

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC139600.7 - phase: 0
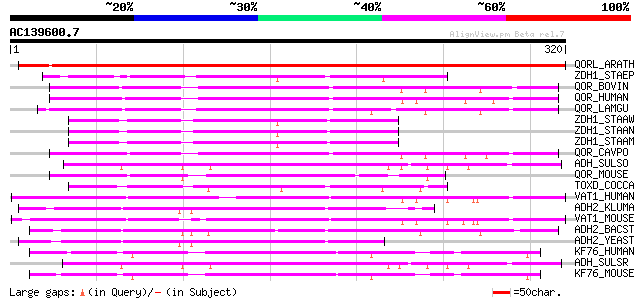
(320 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
QORL_ARATH (Q9ZUC1) Quinone oxidoreductase-like protein At1g2374... 407 e-113
ZDH1_STAEP (Q8CRJ7) Zinc-type alcohol dehydrogenase-like protein... 111 3e-24
QOR_BOVIN (O97764) Zeta-crystallin 107 5e-23
QOR_HUMAN (Q08257) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 102 2e-21
QOR_LAMGU (Q28452) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 100 8e-21
ZDH1_STAAW (Q8NVD1) Zinc-type alcohol dehydrogenase-like protein... 99 2e-20
ZDH1_STAAN (P99173) Zinc-type alcohol dehydrogenase-like protein... 98 3e-20
ZDH1_STAAM (P63475) Zinc-type alcohol dehydrogenase-like protein... 98 3e-20
QOR_CAVPO (P11415) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 96 1e-19
ADH_SULSO (P39462) NAD-dependent alcohol dehydrogenase (EC 1.1.1.1) 95 3e-19
QOR_MOUSE (P47199) Quinone oxidoreductase (EC 1.6.5.5) (NADPH:qu... 94 3e-19
TOXD_COCCA (P54006) TOXD protein 92 2e-18
VAT1_HUMAN (Q99536) Synaptic vesicle membrane protein VAT-1 homo... 91 3e-18
ADH2_KLUMA (Q9P4C2) Alcohol dehydrogenase II (EC 1.1.1.1) 91 4e-18
VAT1_MOUSE (Q62465) Synaptic vesicle membrane protein VAT-1 homo... 89 1e-17
ADH2_BACST (P42327) Alcohol dehydrogenase (EC 1.1.1.1) (ADH) 89 2e-17
ADH2_YEAST (P00331) Alcohol dehydrogenase II (EC 1.1.1.1) (YADH-2) 88 2e-17
KF76_HUMAN (Q9HCJ6) Probable oxidoreductase KIAA1576 (EC 1.-.-.-) 88 3e-17
ADH_SULSR (P50381) NAD-dependent alcohol dehydrogenase (EC 1.1.1.1) 87 4e-17
KF76_MOUSE (Q80TB8) Probable oxidoreductase KIAA1576 (EC 1.-.-.-) 87 6e-17
>QORL_ARATH (Q9ZUC1) Quinone oxidoreductase-like protein At1g23740,
chloroplast precursor (EC 1.-.-.-)
Length = 386
Score = 407 bits (1045), Expect = e-113
Identities = 203/315 (64%), Positives = 245/315 (77%), Gaps = 1/315 (0%)
Query: 6 SIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD 65
SIP KAWVYS YG ++ +LK +SN+ P KEDQVLIKVVAAALNPVD KR G FK
Sbjct: 73 SIPKEMKAWVYSDYGGVD-VLKLESNIVVPEIKEDQVLIKVVAAALNPVDAKRRQGKFKA 131
Query: 66 IDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEK 125
DSPLPTVPGYDVAGVVV VG VK K GDEVY +++E L PK G+L+EYT EEK
Sbjct: 132 TDSPLPTVPGYDVAGVVVKVGSAVKDLKEGDEVYANVSEKALEGPKQFGSLAEYTAVEEK 191
Query: 126 VLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVF 185
+LA KP N+ F +AA LPLAI TA +GL R EFS+GKS+LVL GAGGVGSLVIQLAKHV+
Sbjct: 192 LLALKPKNIDFAQAAGLPLAIETADEGLVRTEFSAGKSILVLNGAGGVGSLVIQLAKHVY 251
Query: 186 EASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAVKAANEG 245
AS++AATAST KL+ +R LGADLAIDYTKEN E+L +K+DVV+DA+G ++AVK EG
Sbjct: 252 GASKVAATASTEKLELVRSLGADLAIDYTKENIEDLPDKYDVVFDAIGMCDKAVKVIKEG 311
Query: 246 GKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYL 305
