVDQKVIQAGMFD 1235
Query: 1393 QRTTHEERRLTLETLLHDEERCQETVH--------------------------------- 1419
Q+++ ERR L+ +L EE+ E+ H
Sbjct: 1236 QKSSSHERRAFLQAILEHEEQ-DESRHCSTGSGSASFAHTAPPPAGVNPDLEEPPLKEED 1294
Query: 1420 DVPSLQEVNRMIARNEEEVELFDQMDEEEDWLE--------EMTRYDQVPDWIRASTREV 1471
+VP + VN+MIAR+EEE +LF +MD + E + D++P WI EV
Sbjct: 1295 EVPDDETVNQMIARHEEEFDLFMRMDLDRRREEARNPKRKPRLMEEDELPSWIIKDDAEV 1354
Score = 61.6 bits (148), Expect = 2e-08
Identities = 59/221 (26%), Positives = 93/221 (41%), Gaps = 30/221 (13%)
Query: 620 RTENLKKIEGLLAVNLERKRIRPDLVLKLQIEEKKLRLLDLQARLRGEIDQQQQEIMAMP 679
R + L+ + G LA DL K IE K LRLL+ Q +LR E+ + A+
Sbjct: 381 RIQELENLPGSLA---------GDLRTKATIELKALRLLNFQRQLRQEVVVCMRRDTALE 431
Query: 680 DRPYRKFVKLCERQRVELAR---QVQTSQKALREKQ--------LKSIFQWRKKLLEVHW 728
K K +RQ + AR +++ QK +E++ L SI Q K E H
Sbjct: 432 TALNAKAYKRSKRQSLREARITEKLEKQQKIEQERKRRQKHQEYLNSILQHAKDFKEYHR 491
Query: 729 AIRDARTARNRGVAKYHEKMLKEFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPG 788
++ + VA YH +E K + +RM L D + YR+++ +++
Sbjct: 492 SVTGKIQKLTKAVATYHANTEREQKKENERIEKERMRRLMAEDEEGYRKLIDQKKDKR-- 549
Query: 789 DAAERYNVLSTFLTQTEEYLQKLGSKITSAKNQQEVEESAK 829
L+ L QT+EY+ L + K Q +E K
Sbjct: 550 --------LAYLLQQTDEYVANLTELVPQHKAAQVAKEKKK 582
Score = 38.9 bits (89), Expect = 0.16
Identities = 39/142 (27%), Positives = 54/142 (37%), Gaps = 32/142 (22%)
Query: 300 NGMPSMH-PQQSSANKSLGADSSLNA-KASSSRSDPEPAKMQYVRQLSQHASLDGGSTKE 357
+ M MH P +S K + D N K RS + QH+ G
Sbjct: 67 DNMHQMHKPMESMHEKGMSDDPRYNQMKGMGMRSGGHAGMGPPPSPMDQHSQ---GYPSP 123
Query: 358 VGSGNYAK---PQGGPSQMPQKLNG-----------------------FTKNQLHVLKAQ 391
+G +A P GPS PQ +G F +NQLH L+AQ
Sbjct: 124 LGGSEHASSPVPASGPSSGPQMSSGPGGAPLDGADPQALGQQNRGPTPFNQNQLHQLRAQ 183
Query: 392 ILAFRRLKKGDGILPQELLEAI 413
I+A++ L +G LP L A+
Sbjct: 184 IMAYKMLARGQP-LPDHLQMAV 204
>ISW2_ARATH (Q8RWY3) Putative chromatin remodelling complex ATPase
chain (EC 3.6.1.-) (ISW2-like) (Sucrose nonfermenting
protein 2 homolog)
Length = 1057
Score = 320 bits (819), Expect = 3e-86
Identities = 217/602 (36%), Positives = 313/602 (51%), Gaps = 92/602 (15%)
Query: 751 EFSKNKDDDRNKRMEALKNNDVDRYREMLLEQQTSLPGDAAERYNVLSTFLTQTEEYLQK 810
E SK ++ R K M+ LK + +EML Q S+ D + +L Q E
Sbjct: 75 EISK-REKARLKEMQKLKKQKI---QEMLESQNASIDADMNNKGKGRLKYLLQQTELFAH 130
Query: 811 LGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRNRFMEMNAPKD 870
S+ + KA ++EEE EE + +D
Sbjct: 131 FAKSDGSSSQK-------KAKGRGRHASKITEEE-------EDEEYL--------KEEED 168
Query: 871 GSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGL 930
G L + N ++L QPS ++ G +R+YQL GL W++ LY N +NGILADEMGL
Sbjct: 169 G---------LTGSGNTRLLTQPSCIQ-GKMRDYQLAGLNWLIRLYENGINGILADEMGL 218
Query: 931 GKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNW--------------KCAFNPENCID 976
GKT+Q ++L+AYL E++G GPH+++ P + L NW K NPE
Sbjct: 219 GKTLQTISLLAYLHEYRGINGPHMVVAPKSTLGNWMNEIRRFCPVLRAVKFLGNPEERRH 278
Query: 977 HAE*--IAYMVTIGVMHFLCWIEGSSVK--------IIFSTQRMKDRESVLARDLDRYRC 1026
E +A I V F I+ + II R+K+ S+L++ + +
Sbjct: 279 IREDLLVAGKFDICVTSFEMAIKEKTALRRFSWRYIIIDEAHRIKNENSLLSKTMRLFST 338
Query: 1027 HRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLET 1086
+ RLL+TGTPLQN+L ELW+LLN LLPE+F + + F++WF + END E
Sbjct: 339 NYRLLITGTPLQNNLHELWALLNFLLPEIFSSAETFDEWFQI---------SGENDQQE- 388
Query: 1087 EKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLR 1146
++ +LH++L PF+LRR +VE LPPK +L+ MS Q Y +
Sbjct: 389 -----VVQQLHKVLRPFLLRRLKSDVEKGLPPKKETILKVGMSQMQKQYYKALLQKDLEA 443
Query: 1147 LNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNY---PFFSDLSKDFMVKCC 1203
+N E+ R L N M+LRK CNHP L P + D ++
Sbjct: 444 VNAGGERKR--------------LLNIAMQLRKCCNHPYLFQGAEPGPPYTTGDHLITNA 489
Query: 1204 GKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAI 1263
GK+ +LD++L KL+ RVL+FS MT+LLDILE+YL +R +Y RIDG T ++R+++I
Sbjct: 490 GKMVLLDKLLPKLKERDSRVLIFSQMTRLLDILEDYLMYRGYLYCRIDGNTGGDERDASI 549
Query: 1264 VDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREV 1323
+N P S+ F+FLLS RA G G+NL +AD V++YD D NP+ + QA RAHRIGQK+EV
