

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.9 - phase: 0 /pseudo
(145 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
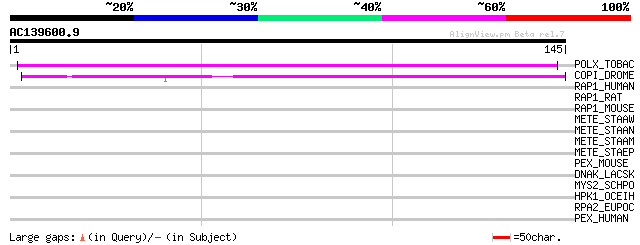
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 96 3e-20
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 50 2e-06
RAP1_HUMAN (Q15276) Rab GTPase binding effector protein 1 (Rabap... 34 0.12
RAP1_RAT (O35550) Rab GTPase binding effector protein 1 (Rabapti... 33 0.21
RAP1_MOUSE (O35551) Rab GTPase binding effector protein 1 (Rabap... 33 0.28
METE_STAAW (Q8NY94) 5-methyltetrahydropteroyltriglutamate--homoc... 32 0.36
METE_STAAN (P65343) 5-methyltetrahydropteroyltriglutamate--homoc... 32 0.36
METE_STAAM (P65342) 5-methyltetrahydropteroyltriglutamate--homoc... 32 0.36
METE_STAEP (Q8CMP5) 5-methyltetrahydropteroyltriglutamate--homoc... 30 1.4
PEX_MOUSE (P70669) Metalloendopeptidase homolog PEX (EC 3.4.24.-... 30 1.8
DNAK_LACSK (O87777) Chaperone protein dnaK (Heat shock protein 7... 29 3.0
MYS2_SCHPO (Q9USI6) Myosin type II heavy chain 1 28 6.8
HPK1_OCEIH (Q8ENK1) HPr kinase/phosphorylase 1 (EC 2.7.1.-) (EC ... 28 6.8
RPA2_EUPOC (P28365) DNA-directed RNA polymerase I second largest... 28 8.9
PEX_HUMAN (P78562) Phosphate regulating neutral endopeptidase (E... 28 8.9
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 95.9 bits (237), Expect = 3e-20
Identities = 51/141 (36%), Positives = 81/141 (57%)
Query: 3 GSKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSV 62
G K+++ KF GDN F + +M +LIQQ K L P TM + ++ ++A S
Sbjct: 3 GVKYEVAKFNGDNGFSTWQRRMRDLLIQQGLHKVLDVDSKKPDTMKAEDWADLDERAASA 62
Query: 63 IVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELL 122
I L L D V+ + E TA +W +LE+LYM+K+LT++ +LK+ LY+ M + + L
Sbjct: 63 IRLHLSDDVVNNIIDEDTARGIWTRLESLYMSKTLTNKLYLKKQLYALHMSEGTNFLSHL 122
Query: 123 VEFNKIIGDLENIEVRLEDVD 143
FN +I L N+ V++E+ D
Sbjct: 123 NVFNGLITQLANLGVKIEEED 143
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 49.7 bits (117), Expect = 2e-06
Identities = 36/144 (25%), Positives = 72/144 (50%), Gaps = 8/144 (5%)
Query: 4 SKWDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALKG--KGSLPVTMSRAEKTEMVDKARS 61
+K +I+ F G+ + + K ++ A+L +Q K + G + + +AE+ A+S
Sbjct: 4 AKRNIKPFDGEK-YAIWKFRIRALLAEQDVLKVVDGLMPNEVDDSWKKAERC-----AKS 57
Query: 62 VIVLCLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMEL 121
I+ L D L + TA + L+A+Y KSL Q L++ L S K+ +++
Sbjct: 58 TIIEYLSDSFLNFATSDITARQILENLDAVYERKSLASQLALRKRLLSLKLSSEMSLLSH 117
Query: 122 LVEFNKIIGDLENIEVRLEDVDAL 145
F+++I +L ++E++D +
Sbjct: 118 FHIFDELISELLAAGAKIEEMDKI 141
>RAP1_HUMAN (Q15276) Rab GTPase binding effector protein 1
(Rabaptin-5) (Rabaptin-5alpha) (Rabaptin-4)
Length = 862
Score = 33.9 bits (76), Expect = 0.12