GKVV + TPP F++TS+G VL+KL PY+E+GKVKP++DPK PFPF + +AFSYL
Sbjct: 312 GKVVALTGAVTPPGFRFVVTSNGDVLKKLNPYIESGKVKPVVDPKGPFPFSRVADAFSYL 371
Query: 306 NTNRAIGKIVIHPIP 320
TN A GK+V++PIP
Sbjct: 372 ETNHATGKVVVYPIP 386
>ZDH1_STAEP (Q8CRJ7) Zinc-type alcohol dehydrogenase-like protein
SE1777
Length = 336
Score = 111 bits (277), Expect = 3e-24
Identities = 81/248 (32%), Positives = 126/248 (50%), Gaps = 31/248 (12%)
Query: 20 GNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVA 79
GN E FD PHP ++L+KV + ++NPVD K+ +D +P V G+D
Sbjct: 15 GNCFEEFNFD----IPHPSGHELLVKVQSISVNPVDTKQRT---MPVDK-VPRVLGFDAV 66
Query: 80 GVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEA 139
GV+ +G+QV F+ GD V+ +P G+ EY + EE ++A P+NL +A
Sbjct: 67 GVIEKIGDQVSMFQEGDVVFYS------GSPNQNGSNEEYQLIEEYLVAKAPTNLKSEQA 120
Query: 140 ASLPLAIITAYQGL-------ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAA 192
ASLPL +TAY+ L + + GKSLL++ GAGGVGS+ Q+AK F ++
Sbjct: 121 ASLPLTGLTAYETLFDVFGISKEPSENKGKSLLIINGAGGVGSIATQIAK--FYGLKVIT 178
Query: 193 TAS-TTKLDFLRKLGADLAIDYT-------KENYEELTEKFDVVYDAVGDSERAVKAANE 244
TAS + + +GAD+ +++ K+N+ E + +D E V
Sbjct: 179 TASREDTIKWSVNMGADVVLNHKKDLSQQFKDNHIEGVDYIFCTFDTDMYYEMMVNLVKP 238
Query: 245 GGKVVTIL 252
G + TI+
Sbjct: 239 RGHIATIV 246
>QOR_BOVIN (O97764) Zeta-crystallin
Length = 330
Score = 107 bits (266), Expect = 5e-23
Identities = 91/322 (28%), Positives = 157/322 (48%), Gaps = 43/322 (13%)
Query: 24 EILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVV 83
E+LK S+V P PK+ QVLIKV A +NPVD G +I LP PG+DVAG++
Sbjct: 20 EVLKLQSDVAVPIPKDHQVLIKVQACGVNPVDTYIRSGTH-NIKPLLPYTPGFDVAGIIE 78
Query: 84 SVGEQVKKFKVGDEVYGDINEITLHNPKTI-GTLSEYTVAEEKVLAHKPSNLSFVEAASL 142
+VGE V FK GD V+ +TI G +EY +A + + P L F + A++
Sbjct: 79 AVGESVSAFKKGDRVF---------TTRTISGGYAEYALAADHTVYTLPEKLDFKQGAAI 129
Query: 143 PLAIITAYQG-LERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTK-LD 200
+ TAY+ L G+S+LV G +GGVG Q+A+ ++ TAST +
Sbjct: 130 GIPYFTAYRALLYSAPVKPGESVLVHGASGGVGIAACQIARAY--GLKVLGTASTEEGQK 187
Query: 201 FLRKLGADLAIDYTKENYEELTEK------FDVVYDAVGDSERA--VKAANEGGKVVTIL 252
+ + GA ++ + NY + +K DV+ + + + + + + GG+V+ +
Sbjct: 188 IVLENGAHKVFNHKEANYIDKIKKSVGEKGVDVIIEMLANVNLSNDLNLLSHGGRVIVVG 247
Query: 253 PPGTPPAIP-FLLTSDGAV----------------LEKLQPYLENGKVKPILDPKSPFPF 295
GT P +T + ++ LQ +E G ++P++ P+ +
Sbjct: 248 SRGTIEINPRDTMTKESSIKGVTLFSSTKEEFQQFAAALQAGMEIGWLRPVIGPQ--YLL 305
Query: 296 YQTLEAF-SYLNTNRAIGKIVI 316
+ +A + ++++ A GK+++
Sbjct: 306 EKATQAHENIIHSSGATGKMIL 327
>QOR_HUMAN (Q08257) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 329
Score = 102 bits (253), Expect = 2e-21
Identities = 90/322 (27%), Positives = 153/322 (46%), Gaps = 43/322 (13%)
Query: 24 EILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVV 83
E+LK S++ P PK+ QVLIKV A +NPV+ G + LP PG DVAGV+