Sbjct: 550 EAYNKPGSEKFVFLLSTRAGGLGINLATADVVILYDSDWNPQVDLQAQDRAHRIGQKKEV 609
Query: 1324 KV 1325
+V
Sbjct: 610 QV 611
>ISWI_DROME (Q24368) Chromatin remodelling complex ATPase chain Iswi
(EC 3.6.1.-) (Imitation swi protein) (Nucleosome
remodeling factor 140 kDa subunit) (NURF-140) (CHRAC 140
kDa subunit)
Length = 1027
Score = 308 bits (789), Expect = 1e-82
Identities = 216/626 (34%), Positives = 313/626 (49%), Gaps = 84/626 (13%)
Query: 893 PSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGP 952
P+ +++G +R+YQ+ GL WM+SLY N +NGILADEMGLGKT+Q ++L+ YL FK GP
Sbjct: 120 PAYIKSGEMRDYQIRGLNWMISLYENGINGILADEMGLGKTLQTISLLGYLKHFKNQAGP 179
Query: 953 HLIIVPNAVLVNWKCAFNP------ENCIDHAE*IAYMVTIGVM-----------HFLCW 995
H++IVP + L NW F C+ + V+ + +C
Sbjct: 180 HIVIVPKSTLQNWVNEFKKWCPSLRAVCLIGDQDTRNTFIRDVLMPGEWDVCVTSYEMCI 239
Query: 996 IEGSSVK-------IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLL 1048
E S K +I R+K+ +S L+ L ++ RLL+TGTPLQN+L ELW+LL
Sbjct: 240 REKSVFKKFNWRYLVIDEAHRIKNEKSKLSEILREFKTANRLLITGTPLQNNLHELWALL 299
Query: 1049 NLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRR 1108
N LLP+VF++ + F++WF+ N L + +I RLH +L+PF+LRR
Sbjct: 300 NFLLPDVFNSSEDFDEWFNT------------NTCLGDDA---LITRLHAVLKPFLLRRL 344
Query: 1109 VEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYK 1168
EVE L PK + + +S Q DW ++ ++EK
Sbjct: 345 KAEVEKRLKPKKEMKIFVGLSKMQR---DWYTKVLLKDIDVVNGAGKVEKM--------- 392
Query: 1169 TLNNRCMELRKTCNHPLL------NYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHR 1222
L N M+LRK NHP L P+ +D +V GK+ +LD++L KLQ G R
Sbjct: 393 RLQNILMQLRKCTNHPYLFDGAEPGPPYTTDTH---LVYNSGKMAILDKLLPKLQEQGSR 449
Query: 1223 VLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRA 1282
VL+FS MT++LDILE+Y WR Y R+DG T EDR I +FN NS F+F+LS RA
Sbjct: 450 VLIFSQMTRMLDILEDYCHWRNYNYCRLDGQTPHEDRNRQIQEFNMDNSAKFLFMLSTRA 509
Query: 1283 AGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM-------EAVVDKI 1335
G G+NL +AD V+IYD D NP+ + QA+ RAHRIGQK++V+V + E +V++
Sbjct: 510 GGLGINLATADVVIIYDSDWNPQMDLQAMDRAHRIGQKKQVRVFRLITESTVEEKIVERA 569
Query: 1336 SSHQKEDEMRI--GGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAGRFDQ 1393
+ D+M I G +D KD + I + D+ DE I+
Sbjct: 570 EVKLRLDKMVIQGGRLVDNRSNQLNKDEMLNIIRFGANQVFSSKETDITDEDIDVILERG 629
Query: 1394 RTTHEERRLTLETLLHDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEEDWLEE 1453
E++ L++L E + D N E E EDW E+
Sbjct: 630 EAKTAEQKAALDSL--GESSLRTFTMDT------------NGEAGTSSVYQFEGEDWREK 675
Query: 1454 MTRYDQVPDWIRASTREVNAAIAASS 1479
+ + + +WI RE A A +
Sbjct: 676 Q-KLNALGNWIEPPKRERKANYAVDA 700
>SMR5_MOUSE (Q91ZW3) SWI/SNF related matrix associated actin dependent
regulator of chromatin, subfamily A member 5 (EC 3.6.1.-)
(Sucrose nonfermenting protein 2 homolog) (mSnf2h)
Length = 1051
Score = 304 bits (778), Expect = 2e-81
Identities = 200/568 (35%), Positives = 298/568 (52%), Gaps = 73/568 (12%)
Query: 859 RNRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNN 918
R+R E ++ + SK N+ + PS ++ G LR+YQ+ GL W++SLY N
Sbjct: 141 RHRRTEQEEDEELLTESSKATNVCTRFEDS----PSYVKWGKLRDYQVRGLNWLISLYEN 196
Query: 919 KLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFNP------E 972
+NGILADEMGLGKT+Q ++L+ Y+ ++ GPH+++VP + L NW F
Sbjct: 197 GINGILADEMGLGKTLQTISLLGYMKHYRNIPGPHMVLVPKSTLHNWMSEFKKWVPTLRS 256
Query: 973 NCI--DHAE*IAYMVTI--------GVMHFLCWIEGSSV--------KIIFSTQRMKDRE 1014
C+ D + A++ + V + I+ SV +I R+K+ +
Sbjct: 257 VCLIGDKEQRAAFVRDVLLPGEWDVCVTSYEMLIKEKSVFKKFNWRYLVIDEAHRIKNEK 316
Query: 1015 SVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKED 1074
S L+ + ++ RLLLTGTPLQN+L ELWSLLN LLP+VF++ F+ WF
Sbjct: 317 SKLSEIVREFKTTNRLLLTGTPLQNNLHELWSLLNFLLPDVFNSADDFDSWFDT------ 370
Query: 1075 PNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSA 1134
N+ L +K ++ RLH +L PF+LRR +VE SLPPK + + +S Q
Sbjct: 371 ------NNCLGDQK---LVERLHMVLRPFLLRRIKADVEKSLPPKKEVKIYVGLSKMQRE 421
Query: 1135 IYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLL------NY 1188
Y I LN + +M L N M+LRK CNHP L