Identities = 30/116 (25%), Positives = 55/116 (46%), Gaps = 5/116 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIVLCLGDKVLRKVAKESTAASM 84
E ++ QK +L+GK SL V++ +AE + D ++ L L K + TAA
Sbjct: 640 EELVRLQKDNDSLQGKHSLHVSLQQAEDFILPDTTEALRELVL--KYREDIINVRTAADH 697
Query: 85 WP---KLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEV 137
K E L++ + + +Q LK+ L ++ + E + + + +LE I+V
Sbjct: 698 VEEKLKAEILFLKEQIQAEQCLKENLEETLQLEIENCKEEIASISSLKAELERIKV 753
>RAP1_RAT (O35550) Rab GTPase binding effector protein 1
(Rabaptin-5) (Rabaptin-5alpha)
Length = 862
Score = 33.1 bits (74), Expect = 0.21
Identities = 30/116 (25%), Positives = 54/116 (45%), Gaps = 5/116 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIVLCLGDKVLRKVAKESTAASM 84
E ++ QK +L+GK SL V++ AE + D + + L L K + TAA
Sbjct: 640 EELVRLQKDNDSLQGKHSLHVSLQLAEDFILPDTVQVLRELVL--KYRENIVHVRTAADH 697
Query: 85 WP---KLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEV 137
K E L++ + + +Q LK+ L ++ + E + + + +LE I+V
Sbjct: 698 MEEKLKAEILFLKEQIQAEQCLKENLEETLQLEIENCKEEIASISSLKAELERIKV 753
>RAP1_MOUSE (O35551) Rab GTPase binding effector protein 1
(Rabaptin-5) (Rabaptin-5alpha)
Length = 862
Score = 32.7 bits (73), Expect = 0.28
Identities = 30/116 (25%), Positives = 53/116 (44%), Gaps = 5/116 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSVIVLCLGDKVLRKVAKESTAASM 84
E ++ QK +L+GK SL V++ AE + D + L L K + TAA
Sbjct: 640 EELVRLQKDNDSLQGKHSLHVSLQLAEDFILPDTVEVLRELVL--KYRENIVHVRTAADH 697
Query: 85 WP---KLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEV 137
K E L++ + + +Q LK+ L ++ + E + + + +LE I+V
Sbjct: 698 MEEKLKAEILFLKEQIQAEQCLKENLEETLQLEIENCKEEIASISSLKAELERIKV 753
>METE_STAAW (Q8NY94)
5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (EC 2.1.1.14) (Methionine synthase,
vitamin-B12 independent isozyme) (Cobalamin-independent
methionine synthase)
Length = 742
Score = 32.3 bits (72), Expect = 0.36
Identities = 34/125 (27%), Positives = 52/125 (41%), Gaps = 6/125 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSV----IVLCLGDKVLRKVAKEST 80
E + Q +K +KG + PVT+ E VD R V I L + ++VL A
Sbjct: 525 ETVYAQSLTDKPVKGMLTGPVTILNWS-FERVDLPRKVVQDQIALAINEEVLALEAAGIK 583
Query: 81 AASM-WPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEVRL 139
+ P L +S H+Q+LK + SFK+ S E + + I +
Sbjct: 584 VIQVDEPALREGLPLRSEYHEQYLKDAVLSFKLATSSVRDETQIHTHMCYSQFGQIIHAI 643
Query: 140 EDVDA 144
D+DA
Sbjct: 644 HDLDA 648
>METE_STAAN (P65343)
5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (EC 2.1.1.14) (Methionine synthase,
vitamin-B12 independent isozyme) (Cobalamin-independent
methionine synthase)
Length = 742
Score = 32.3 bits (72), Expect = 0.36