Sbjct: 20 EVLKLRSDIAVPIPKDHQVLIKVHACGVNPVETYIRSGTYSR-KPLLPYTPGSDVAGVIE 78
Query: 84 SVGEQVKKFKVGDEVYGDINEITLHNPKTI-GTLSEYTVAEEKVLAHKPSNLSFVEAASL 142
+VG+ FK GD V+ TI G +EY +A + + P L F + A++
Sbjct: 79 AVGDNASAFKKGDRVF---------TSSTISGGYAEYALAADHTVYKLPEKLDFKQGAAI 129
Query: 143 PLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTK-LD 200
+ TAY+ L +G+S+LV G +GGVG Q+A+ +I TA T +
Sbjct: 130 GIPYFTAYRALIHSACVKAGESVLVHGASGGVGLAACQIARAY--GLKILGTAGTEEGQK 187
Query: 201 FLRKLGADLAIDYTKENYEELTEKF------DVVYDAVG--DSERAVKAANEGGKVVTIL 252
+ + GA ++ + NY + +K+ D++ + + + + + + GG+V+ +
Sbjct: 188 IVLQNGAHEVFNHREVNYIDKIKKYVGEKGIDIIIEMLANVNLSKDLSLLSHGGRVIVVG 247
Query: 253 PPGTPPAIP----------FLLTSDGAVLEKLQPY-------LENGKVKPILDPKSPFPF 295
GT P +T + E+ Q Y +E G +KP++ S +P
Sbjct: 248 SRGTIEINPRDTMAKESSIIGVTLFSSTKEEFQQYAAALQAGMEIGWLKPVIG--SQYPL 305
Query: 296 YQTLEAF-SYLNTNRAIGKIVI 316
+ EA + ++ + A GK+++
Sbjct: 306 EKVAEAHENIIHGSGATGKMIL 327
>QOR_LAMGU (Q28452) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 330
Score = 99.8 bits (247), Expect = 8e-21
Identities = 93/330 (28%), Positives = 157/330 (47%), Gaps = 46/330 (13%)
Query: 17 SQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGY 76
S++G E+LK S+V P P+E QVLIKV A +NPVD G + LP PG
Sbjct: 14 SEFGG-PEVLKLQSDVAVPIPEEHQVLIKVQACGVNPVDTYIRSGTYSR-KPRLPYTPGL 71
Query: 77 DVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSF 136
DVAG++ +VGE+V FK GD V+ G +EY +A + + P L F
Sbjct: 72 DVAGLIEAVGERVSAFKKGDRVF--------TTSTVSGGYAEYALAADHTVYKLPGELDF 123
Query: 137 VEAASLPLAIITAYQG-LERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATAS 195
+ A++ + TAY+ L +G+S+LV G +GGVG Q+A+ ++ TA
Sbjct: 124 QKGAAIGVPYFTAYRALLHSACAKAGESVLVHGASGGVGLAACQIARAC--CFKVLGTAG 181
Query: 196 TTK-LDFLRKLGA--------DLAIDYTKENYEELTEKFDVVYDAVGDSERA--VKAANE 244
T + + + GA D+ ID K++ E + DV+ + + + + + ++
Sbjct: 182 TEEGQRVVLQNGAHEVFNHREDINIDKIKKSVGE--KGIDVIIEMLANVNLSNDLNLLSQ 239
Query: 245 GGKVVTILPPGTPPAIP-FLLTSDGAV----------------LEKLQPYLENGKVKPIL 287
GG+V+ + G P +T + ++ LQ +E G ++P++
Sbjct: 240 GGRVIIVGSKGPVEINPRDTMTKESSIKGVTLFSSTKEEFQQFAAALQAGMEIGWLRPVI 299
Query: 288 DPKSPFPFYQTLEAFSYL-NTNRAIGKIVI 316
S +P + +A L +++ A GK+V+
Sbjct: 300 G--SQYPLEKVAQAHEDLTHSSGAAGKVVL 327
>ZDH1_STAAW (Q8NVD1) Zinc-type alcohol dehydrogenase-like protein
MW2112
Length = 335
Score = 98.6 bits (244), Expect = 2e-20
Identities = 62/197 (31%), Positives = 101/197 (50%), Gaps = 18/197 (9%)
Query: 35 PHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKV 94
P P+ D +L+KV + ++NPVD K+ + P V G+D G V ++G V F
Sbjct: 26 PTPENDDILVKVNSISVNPVDTKQRQMEV----TQAPRVLGFDAIGTVEAIGPDVTLFSP 81
Query: 95 GDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGL- 153
GD V+ +P G+ + Y + E ++A P N+S EA SLPL ITAY+