Sbjct: 422 WYTRILMKDIDILNSAGKMDKMR------------LLNILMQLRKCCNHPYLFDGAEPGP 469
Query: 1189 PFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYR 1248
P+ +D+ +V GK+ +LD++L KL+ G RVL+FS MT++LDILE+Y WR Y
Sbjct: 470 PYTTDMH---LVTNSGKMVVLDKLLPKLKEQGSRVLIFSQMTRVLDILEDYCMWRNYEYC 526
Query: 1249 RIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEE 1308
R+DG T ++R+ +I +N PNS F+F+LS RA G G+NL +AD V++YD D NP+ +
Sbjct: 527 RLDGQTPHDERQDSINAYNEPNSTKFVFMLSTRAGGLGINLATADVVILYDSDWNPQVDL 586
Query: 1309 QAVARAHRIGQKREVKVIYM-------EAVVDKISSHQKEDEMRI--GGTIDMEDELAGK 1359
QA+ RAHRIGQ + V+V E +V++ + D + I G +D GK
Sbjct: 587 QAMDRAHRIGQTKTVRVFRFITDNTVEERIVERAEMKLRLDSIVIQQGRLVDQNLNKIGK 646
Query: 1360 DRYIGSIESLIRSNIQQYKIDMADEVIN 1387
D + I + ++ DE I+
Sbjct: 647 DEMLQMIRHGATHVFASKESEITDEDID 674
>SMR5_HUMAN (O60264) SWI/SNF related matrix associated actin dependent
regulator of chromatin subfamily A member 5 (EC 3.6.1.-)
(SWI/SNF-related matrix-associated actin-dependent
regulator of chromatin A5) (Sucrose nonfermenting protein
2 homolog) (hSNF2H
Length = 1052
Score = 303 bits (777), Expect = 3e-81
Identities = 200/568 (35%), Positives = 298/568 (52%), Gaps = 73/568 (12%)
Query: 859 RNRFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGTLREYQLVGLQWMLSLYNN 918
R+R E ++ + SK N+ + PS ++ G LR+YQ+ GL W++SLY N
Sbjct: 142 RHRRTEQEEDEELLTESSKATNVCTRFEDS----PSYVKWGKLRDYQVRGLNWLISLYEN 197
Query: 919 KLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFNP------E 972
+NGILADEMGLGKT+Q ++L+ Y+ ++ GPH+++VP + L NW F
Sbjct: 198 GINGILADEMGLGKTLQTISLLGYMKHYRNIPGPHMVLVPKSTLHNWMSEFKRWVPTLRS 257
Query: 973 NCI--DHAE*IAYMVTI--------GVMHFLCWIEGSSV--------KIIFSTQRMKDRE 1014
C+ D + A++ + V + I+ SV +I R+K+ +
Sbjct: 258 VCLIGDKEQRAAFVRDVLLPGEWDVCVTSYEMLIKEKSVFKKFNWRYLVIDEAHRIKNEK 317
Query: 1015 SVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKED 1074
S L+ + ++ RLLLTGTPLQN+L ELWSLLN LLP+VF++ F+ WF
Sbjct: 318 SKLSEIVREFKTTNRLLLTGTPLQNNLHELWSLLNFLLPDVFNSADDFDSWFDT------ 371
Query: 1075 PNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSA 1134
N+ L +K ++ RLH +L PF+LRR +VE SLPPK + + +S Q
Sbjct: 372 ------NNCLGDQK---LVERLHMVLRPFLLRRIKADVEKSLPPKKEVKIYVGLSKMQRE 422
Query: 1135 IYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLL------NY 1188
Y I LN + +M L N M+LRK CNHP L
Sbjct: 423 WYTRILMKDIDILNSAGKMDKMR------------LLNILMQLRKCCNHPYLFDGAEPGP 470
Query: 1189 PFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYR 1248
P+ +D+ +V GK+ +LD++L KL+ G RVL+FS MT++LDILE+Y WR Y
Sbjct: 471 PYTTDMH---LVTNSGKMVVLDKLLPKLKEQGSRVLIFSQMTRVLDILEDYCMWRNYEYC 527
Query: 1249 RIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEE 1308
R+DG T ++R+ +I +N PNS F+F+LS RA G G+NL +AD V++YD D NP+ +
Sbjct: 528 RLDGQTPHDERQDSINAYNEPNSTKFVFMLSTRAGGLGINLATADVVILYDSDWNPQVDL 587
Query: 1309 QAVARAHRIGQKREVKVIYM-------EAVVDKISSHQKEDEMRI--GGTIDMEDELAGK 1359
QA+ RAHRIGQ + V+V E +V++ + D + I G +D GK
Sbjct: 588 QAMDRAHRIGQTKTVRVFRFITDNTVEERIVERAEMKLRLDSIVIQQGRLVDQNLNKIGK 647
Query: 1360 DRYIGSIESLIRSNIQQYKIDMADEVIN 1387
D + I + ++ DE I+
Sbjct: 648 DEMLQMIRHGATHVFASKESEITDEDID 675
>SN21_HUMAN (P28370) Possible global transcription activator SNF2L1
(SWI/SNF related matrix associated actin dependent
regulator of chromatin subfamily A member 1)
Length = 976
Score = 303 bits (775), Expect = 4e-81
Identities = 204/563 (36%), Positives = 290/563 (51%), Gaps = 83/563 (14%)
Query: 893 PSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGP 952
PS ++ G LR+YQ+ GL W++SLY N +NGILADEMGLGKT+Q +AL+ YL ++ GP
Sbjct: 97 PSYVKGGPLRDYQIRGLNWLISLYENGVNGILADEMGLGKTLQTIALLGYLKHYRNIPGP 156
Query: 953 HLIIVPNAVLVNWKCAF----------------NPENCIDHAE*IAYMVTIGVMHFLCWI 996
H+++VP + L NW F + E + + V + I
Sbjct: 157 HMVLVPKSTLHNWMNEFKRWVPSLRVICFVGDKDARAAFIRDEMMPGEWDVCVTSYEMVI 216
Query: 997 EGSSV--------KIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLL 1048
+ SV +I R+K+ +S L+ + ++ RLLLTGTPLQN+L ELW+LL
Sbjct: 217 KEKSVFKKFHWRYLVIDEAHRIKNEKSKLSEIVREFKSTNRLLLTGTPLQNNLHELWALL 276