Identities = 34/125 (27%), Positives = 52/125 (41%), Gaps = 6/125 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSV----IVLCLGDKVLRKVAKEST 80
E + Q +K +KG + PVT+ E VD R V I L + ++VL A
Sbjct: 525 ETVYAQSLTDKPVKGMLTGPVTILNWS-FERVDLPRKVVQDQIALAINEEVLALEAAGIK 583
Query: 81 AASM-WPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEVRL 139
+ P L +S H+Q+LK + SFK+ S E + + I +
Sbjct: 584 VIQVDEPALREGLPLRSEYHEQYLKDAVLSFKLATSSVRDETQIHTHMCYSQFGQIIHAI 643
Query: 140 EDVDA 144
D+DA
Sbjct: 644 HDLDA 648
>METE_STAAM (P65342)
5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (EC 2.1.1.14) (Methionine synthase,
vitamin-B12 independent isozyme) (Cobalamin-independent
methionine synthase)
Length = 742
Score = 32.3 bits (72), Expect = 0.36
Identities = 34/125 (27%), Positives = 52/125 (41%), Gaps = 6/125 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSV----IVLCLGDKVLRKVAKEST 80
E + Q +K +KG + PVT+ E VD R V I L + ++VL A
Sbjct: 525 ETVYAQSLTDKPVKGMLTGPVTILNWS-FERVDLPRKVVQDQIALAINEEVLALEAAGIK 583
Query: 81 AASM-WPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEVRL 139
+ P L +S H+Q+LK + SFK+ S E + + I +
Sbjct: 584 VIQVDEPALREGLPLRSEYHEQYLKDAVLSFKLATSSVRDETQIHTHMCYSQFGQIIHAI 643
Query: 140 EDVDA 144
D+DA
Sbjct: 644 HDLDA 648
>METE_STAEP (Q8CMP5)
5-methyltetrahydropteroyltriglutamate--homocysteine
methyltransferase (EC 2.1.1.14) (Methionine synthase,
vitamin-B12 independent isozyme) (Cobalamin-independent
methionine synthase)
Length = 748
Score = 30.4 bits (67), Expect = 1.4
Identities = 33/125 (26%), Positives = 53/125 (42%), Gaps = 6/125 (4%)
Query: 25 EAMLIQQKCEKALKGKGSLPVTMSRAEKTEMVDKARSV----IVLCLGDKVLR-KVAKES 79
E + Q +K +KG + PVT+ E VD R V I L + ++VL + A
Sbjct: 525 ETVYAQSLTDKPVKGMLTGPVTILNWS-FERVDVPRKVVQDQIALAIDEEVLALEEAGIK 583
Query: 80 TAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFNKIIGDLENIEVRL 139
P L +S H+Q+L+ ++SFK+ S E + + I +
Sbjct: 584 VIQVDEPALREGLPLRSEYHEQYLEDAVHSFKLATSSVHDETQIHTHMCYSQFGQIIHAI 643
Query: 140 EDVDA 144
D+DA
Sbjct: 644 HDLDA 648
>PEX_MOUSE (P70669) Metalloendopeptidase homolog PEX (EC 3.4.24.-)
(Phosphate regulating neutral endopeptidase) (X-linked
hypophosphatemia protein) (HYP) (Vitamin D-resistant
hypophosphatemic rickets protein)
Length = 749
Score = 30.0 bits (66), Expect = 1.8
Identities = 19/60 (31%), Positives = 28/60 (46%), Gaps = 6/60 (10%)
Query: 37 LKGKGSLPVTMSRAEKTEMVDKARSVIVLCLGDKVLRKVAKEST------AASMWPKLEA 90
LK K L ++SR TE V KA+ + C+ +K + K + + WP LEA
Sbjct: 113 LKLKALLEKSVSRRRDTEAVQKAKILYSSCMNEKAIEKADAKPLLHILRHSPFRWPVLEA 172
>DNAK_LACSK (O87777) Chaperone protein dnaK (Heat shock protein 70)
(Heat shock 70 kDa protein) (HSP70)
Length = 613
Score = 29.3 bits (64), Expect = 3.0
Identities = 23/88 (26%), Positives = 36/88 (40%), Gaps = 24/88 (27%)