Sbjct: 82 GDVVF------YAGSPNRQGSNATYQLVSEAIVAKAPHNISANEAVSLPLTGITAYETFF 135
Query: 154 ------ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGA 207
+ GKS+L++ GAGGVGS+ Q+AK + + I + ++ K+GA
Sbjct: 136 DTFKISHNPAENEGKSVLIINGAGGVGSIATQIAKR-YGLTVITTASRQETTEWCEKMGA 194
Query: 208 DLAIDYTKENYEELTEK 224
D+ +++ ++ + EK
Sbjct: 195 DIVLNHKEDLVRQFKEK 211
>ZDH1_STAAN (P99173) Zinc-type alcohol dehydrogenase-like protein
SA1988
Length = 335
Score = 97.8 bits (242), Expect = 3e-20
Identities = 62/197 (31%), Positives = 101/197 (50%), Gaps = 18/197 (9%)
Query: 35 PHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKV 94
P P+ D +L+KV + ++NPVD K+ + P V G+D G V ++G V F
Sbjct: 26 PTPENDDILVKVNSISVNPVDTKQRQMKV----TQAPRVLGFDAIGTVEAIGPDVTLFSP 81
Query: 95 GDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGL- 153
GD V+ +P G+ + Y + E ++A P N+S EA SLPL ITAY+
Sbjct: 82 GDVVF------YAGSPNRQGSNATYQLVSEAIVAKAPHNISANEAVSLPLTGITAYETFF 135
Query: 154 ------ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGA 207
+ GKS+L++ GAGGVGS+ Q+AK + + I + ++ K+GA
Sbjct: 136 DTFKISHNPSENVGKSVLIINGAGGVGSIATQIAKR-YGLTVITTASRQETTEWCEKMGA 194
Query: 208 DLAIDYTKENYEELTEK 224
D+ +++ ++ + EK
Sbjct: 195 DIVLNHKEDLVRQFKEK 211
>ZDH1_STAAM (P63475) Zinc-type alcohol dehydrogenase-like protein
SAV2186
Length = 335
Score = 97.8 bits (242), Expect = 3e-20
Identities = 62/197 (31%), Positives = 101/197 (50%), Gaps = 18/197 (9%)
Query: 35 PHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKV 94
P P+ D +L+KV + ++NPVD K+ + P V G+D G V ++G V F
Sbjct: 26 PTPENDDILVKVNSISVNPVDTKQRQMKV----TQAPRVLGFDAIGTVEAIGPDVTLFSP 81
Query: 95 GDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGL- 153
GD V+ +P G+ + Y + E ++A P N+S EA SLPL ITAY+
Sbjct: 82 GDVVF------YAGSPNRQGSNATYQLVSEAIVAKAPHNISANEAVSLPLTGITAYETFF 135
Query: 154 ------ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGA 207
+ GKS+L++ GAGGVGS+ Q+AK + + I + ++ K+GA
Sbjct: 136 DTFKISHNPSENVGKSVLIINGAGGVGSIATQIAKR-YGLTVITTASRQETTEWCEKMGA 194
Query: 208 DLAIDYTKENYEELTEK 224
D+ +++ ++ + EK
Sbjct: 195 DIVLNHKEDLVRQFKEK 211
>QOR_CAVPO (P11415) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 329
Score = 95.9 bits (237), Expect = 1e-19
Identities = 91/322 (28%), Positives = 152/322 (46%), Gaps = 43/322 (13%)
Query: 24 EILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVV 83
E+LK S+V P PK+ QVLIKV A +NPV+ G + I LP PG DVAGVV
Sbjct: 20 EVLKVQSDVAVPIPKDHQVLIKVHACGINPVETYIRSGTYTRIPL-LPYTPGTDVAGVVE 78
Query: 84 SVGEQVKKFKVGDEVYGDINEITLHNPKTI-GTLSEYTVAEEKVLAHKPSNLSFVEAASL 142
S+G V FK GD V+ TI G +EY +A + + P L F + A++
Sbjct: 79 SIGNDVSAFKKGDRVF---------TTSTISGGYAEYALASDHTVYRLPEKLDFRQGAAI 129
Query: 143 PLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTK-LD 200
+ TA + L +G+S+LV G +GGVG Q+A+ ++ TA T +