Query: 1049 NLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRR 1108
N LLP+VF++ F+ WF + L +K ++ RLH +L+PF+LRR
Sbjct: 277 NFLLPDVFNSADDFDSWFDT------------KNCLGDQK---LVERLHAVLKPFLLRRI 321
Query: 1109 VEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYK 1168
+VE SLPPK I + +S Q Y I LN + +M
Sbjct: 322 KTDVEKSLPPKKEIKIYLGLSKMQREWYTKILMKDIDVLNSSGKMDKMR----------- 370
Query: 1169 TLNNRCMELRKTCNHPLL------NYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHR 1222
L N M+LRK CNHP L P+ +D + +V GK+ +LD++L KL+ G R
Sbjct: 371 -LLNILMQLRKCCNHPYLFDGAEPGPPYTTD---EHIVSNSGKMVVLDKLLAKLKEQGSR 426
Query: 1223 VLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRES------------AIVDFNSPN 1270
VL+FS MT+LLDILE+Y WR Y R+DG T E+RE AI FN+PN
Sbjct: 427 VLIFSQMTRLLDILEDYCMWRGYEYCRLDGQTPHEEREDKFLEVEFLGQREAIEAFNAPN 486
Query: 1271 SDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM-- 1328
S FIF+LS RA G G+NL SAD V++YD D NP+ + QA+ RAHRIGQK+ V+V +
Sbjct: 487 SSKFIFMLSTRAGGLGINLASADVVILYDSDWNPQVDLQAMDRAHRIGQKKPVRVFRLIT 546
Query: 1329 -----EAVVDKISSHQKEDEMRI--GGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDM 1381
E +V++ + D + I G ID K+ + I + ++
Sbjct: 547 DNTVEERIVERAEIKLRLDSIVIQQGRLIDQRSNKLAKEEMLQMIRHGATHVFASKESEL 606
Query: 1382 ADEVINA--GRFDQRTTHEERRL 1402
DE I R +++T RL
Sbjct: 607 TDEDITTILERGEKKTAEMNERL 629
>ISW1_YEAST (P38144) Chromatin remodelling complex ATPase chain ISW1
(EC 3.6.1.-)
Length = 1129
Score = 296 bits (758), Expect = 4e-79
Identities = 190/487 (39%), Positives = 271/487 (55%), Gaps = 68/487 (13%)
Query: 899 GTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVP 958
G LR YQ+ G+ W++SL+ NK+ GILADEMGLGKT+Q ++ + YL + GP L+I P
Sbjct: 194 GQLRPYQIQGVNWLVSLHKNKIAGILADEMGLGKTLQTISFLGYLRYIEKIPGPFLVIAP 253
Query: 959 NAVLVNWKCAFN---PE----------------------NCIDHAE*IAYMVTIGVMHFL 993
+ L NW N P+ C +Y + I L
Sbjct: 254 KSTLNNWLREINRWTPDVNAFILQGDKEERAELIQKKLLGCDFDVVIASYEIIIREKSPL 313
Query: 994 CWIEGSSVKIIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLP 1053
I + II R+K+ ES+L++ L + RLL+TGTPLQN+L ELW+LLN LLP
Sbjct: 314 KKINWEYI-IIDEAHRIKNEESMLSQVLREFTSRNRLLITGTPLQNNLHELWALLNFLLP 372
Query: 1054 EVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVE 1113
++F + + F+DWFS +ED ++ I+ +LH +L+PF+LRR +VE
Sbjct: 373 DIFSDAQDFDDWFSSESTEEDQDK--------------IVKQLHTVLQPFLLRRIKSDVE 418
Query: 1114 GSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKT-LNN 1172
SL PK + L MS+ Q Y I +N +K+ KT L N
Sbjct: 419 TSLLPKKELNLYVGMSSMQKKWYKKILEKDLDAVNGSNG-----------SKESKTRLLN 467
Query: 1173 RCMELRKTCNHPLL------NYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRVLLF 1226
M+LRK CNHP L P+ +D + +V KL +LD++L KL+ G RVL+F
Sbjct: 468 IMMQLRKCCNHPYLFDGAEPGPPYTTD---EHLVYNAAKLQVLDKLLKKLKEEGSRVLIF 524
Query: 1227 STMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRG 1286
S M++LLDILE+Y +R Y RIDG+TA EDR AI D+N+P+S F+FLL+ RA G G
Sbjct: 525 SQMSRLLDILEDYCYFRNYEYCRIDGSTAHEDRIQAIDDYNAPDSKKFVFLLTTRAGGLG 584
Query: 1287 LNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKVIYM-------EAVVDKISSHQ 1339
+NL SAD VV+YD D NP+ + QA+ RAHRIGQK++VKV + E ++++ +
Sbjct: 585 INLTSADVVVLYDSDWNPQADLQAMDRAHRIGQKKQVKVFRLVTDNSVEEKILERATQKL 644
Query: 1340 KEDEMRI 1346
+ D++ I
Sbjct: 645 RLDQLVI 651
>ISW1_CAEEL (P41877) Chromatin remodelling complex ATPase chain isw-1
(EC 3.6.1.-)
Length = 1009
Score = 291 bits (745), Expect = 1e-77
Identities = 181/520 (34%), Positives = 281/520 (53%), Gaps = 64/520 (12%)
Query: 891 RQPSMLRAGTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNY 950
+ P + G +R+YQ+ GL W+ SL +NK+NGILADEMGLGKT+Q +++I Y+ +K
Sbjct: 122 KSPFYIENGEMRDYQVRGLNWLASLQHNKINGILADEMGLGKTLQTISMIGYMKHYKNKA 181
Query: 951 GPHLIIVPNAVLVNWKCAFNPE----NCI----DHA---E*IAYMVTIGVMHFLCWIEGS 999
PHL+IVP + L NW F N + D A + + ++ C
Sbjct: 182 SPHLVIVPKSTLQNWANEFKKWCPSINAVVLIGDEAARNQVLRDVILPQKFDVCCTTYEM 241
Query: 1000 SVK-------------IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWS 1046
+K II R+K+ +S L+ + RLL+TGTPLQN+L ELW+
Sbjct: 242 MLKVKTQLKKLNWRYIIIDEAHRIKNEKSKLSETVRELNSENRLLITGTPLQNNLHELWA 301