Query: 6 WDIEKFTGDNDFGLLKVKMEAMLIQQKCEKALK--------------------GKGSLPV 45
W +E F DN L K KM ++ EKA K G L +
Sbjct: 210 WLVENFKSDNGIDLSKDKMAMQRLKDAAEKAKKDLSGVSSTQISLPFISAGENGPLHLEM 269
Query: 46 TMSRAE----KTEMVDKARSVIVLCLGD 69
T+SR E +++VD+ ++ ++ L D
Sbjct: 270 TLSRTEFDRLTSDLVDRTKAPVMNALKD 297
>MYS2_SCHPO (Q9USI6) Myosin type II heavy chain 1
Length = 1526
Score = 28.1 bits (61), Expect = 6.8
Identities = 16/50 (32%), Positives = 31/50 (62%), Gaps = 4/50 (8%)
Query: 54 EMVDKARSVIVL-CLGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQF 102
++++KA + +L CL ++ + A ++T S KL+AL+ KSL ++ F
Sbjct: 506 DLIEKANPIGILSCLDEECVMPKATDATFTS---KLDALWRNKSLKYKPF 552
>HPK1_OCEIH (Q8ENK1) HPr kinase/phosphorylase 1 (EC 2.7.1.-) (EC
2.7.4.-) (HPrK/P 1) (HPr(Ser) kinase/phosphorylase 1)
Length = 310
Score = 28.1 bits (61), Expect = 6.8
Identities = 12/32 (37%), Positives = 23/32 (71%), Gaps = 1/32 (3%)
Query: 114 KSKAIMELLVEFNKIIGDLENIEVRLEDVDAL 145
KS+ +EL+ ++++ D +N+E+R ED D+L
Sbjct: 160 KSETALELVKRGHRLVAD-DNVEIRQEDYDSL 190
>RPA2_EUPOC (P28365) DNA-directed RNA polymerase I second largest
subunit (EC 2.7.7.6) (RNA polymerase I subunit 2)
Length = 1166
Score = 27.7 bits (60), Expect = 8.9
Identities = 13/79 (16%), Positives = 34/79 (42%), Gaps = 2/79 (2%)
Query: 67 LGDKVLRKVAKESTAASMWPKLEALYMTKSLTHQQFLKQ*LYSFKMVKSKAIMELLVEFN 126
+GDK + ++ + +WP+++ + +T + + +M I + +
Sbjct: 890 IGDKFSSRHGQKGVLSVLWPQVDMPFTENGITPDLIINPHAFPSRMTMGMLIQSMAAKSG 949
Query: 127 KIIGDLENIEV--RLEDVD 143
+ G+ + +E R +D D
Sbjct: 950 SLRGEFKTVETFQRYDDND 968
>PEX_HUMAN (P78562) Phosphate regulating neutral endopeptidase (EC
3.4.24.-) (Metalloendopeptidase homolog PEX) (X-linked
hypophosphatemia protein) (HYP) (Vitamin D-resistant
hypophosphatemic rickets protein)
Length = 749
Score = 27.7 bits (60), Expect = 8.9
Identities = 17/60 (28%), Positives = 28/60 (46%), Gaps = 6/60 (10%)
Query: 37 LKGKGSLPVTMSRAEKTEMVDKARSVIVLCLGDKVLRKVAKEST------AASMWPKLEA 90
LK K L ++SR TE + KA+ + C+ +K + K + + WP LE+
Sbjct: 113 LKLKELLEKSISRRRDTEAIQKAKILYSSCMNEKAIEKADAKPLLHILRHSPFRWPVLES 172
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.135 0.377
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,105,804
Number of Sequences: 164201
Number of extensions: 450788
Number of successful extensions: 1220
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 12
Number of HSP's that attempted gapping in prelim test: 1216
Number of HSP's gapped (non-prelim): 16
length of query: 145
length of database: 59,974,054
effective HSP length: 100
effective length of query: 45
effective length of database: 43,553,954
effective search space: 1959927930
effective search space used: 1959927930
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC139600.9