Sbjct: 130 GIPYFTACRALFHSARAKAGESVLVHGASGGVGLAACQIARAY--GLKVLGTAGTEEGQK 187
Query: 201 FLRKLGADLAIDYTKENYEELTEK------FDVVYDAVGDSERA--VKAANEGGKVVTIL 252
+ + GA ++ +Y + +K DV+ + + + + +K + GG+V+ +
Sbjct: 188 VVLQNGAHEVFNHRDAHYIDEIKKSIGEKGVDVIIEMLANVNLSNDLKLLSCGGRVIIVG 247
Query: 253 PPGTPPAIP-------------FLLTSDGAVLEK----LQPYLENGKVKPILDPKSPFPF 295
G+ P L +S ++ +Q +E G VKP++ S +P
Sbjct: 248 CRGSIEINPRDTMAKESTISGVSLFSSTKEEFQQFASTIQAGMELGWVKPVIG--SQYPL 305
Query: 296 YQTLEAF-SYLNTNRAIGKIVI 316
+ +A + ++++ +GK V+
Sbjct: 306 EKASQAHENIIHSSGTVGKTVL 327
>ADH_SULSO (P39462) NAD-dependent alcohol dehydrogenase (EC 1.1.1.1)
Length = 347
Score = 94.7 bits (234), Expect = 3e-19
Identities = 93/333 (27%), Positives = 149/333 (43%), Gaps = 49/333 (14%)
Query: 32 VPTPHPKEDQVLIKVVAAALNPVDIKRALGHF------KDIDSPLPTVPGYDVAGVVVSV 85
+ P PK QVLIKV AA + D+ G F +D+ LP G+++AG + V
Sbjct: 18 IGVPKPKGPQVLIKVEAAGVCHSDVHMRQGRFGNLRIVEDLGVKLPVTLGHEIAGKIEEV 77
Query: 86 GEQVKKFKVGDEV------------YGDINEITL-HNPKTIG-----TLSEYTVAEEKVL 127
G++V + GD V Y I E L +P+ +G +EY +
Sbjct: 78 GDEVVGYSKGDLVAVNPWQGEGNCYYCRIGEEHLCDSPRWLGINFDGAYAEYVIVPHYKY 137
Query: 128 AHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEA 187
+K L+ VEAA L + IT Y+ + + K+LLV+G GG+G++ +Q+AK V A
Sbjct: 138 MYKLRRLNAVEAAPLTCSGITTYRAVRKASLDPTKTLLVVGAGGGLGTMAVQIAKAVSGA 197
Query: 188 SRIAATASTTKLDFLRKLGADLAIDYTKEN----YEELTEK--FDVVYDAVGDSERAV-- 239
+ I ++ ++ GAD I+ + ++ +TE D V D + +SE+ +
Sbjct: 198 TIIGVDVREEAVEAAKRAGADYVINASMQDPLAEIRRITESKGVDAVID-LNNSEKTLSV 256
Query: 240 --KAANEGGKVVTI----------LPPGTPPAIPFL--LTSDGAVLEKLQPYLENGKVKP 285
KA + GK V + P T I F+ L + + + E GKVKP
Sbjct: 257 YPKALAKQGKYVMVGLFGADLHYHAPLITLSEIQFVGSLVGNQSDFLGIMRLAEAGKVKP 316
Query: 286 ILDPKSPFPFYQTLEAFSYLNTNRAIGKIVIHP 318
++ + EA L +AIG+ V+ P
Sbjct: 317 MI--TKTMKLEEANEAIDNLENFKAIGRQVLIP 347
>QOR_MOUSE (P47199) Quinone oxidoreductase (EC 1.6.5.5)
(NADPH:quinone reductase) (Zeta-crystallin)
Length = 331
Score = 94.4 bits (233), Expect = 3e-19
Identities = 76/248 (30%), Positives = 119/248 (47%), Gaps = 39/248 (15%)
Query: 24 EILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVV 83
E+LK S+V P P+ QVLIKV A +NPV+ G + + LP PG DVAG++
Sbjct: 20 EVLKLQSDVVVPVPQSHQVLIKVHACGVNPVETYIRSGAYSRKPA-LPYTPGSDVAGIIE 78
Query: 84 SVGEQVKKFKVGDEV--YGDINEITLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAAS 141
SVG++V FK GD V Y ++ G +E+ +A + + P L+F + A+
Sbjct: 79 SVGDKVSAFKKGDRVFCYSTVS----------GGYAEFALAADDTIYPLPETLNFRQGAA 128
Query: 142 LPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAK-HVFEASRIAATASTTKL 199
L + TA + L +G+S+LV G +GGVG Q+A+ H + A + KL
Sbjct: 129 LGIPYFTACRALFHSARARAGESVLVHGASGGVGLATCQIARAHGLKVLGTAGSEEGKKL 188
Query: 200 DFLRKLGADLAIDYTKENYEELTEKFDVVYDAVGDSERAV----------------KAAN 243
+ + GA ++ + NY D + +VGD ++ V K +