Query: 1047 LLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLR 1106
LLN LLP++F + F+ WFS + + ++ RLH++L+PF+LR
Sbjct: 302 LLNFLLPDIFTSSDDFDSWFSNDAMSGNTD---------------LVQRLHKVLQPFLLR 346
Query: 1107 RRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQ 1166
R +VE SL PK + + +S Q +W ++ ++EK+ L
Sbjct: 347 RIKSDVEKSLLPKKEVKVYVGLSKMQR---EWYTKVLMKDIDIINGAGKVEKARLM---- 399
Query: 1167 YKTLNNRCMELRKTCNHPLL------NYPFFSDLSKDFMVKCCGKLWMLDRILIKLQRTG 1220
N M LRK NHP L PF +D +V GK+ +LD++L+K + G
Sbjct: 400 -----NILMHLRKCVNHPYLFDGAEPGPPFTTD---QHLVDNSGKMVVLDKLLMKFKEQG 451
Query: 1221 HRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSI 1280
RVL+FS +++LD+LE++ WR Y R+DG+T EDR +AI +N+P+S FIF+L+
Sbjct: 452 SRVLIFSQFSRMLDLLEDFCWWRHYEYCRLDGSTPHEDRSNAIEAYNAPDSKKFIFMLTT 511
Query: 1281 RAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV--IYMEAVVDKISSH 1338
RA G G+NL +AD V+IYD D NP+++ QA+ RAHRIGQK++V+V + E VD+
Sbjct: 512 RAGGLGINLATADVVIIYDSDWNPQSDLQAMDRAHRIGQKKQVRVFRLITENTVDERIIE 571
Query: 1339 QKEDEMRIGGTIDMEDELAGKDRYI--GSIESLIRSNIQQ 1376
+ E ++R+ + + ++ + + G + S+IR +Q
Sbjct: 572 KAEAKLRLDNIVIQQGRMSEAQKTLGKGDMISMIRHGAEQ 611
>CHD2_HUMAN (O14647) Chromodomain-helicase-DNA-binding protein 2
(CHD-2)
Length = 1739
Score = 277 bits (708), Expect = 3e-73
Identities = 215/701 (30%), Positives = 341/701 (47%), Gaps = 106/701 (15%)
Query: 891 RQPSMLRAGTL--REYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKG 948
+QP+ L L R+YQL GL W+ + + ILADEMGLGKT+Q ++ ++YL
Sbjct: 472 KQPAYLGGENLELRDYQLEGLNWLAHSWCKNNSVILADEMGLGKTIQTISFLSYLFHQHQ 531
Query: 949 NYGPHLIIVPNAVLVNWKCAFN---PENCIDHAE*IAYMVTIGVMHFLCWIEGSSVKIIF 1005
YGP LI+VP + L +W+ F PE I+ I +++ + WI + ++ F
Sbjct: 532 LYGPFLIVVPLSTLTSWQREFEIWAPE--INVVVYIGDLMSRNTIREYEWIHSQTKRLKF 589
Query: 1006 ST----------------------------QRMKDRESVLARDLDRYRCHRRLLLTGTPL 1037
+ R+K+ +S+L + L ++ + RLL+TGTPL
Sbjct: 590 NALITTYEILLKDKTVLGSINWAFLGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPL 649
Query: 1038 QNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLH 1097
QN LKELWSLL+ ++PE F+ + F +ED + EN + LH
Sbjct: 650 QNSLKELWSLLHFIMPEKFEFWEDF---------EEDHGKGRENGY----------QSLH 690
Query: 1098 QILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRME 1157
++LEPF+LRR ++VE SLP KV +LR MSA Q Y WI + L S
Sbjct: 691 KVLEPFLLRRVKKDVEKSLPAKVEQILRVEMSALQKQYYKWILTRNYKALAKGTRGST-- 748
Query: 1158 KSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDLSKD------FMVKCCGKLWMLDR 1211
N MEL+K CNH L P + ++ +++ GKL +LD+
Sbjct: 749 ----------SGFLNIVMELKKCCNHCYLIKPPEENERENGQEILLSLIRSSGKLILLDK 798
Query: 1212 ILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNS 1271
+L +L+ G+RVL+FS M ++LDIL EYL + ++R+DG+ E R+ A+ FN+ S
Sbjct: 799 LLTRLRERGNRVLIFSQMVRMLDILAEYLTIKHYPFQRLDGSIKGEIRKQALDHFNADGS 858
Query: 1272 DCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV--IYME 1329
+ F FLLS RA G G+NL SADTVVI+D D NP+N+ QA ARAHRIGQK++V + + +
Sbjct: 859 EDFCFLLSTRAGGLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNIYRLVTK 918
Query: 1330 AVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIESLIRSNIQQYKIDMADEVINAG 1389
V++ + + +M + + + G+ + RSN + + ++ G
Sbjct: 919 GTVEEEIIERAKKKMVLDHLVIQRMDTTGRTILENNSG---RSNSNPFNKEELTAILKFG 975
Query: 1390 RFD-----QRTTHEERRLTLETLL------HDEERCQETVHDVPSLQEVNRMIARNEEEV 1438
D + E + + ++ +L +E T + + N +EEE+
Sbjct: 976 AEDLFKELEGEESEPQEMDIDEILRLAETRENEVSTSATDELLSQFKVANFATMEDEEEL 1035
Query: 1439 ELFDQMD-------------EEEDWLEEMTRYDQVPDWIRASTREVNAAIAAS---SKRP 1482
E D EEE+ +E+ +P IR+ST++ + S SKR
Sbjct: 1036 EERPHKDWDEIIPEEQRKKVEEEERQKELEEIYMLPR-IRSSTKKAQTNDSDSDTESKRQ 1094
Query: 1483 SKKNALSGGNVVLDSTEIGSERRRGRPKGKKNPSYKELEDS 1523
+++++ S DS + +RRGRP+ + + D+
Sbjct: 1095 AQRSSASESETE-DSDDDKKPKRRGRPRSVRKDLVEGFTDA 1134
>CHD1_MOUSE (P40201) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1711
Score = 265 bits (676), Expect = 1e-69
Identities = 167/462 (36%), Positives = 249/462 (53%), Gaps = 70/462 (15%)
Query: 901 LREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNA 960