Sbjct: 189 --VLQNGAHEVFNHKEANY------IDKIKMSVGDKDKGVDVIIEMLANENLSNDLKLLS 240
Query: 244 EGGKVVTI 251
GG+VV +
Sbjct: 241 HGGRVVVV 248
>TOXD_COCCA (P54006) TOXD protein
Length = 297
Score = 92.0 bits (227), Expect = 2e-18
Identities = 78/242 (32%), Positives = 113/242 (46%), Gaps = 38/242 (15%)
Query: 35 PHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLPTVPGYDVAGVVVSVGEQVKK-FK 93
P ++D +L++ V+ ALNP D K L SP + G D AG+V VG VKK FK
Sbjct: 24 PKLRDDYILVRTVSVALNPTDWKHILRL-----SPPGCLVGCDYAGIVEEVGRSVKKPFK 78
Query: 94 VGDEVYGDINEITLHNPKTI----GTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITA 149
GD V G H + GT +E + + A P NLSF EAA+L + I T
Sbjct: 79 KGDRVCG-----FAHGGNAVFSDDGTFAEVITVKGDIQAWIPENLSFQEAATLGVGIKTV 133
Query: 150 YQGLER--------VEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDF 201
QGL + +L+ GG+ G+L IQLAK R+ T S +
Sbjct: 134 GQGLYQSLKLSWPTTPIEHAVPILIYGGSTATGTLAIQLAK--LSGYRVITTCSPHHFEL 191
Query: 202 LRKLGADLAIDY----TKENYEELTE-KFDVVYDAVGDS------ERAVKAANEGGKVVT 250
++ LGADL DY + ++ T+ K +V+D + +RA+ + EGG+
Sbjct: 192 MKSLGADLVFDYHEITSADHIRRCTQNKLKLVFDTISIDVSARFCDRAM--STEGGEYSA 249
Query: 251 IL 252
+L
Sbjct: 250 LL 251
>VAT1_HUMAN (Q99536) Synaptic vesicle membrane protein VAT-1 homolog
(EC 1.-.-.-)
Length = 393
Score = 91.3 bits (225), Expect = 3e-18
Identities = 94/364 (25%), Positives = 152/364 (40%), Gaps = 58/364 (15%)
Query: 2 ASTPSIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALG 61
A+ + P + V + +G +++ P P Q+ +++ A LN D+ G
Sbjct: 39 AAAAASPPLLRCLVLTGFGGYDKVKLQSRPAAPPAPGPGQLTLRLRACGLNFADLMARQG 98
Query: 62 HFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTV 121
+ + PLP PG + AGVV++VGE V K GD V T+ ++ + +
Sbjct: 99 LYDRLP-PLPVTPGMEGAGVVIAVGEGVSDRKAGDRVMVLNRSGMWQEEVTVPSVQTFLI 157
Query: 122 AEEKVLAHKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQL 180
P ++F EAA+L + ITAY L + G S+LV AGGVG +QL
Sbjct: 158 ---------PEAMTFEEAAALLVNYITAYMVLFDFGNLQPGHSVLVHMAAGGVGMAAVQL 208
Query: 181 AKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKF-----DVVYDAVG-- 233
+ V E + TAS +K + L++ G IDY +Y + +K D+V D +G
Sbjct: 209 CRTV-ENVTVFGTASASKHEALKENGVTHPIDYHTTDYVDEIKKISPKGVDIVMDPLGGS 267
Query: 234 DSERAVKAANEGGKVVTI----LPPGTPPAIPFLLTS----------------------- 266
D+ + GKVVT L G + L +
Sbjct: 268 DTAKGYNLLKPMGKVVTYGMANLLTGPKRNLMALARTWWNQFSVTALQLLQANRAVCGFH 327
Query: 267 ----DG------AVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
DG V+ +L G +KP +D S +PF + +A + + +GK+++
Sbjct: 328 LGYLDGEVELVSGVVARLLALYNQGHIKPHID--SVWPFEKVADAMKQMQEKKNVGKVLL 385
Query: 317 HPIP 320
P P
Sbjct: 386 VPGP 389
>ADH2_KLUMA (Q9P4C2) Alcohol dehydrogenase II (EC 1.1.1.1)
Length = 347
Score = 90.9 bits (224), Expect = 4e-18
Identities = 70/259 (27%), Positives = 118/259 (45%), Gaps = 40/259 (15%)