LR+YQL GL W+ + + ILADEMGLGKT+Q ++ + YL YGP L++VP +
Sbjct: 479 LRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLS 538
Query: 961 VLVNWKCAFNPENCIDHAE*IAYMVTIGVMHFLC---WIEGSSVKIIFS----------- 1006
L +W+ + + Y+ I + + W+ + ++ F+
Sbjct: 539 TLTSWQREI--QTWASQMNAVVYLGDINSRNMIRTHEWMHPQTKRLKFNILLTTYEILLK 596
Query: 1007 -----------------TQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLN 1049
R+K+ +S+L + L ++ + RLL+TGTPLQN LKELWSLL+
Sbjct: 597 DKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLH 656
Query: 1050 LLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRV 1109
++PE F + + F +E+ + E + LH+ LEPF+LRR
Sbjct: 657 FIMPEKFSSWEDF---------EEEHGKGREYGYAS----------LHKELEPFLLRRVK 697
Query: 1110 EEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKT 1169
++VE SLP KV +LR MSA Q Y WI + L+ + S
Sbjct: 698 KDVEKSLPAKVEQILRMEMSALQKQYYKWILTRNYKALSKGSKGST------------SG 745
Query: 1170 LNNRCMELRKTCNHPLLNYP------FFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRV 1223
N MEL+K CNH L P + + +++ GKL +LD++LI+L+ G+RV
Sbjct: 746 FLNIMMELKKCCNHCYLIKPPDNNEFYNKQEALQHLIRSSGKLILLDKLLIRLRERGNRV 805
Query: 1224 LLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAA 1283
L+FS M ++LDIL EYL++R+ ++R+DG+ E R+ A+ FN+ S+ F FLLS RA
Sbjct: 806 LIFSQMVRMLDILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAG 865
Query: 1284 GRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
G G+NL SADTVVI+D D NP+N+ QA ARAHRIGQK++V +
Sbjct: 866 GLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNI 907
>CHD1_HUMAN (O14646) Chromodomain-helicase-DNA-binding protein 1
(CHD-1)
Length = 1709
Score = 264 bits (674), Expect = 2e-69
Identities = 167/462 (36%), Positives = 248/462 (53%), Gaps = 70/462 (15%)
Query: 901 LREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNA 960
LR+YQL GL W+ + + ILADEMGLGKT+Q ++ + YL YGP L++VP +
Sbjct: 481 LRDYQLNGLNWLAHSWCKGNSCILADEMGLGKTIQTISFLNYLFHEHQLYGPFLLVVPLS 540
Query: 961 VLVNWKCAFNPENCIDHAE*IAYMVTIGVMHFLC---WIEGSSVKIIFS----------- 1006
L +W+ + + Y+ I + + W + ++ F+
Sbjct: 541 TLTSWQREI--QTWASQMNAVVYLGDINSRNMIRTHEWTHHQTKRLKFNILLTTYEILLK 598
Query: 1007 -----------------TQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLN 1049
R+K+ +S+L + L ++ + RLL+TGTPLQN LKELWSLL+
Sbjct: 599 DKAFLGGLNWAFIGVDEAHRLKNDDSLLYKTLIDFKSNHRLLITGTPLQNSLKELWSLLH 658
Query: 1050 LLLPEVFDNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRV 1109
++PE F + + F +E+ + E + LH+ LEPF+LRR
Sbjct: 659 FIMPEKFSSWEDF---------EEEHGKGREYGYAS----------LHKELEPFLLRRVK 699
Query: 1110 EEVEGSLPPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKT 1169
++VE SLP KV +LR MSA Q Y WI + L+ + S
Sbjct: 700 KDVEKSLPAKVEQILRMEMSALQKQYYKWILTRNYKALSKGSKGST------------SG 747
Query: 1170 LNNRCMELRKTCNHPLLNYP------FFSDLSKDFMVKCCGKLWMLDRILIKLQRTGHRV 1223
N MEL+K CNH L P + + +++ GKL +LD++LI+L+ G+RV
Sbjct: 748 FLNIMMELKKCCNHCYLIKPPDNNEFYNKQEALQHLIRSSGKLILLDKLLIRLRERGNRV 807
Query: 1224 LLFSTMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAA 1283
L+FS M ++LDIL EYL++R+ ++R+DG+ E R+ A+ FN+ S+ F FLLS RA
Sbjct: 808 LIFSQMVRMLDILAEYLKYRQFPFQRLDGSIKGELRKQALDHFNAEGSEDFCFLLSTRAG 867
Query: 1284 GRGLNLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
G G+NL SADTVVI+D D NP+N+ QA ARAHRIGQK++V +
Sbjct: 868 GLGINLASADTVVIFDSDWNPQNDLQAQARAHRIGQKKQVNI 909
Score = 35.4 bits (80), Expect = 1.7
Identities = 34/142 (23%), Positives = 60/142 (41%), Gaps = 14/142 (9%)
Query: 667 EIDQQQQEIMAMPDRPYRKFVK--------LCERQRVELARQVQTS-----QKALRE-KQ 712
E+DQ+ I RP + +K L ER+++E RQ + L+E
Sbjct: 1408 ELDQKTFSICKERMRPVKAALKQLDRPEKGLSEREQLEHTRQCLIKIGDHITECLKEYTN 1467
Query: 713 LKSIFQWRKKLLEVHWAIRDARTARNRGVAKYHEKMLKEFSKNKDDDRNKRMEALKNNDV 772
+ I QWRK L + + + K+ K +E +N D + N ++N DV
Sbjct: 1468 PEQIKQWRKNLWIFVSKFTEFDARKLHKLYKHAIKKRQESQQNSDQNSNLNPHVIRNPDV 1527
Query: 773 DRYREMLLEQQTSLPGDAAERY 794
+R +E +S +++R+
Sbjct: 1528 ERLKENTNHDDSSRDSYSSDRH 1549
>CHDM_DROME (O97159) Chromodomain helicase-DNA-binding protein Mi-2
homolog (dMi-2)