Query: 6 SIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKD 65
SIP+ K ++ + G + ++P P PK +++LI V + + D+ G +
Sbjct: 1 SIPTTQKGVIFYENGG----QLYYKDIPVPKPKSNELLINVKYSGVCHTDLHAWKGDWP- 55
Query: 66 IDSPLPTVPGYDVAGVVVSVGEQVKKFKVGD-----------------EVYGDIN--EIT 106
+D+ LP V G++ AGVVV++G+ VK +K+GD E+ + N +
Sbjct: 56 LDTKLPLVGGHEGAGVVVAMGDNVKGWKIGDLAGIKWLNGSCMNCEECELSNESNCPDAD 115
Query: 107 LHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLV 166
L G+ +Y A+ AH P+ + A + A +T Y+ L+ E +G + +
Sbjct: 116 LSGYTHDGSFQQYATADAVQAAHIPAGTDLAQVAPILCAGVTVYKALKTAEMKAGDWVAI 175
Query: 167 LGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKFD 226
G AGG+GSL +Q AK + K + + LG ++ ID+TK
Sbjct: 176 SGAAGGLGSLAVQYAK-AMGFRVLGIDGGEGKEELFKSLGGEVFIDFTKSK--------- 225
Query: 227 VVYDAVGDSERAVKAANEG 245
D VG+ +KA N G
Sbjct: 226 ---DIVGE---VIKATNGG 238
>VAT1_MOUSE (Q62465) Synaptic vesicle membrane protein VAT-1 homolog
(EC 1.-.-.-)
Length = 406
Score = 89.0 bits (219), Expect = 1e-17
Identities = 101/364 (27%), Positives = 154/364 (41%), Gaps = 62/364 (17%)
Query: 2 ASTPSIPSHTKAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALG 61
AS P + + V + +G +++ P P Q+ ++V A LN D+ G
Sbjct: 56 ASAPPL----RCLVLTGFGGYDKVKLQSRPAVPPAPGPGQLTLRVRACGLNFADLMGRQG 111
Query: 62 HFKDIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTV 121
+ + PLP PG + AGVVV+VGE V K GD V + L+ G E
Sbjct: 112 LYDRLP-PLPVTPGMEGAGVVVAVGEGVGDRKAGDRV------MVLNRS---GMWQEEVT 161
Query: 122 AEEKVLAHKPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQL 180
P ++F EAA+L + ITAY L + G S+LV AGGVG +QL
Sbjct: 162 VPSAQTFLMPEAMTFEEAAALLVNYITAYMVLFDFGNLRPGHSVLVHMAAGGVGMAALQL 221
Query: 181 AKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKENYEELTEKF-----DVVYDAVG-- 233
+ V E + TAS +K + L++ G IDY +Y + +K D+V D +G
Sbjct: 222 CRTV-ENVTVFGTASASKHEVLKENGVTHPIDYHTTDYVDEIKKISPKGVDIVMDPLGGS 280
Query: 234 DSERAVKAANEGGKVVTI----LPPGTPPAI---------PFLLTS-------------- 266
D+ + GKVVT L G + F +T+
Sbjct: 281 DTAKGYHLLKPMGKVVTYGMANLLTGPKRNLMAMARTWWNQFSVTALQLLQANRAVCGFH 340
Query: 267 ----DG------AVLEKLQPYLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
DG +V+ +L G +KP +D S +PF + +A + + IGK+++
Sbjct: 341 LGYLDGEVELVNSVVTRLVALYNQGHIKPRID--SVWPFEKVADAMKQMQEKKNIGKVLL 398
Query: 317 HPIP 320
P P
Sbjct: 399 VPGP 402
>ADH2_BACST (P42327) Alcohol dehydrogenase (EC 1.1.1.1) (ADH)
Length = 339
Score = 88.6 bits (218), Expect = 2e-17
Identities = 91/343 (26%), Positives = 150/343 (43%), Gaps = 48/343 (13%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSPLP 71
KA V +++ EI V P +E +VL+K+ A + D+ A G + I LP
Sbjct: 2 KAAVVNEFKKALEI----KEVERPKLEEGEVLVKIEACGVCHTDLHAAHGDWP-IKPKLP 56
Query: 72 TVPGYDVAGVVVSVGEQVKKFKVGDEV-----YGDIN--EITLHNPKTI----------- 113
+PG++ G+VV V + VK KVGD V Y E L +T+
Sbjct: 57 LIPGHEGVGIVVEVAKGVKSIKVGDRVGIPWLYSACGECEYCLTGQETLCPHQLNGGYSV 116