Length = 1982
Score = 237 bits (605), Expect = 2e-61
Identities = 218/793 (27%), Positives = 348/793 (43%), Gaps = 130/793 (16%)
Query: 801 LTQTEEYLQKLGSKITSAKNQQEVEESAKAAAAAARLQGLSEEEVRAAAACAGEEVMIRN 860
L Q +Y Q L + TS Q ++S K + +++ + V+
Sbjct: 654 LRQAIDYYQDLRAVCTSETTQSRSKKSKKGRKSKLKVEDDEDRPVKHYTP---------- 703
Query: 861 RFMEMNAPKDGSSSVSKYYNLAHAVNEKVLRQPSMLRAGT---LREYQLVGLQWMLSLYN 917
P+ ++ + K Y QP+ L GT L YQ+ G+ W+ +
Sbjct: 704 ------PPEKPTTDLKKKYE----------DQPAFLE-GTGMQLHPYQIEGINWLRYSWG 746
Query: 918 NKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVPNAVLVNWKCAFN---PE-N 973
++ ILADEMGLGKT+Q + + L + GP L+ VP + LVNW+ F P+
Sbjct: 747 QGIDTILADEMGLGKTIQTVTFLYSLYKEGHCRGPFLVAVPLSTLVNWEREFELWAPDFY 806
Query: 974 CIDH---AE*IAYMVTIGVMHFLCWIEGSSVKIIFSTQ---------------------- 1008
CI + + A + + I GS V + +TQ
Sbjct: 807 CITYIGDKDSRAVIRENELSFEEGAIRGSKVSRLRTTQYKFNVLLTSYELISMDAACLGS 866
Query: 1009 ------------RMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVF 1056
R+K +S R L+ Y +LLLTGTPLQN+L+EL+ LLN L + F
Sbjct: 867 IDWAVLVVDEAHRLKSNQSKFFRILNSYTIAYKLLLTGTPLQNNLEELFHLLNFLSRDKF 926
Query: 1057 DNKKAFNDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSL 1116
++ +AF F+ ++E + RLH++L P MLRR +V ++
Sbjct: 927 NDLQAFQGEFADVSKEEQ------------------VKRLHEMLGPHMLRRLKTDVLKNM 968
Query: 1117 PPKVSIVLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCME 1176
P K ++R +SA Q Y +I + LN + +L N M+
Sbjct: 969 PSKSEFIVRVELSAMQKKFYKFILTKNYEALNSKSGGGSC------------SLINIMMD 1016
Query: 1177 LRKTCNHPLLNYPFFSDLSK---------DFMVKCCGKLWMLDRILIKLQRTGHRVLLFS 1227
L+K CNHP L +P ++ + + + K GKL +L ++L +L+ HRVL+FS
Sbjct: 1017 LKKCCNHPYL-FPSAAEEATTAAGGLYEINSLTKAAGKLVLLSKMLKQLKAQNHRVLIFS 1075
Query: 1228 TMTKLLDILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGL 1287
MTK+LDILE++L+ + Y RIDG R+ AI FN+P + F+FLLS RA G G+
Sbjct: 1076 QMTKMLDILEDFLEGEQYKYERIDGGITGTLRQEAIDRFNAPGAQQFVFLLSTRAGGLGI 1135
Query: 1288 NLQSADTVVIYDPDPNPKNEEQAVARAHRIGQKREVKV---IYMEAVVDKISSHQKEDEM 1344
NL +ADTV+IYD D NP N+ QA +RAHRIGQ +V + + +V ++++ K M
Sbjct: 1136 NLATADTVIIYDSDWNPHNDIQAFSRAHRIGQANKVMIYRFVTRNSVEERVTQVAKRKMM 1195
Query: 1345 RIGGTIDMEDELAGKDRYIGSIESLIRSNIQQ-YKIDMADEVINAGRFDQRTTHEERRLT 1403
+ G + ++ ++R + +K D +E I+ +D + E T
Sbjct: 1196 LTHLVVRPGMGGKGANFTKQELDDILRFGTEDLFKEDDKEEAIH---YDDKAVAELLDRT 1252
Query: 1404 LETLLHDEERCQETVHDVPSLQEVNRMIARNEEEVELFDQMDEEED---WLEEMT-RYDQ 1459
+ E E + + EEE E+ Q E D W++ + Y+Q
Sbjct: 1253 NRGIEEKESWANEYLSSFKVASYATKE-EEEEEETEIIKQDAENSDPAYWVKLLRHHYEQ 1311
Query: 1460 VPDWIRASTREVNAAIAASSKRPSKKNALSGGNVVLDSTEIGSERRRGRPKGKKNPSYKE 1519
+V ++ + + N GG V D+T S + + S
Sbjct: 1312 -------HQEDVGRSLGKGKRVRKQVNYTDGGVVAADTTRDDSNWQDNGSEYNSEYSAGS 1364
Query: 1520 LEDSSEEISEDRN 1532
ED ++ +D+N
Sbjct: 1365 DEDGGDDDFDDQN 1377
>CHD3_HUMAN (Q12873) Chromodomain helicase-DNA-binding protein 3
(CHD-3) (Mi-2 autoantigen 240 kDa protein) (Mi2-alpha)
Length = 1944
Score = 225 bits (574), Expect = 9e-58
Identities = 206/710 (29%), Positives = 324/710 (45%), Gaps = 136/710 (19%)
Query: 892 QPSMLRA--GTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTV--------------- 934
QP + A GTL YQL GL W+ + + ILADEMGLGKT+
Sbjct: 725 QPRFITATGGTLHMYQLEGLNWLRFSWAQGTDTILADEMGLGKTIQTIVFLYSLYKEGHT 784
Query: 935 --------------------QVMALIAYLMEFKGNYGPHLIIVPNA------VLVNWKCA 968
Q+ A Y++ + G+ II N + K A
Sbjct: 785 KGPFLVSAPLSTIINWEREFQMWAPKFYVVTYTGDKDSRAIIRENEFSFEDNAIKGGKKA 844
Query: 969 FNPENCID---HAE*IAY-MVTI-----GVMHFLCWIEGSSVKIIFSTQRMKDRESVLAR 1019
F + H +Y ++TI G + + C + + R+K+ +S R
Sbjct: 845 FKMKREAQVKFHVLLTSYELITIDQAALGSIRWACLV-------VDEAHRLKNNQSKFFR 897
Query: 1020 DLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAFNDWFSKPFQKEDPNQNA 1079
L+ Y+ +LLLTGTPLQN+L+EL+ LLN L PE F+N + F + F+ KED
Sbjct: 898 VLNGYKIDHKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGFLEEFAD-ISKEDQ---- 952
Query: 1080 ENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSIVLRCRMSAFQSAIYDWI 1139
I +LH +L P MLRR +V ++P K +++R +S Q Y +I
Sbjct: 953 -------------IKKLHDLLGPHMLRRLKADVFKNMPAKTELIVRVELSPMQKKYYKYI 999
Query: 1140 KSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCNHPLLNYPFFSDLSKDF- 1198
+ LN +++ +L N M+L+K CNHP L +P + S
Sbjct: 1000 LTRNFEALNSRGGGNQV------------SLLNIMMDLKKCCNHPYL-FPVAAMESPKLP 1046
Query: 1199 --------MVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLDILEEYLQWRRLVYRRI 1250
++K GKL +L ++L KL+ GHRVL+FS MTK+LD+LE++L + Y RI
Sbjct: 1047 SGAYEGGALIKSSGKLMLLQKMLRKLKEQGHRVLIFSQMTKMLDLLEDFLDYEGYKYERI 1106
Query: 1251 DGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADTVVIYDPDPNPKNEEQA 1310
DG R+ AI FN+P + F FLLS RA G G+NL +ADTV+I+D D NP N+ QA
Sbjct: 1107 DGGITGALRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATADTVIIFDSDWNPHNDIQA 1166
Query: 1311 VARAHRIGQKREVKV---IYMEAVVDKISSHQKEDEMRIGGTIDMEDELAGKDRYIGSIE 1367
+RAHRIGQ +V + + +V ++I+ K M + G GS+
Sbjct: 1167 FSRAHRIGQANKVMIYRFVTRASVEERITQVAKRKMMLTHLVV-----RPGLGSKAGSMS 1221
Query: 1368 SLIRSNIQQYKIDMADEVINAG---RFDQRTTHEERRLTLETLLHDEERCQETVHDVPSL 1424
+I ++ + + N G D H + L +++ ++T DV ++
Sbjct: 1222 KQELDDILKFGTEELFKDENEGENKEEDSSVIHYDNEAIARLLDRNQDATEDT--DVQNM 1279
Query: 1425 QE------VNRMIARNEEEVELFDQ--MDEEE----DWLEEMTR--YDQVPDWI------ 1464
E V + + R E+++E ++ + +EE D+ E++ R Y+Q + +
Sbjct: 1280 NEYLSSFKVAQYVVREEDKIEEIEREIIKQEENVDPDYWEKLLRHHYEQQQEDLARNLGK 1339
Query: 1465 -RASTREVNAAIAASSKRPSKKNALSGGNVVLDSTEIGSERRRGRPKGKK 1513
+ ++VN AA + ++ G + E ER GR + K+
Sbjct: 1340 GKRVRKQVNYNDAAQEDQDNQSEYSVGSE---EEDEDFDERPEGRRQSKR 1386
>CHD5_HUMAN (Q8TDI0) Chromodomain helicase-DNA-binding protein 5
(CHD-5)
Length = 1954
Score = 212 bits (540), Expect = 8e-54
Identities = 131/331 (39%), Positives = 188/331 (56%), Gaps = 38/331 (11%)
Query: 1003 IIFSTQRMKDRESVLARDLDRYRCHRRLLLTGTPLQNDLKELWSLLNLLLPEVFDNKKAF 1062
++ R+K+ +S R L+ Y+ +LLLTGTPLQN+L+EL+ LLN L PE F+N + F
Sbjct: 845 VVDEAHRLKNNQSKFFRVLNSYKIDYKLLLTGTPLQNNLEELFHLLNFLTPERFNNLEGF 904
Query: 1063 NDWFSKPFQKEDPNQNAENDWLETEKKVIIIHRLHQILEPFMLRRRVEEVEGSLPPKVSI 1122
+ F+ KED I +LH +L P MLRR +V ++P K +
Sbjct: 905 LEEFAD-ISKEDQ-----------------IKKLHDLLGPHMLRRLKADVFKNMPAKTEL 946
Query: 1123 VLRCRMSAFQSAIYDWIKSTGTLRLNPEEEQSRMEKSPLYQAKQYKTLNNRCMELRKTCN 1182
++R +S Q Y +I + LN + +++ +L N M+L+K CN
Sbjct: 947 IVRVELSQMQKKYYKFILTRNFEALNSKGGGNQV------------SLLNIMMDLKKCCN 994
Query: 1183 HPLL------NYPFFSDLSKD--FMVKCCGKLWMLDRILIKLQRTGHRVLLFSTMTKLLD 1234
HP L P + S D +VK GKL +L ++L KL+ GHRVL+FS MTK+LD
Sbjct: 995 HPYLFPVAAVEAPVLPNGSYDGSSLVKSSGKLMLLQKMLKKLRDEGHRVLIFSQMTKMLD 1054
Query: 1235 ILEEYLQWRRLVYRRIDGTTALEDRESAIVDFNSPNSDCFIFLLSIRAAGRGLNLQSADT 1294
+LE++L++ Y RIDG R+ AI FN+P + F FLLS RA G G+NL +ADT
Sbjct: 1055 LLEDFLEYEGYKYERIDGGITGGLRQEAIDRFNAPGAQQFCFLLSTRAGGLGINLATADT 1114
Query: 1295 VVIYDPDPNPKNEEQAVARAHRIGQKREVKV 1325
V+IYD D NP N+ QA +RAHRIGQ ++V +
Sbjct: 1115 VIIYDSDWNPHNDIQAFSRAHRIGQNKKVMI 1145
Score = 62.0 bits (149), Expect = 2e-08
Identities = 30/71 (42%), Positives = 43/71 (60%)
Query: 899 GTLREYQLVGLQWMLSLYNNKLNGILADEMGLGKTVQVMALIAYLMEFKGNYGPHLIIVP 958
GTL YQL GL W+ + + ILADEMGLGKTVQ + + L + + GP+L+ P
Sbjct: 698 GTLHPYQLEGLNWLRFSWAQGTDTILADEMGLGKTVQTIVFLYSLYKEGHSKGPYLVSAP 757
Query: 959 NAVLVNWKCAF 969
+ ++NW+ F
Sbjct: 758 LSTIINWEREF 768
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.130 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 246,107,924
Number of Sequences: 164201
Number of extensions: 10766078
Number of successful extensions: 31878
Number of sequences better than 10.0: 430
Number of HSP's better than 10.0 without gapping: 112
Number of HSP's successfully gapped in prelim test: 327
Number of HSP's that attempted gapping in prelim test: 29988
Number of HSP's gapped (non-prelim): 1586
length of query: 2139
length of database: 59,974,054
effective HSP length: 126
effective length of query: 2013
effective length of database: 39,284,728
effective search space: 79080157464
effective search space used: 79080157464
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 74 (33.1 bits)
Medicago: description of AC139601.1