Query: 114 -GTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGG 172
G +EY A +A P NL VE A + A +T Y+ L +V + + + G GG
Sbjct: 117 DGGYAEYCKAPADYVAKIPDNLDPVEVAPILCAGVTTYKAL-KVSGARPGEWVAIYGIGG 175
Query: 173 VGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKEN-YEELTEKFDVVYDA 231
+G + +Q AK + +A S K + LGAD+AI+ KE+ + + ++ V+ A
Sbjct: 176 LGHIALQYAK-AMGLNVVAVDISDEKSKLAKDLGADIAINGLKEDPVKAIHDQVGGVHAA 234
Query: 232 VGDS------ERAVKAANEGGKVVTILPPGTPPAIPFLLTSDGAV------------LEK 273
+ + E+A ++ GG +V + P IP T V +++
Sbjct: 235 ISVAVNKKAFEQAYQSVKRGGTLVVVGLPNADLPIPIFDTVLNGVSVKGSIVGTRKDMQE 294
Query: 274 LQPYLENGKVKPILDPKSPFPFYQTLEAFSYLNTNRAIGKIVI 316
+ GKV+PI++ + E F + + G+IV+
Sbjct: 295 ALDFAARGKVRPIVETAE---LEEINEVFERMEKGKINGRIVL 334
>ADH2_YEAST (P00331) Alcohol dehydrogenase II (EC 1.1.1.1) (YADH-2)
Length = 347
Score = 88.2 bits (217), Expect = 2e-17
Identities = 69/231 (29%), Positives = 103/231 (43%), Gaps = 27/231 (11%)
Query: 6 SIPSHTKAWV-YSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFK 64
SIP KA + Y G +E ++P P PK +++LI V + + D+ G +
Sbjct: 1 SIPETQKAIIFYESNGKLEH-----KDIPVPKPKPNELLINVKYSGVCHTDLHAWHGDWP 55
Query: 65 DIDSPLPTVPGYDVAGVVVSVGEQVKKFKVGD-----------------EVYGDIN--EI 105
+ + LP V G++ AGVVV +GE VK +K+GD E+ + N
Sbjct: 56 -LPTKLPLVGGHEGAGVVVGMGENVKGWKIGDYAGIKWLNGSCMACEYCELGNESNCPHA 114
Query: 106 TLHNPKTIGTLSEYTVAEEKVLAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLL 165
L G+ EY A+ AH P E A + A IT Y+ L+ +G
Sbjct: 115 DLSGYTHDGSFQEYATADAVQAAHIPQGTDLAEVAPILCAGITVYKALKSANLRAGHWAA 174
Query: 166 VLGGAGGVGSLVIQLAKHVFEASRIAATASTTKLDFLRKLGADLAIDYTKE 216
+ G AGG+GSL +Q AK + K + LG ++ ID+TKE
Sbjct: 175 ISGAAGGLGSLAVQYAK-AMGYRVLGIDGGPGKEELFTSLGGEVFIDFTKE 224
>KF76_HUMAN (Q9HCJ6) Probable oxidoreductase KIAA1576 (EC 1.-.-.-)
Length = 419
Score = 87.8 bits (216), Expect = 3e-17
Identities = 84/305 (27%), Positives = 138/305 (44%), Gaps = 37/305 (12%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSP-- 69
+A V + +G + ++ F +P P++ ++ I+V A LN +D+ G+ ID+P
Sbjct: 43 RAVVLAGFGGLNKLRLFRKAMP--EPQDGELKIRVKACGLNFIDLMVRQGN---IDNPPK 97
Query: 70 LPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAH 129
P VPG++ +G+V ++G+ VK +++GD V +N +E + +
Sbjct: 98 TPLVPGFECSGIVEALGDSVKGYEIGDRVMAFVN---------YNAWAEVVCTPVEFVYK 148
Query: 130 KPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEAS 188
P ++SF EAA+ P+ +TAY L E G S+LV GGVG V QL V +
Sbjct: 149 IPDDMSFSEAAAFPMNFVTAYVMLFEVANLREGMSVLVHSAGGGVGQAVAQLCSTVPNVT 208
Query: 189 RIAATASTTKLDFLRKLGA---DLAIDYTKENYEELTEKFDVVYDAV-GDSERAVKAANE 244
+ TAST K + ++ D DY +E E D+V D + GD N
Sbjct: 209 -VFGTASTFKHEAIKDSVTHLFDRNADYVQEVKRISAEGVDIVLDCLCGD--------NT 259
Query: 245 GGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQ---TLEA 301
G + + P GT L S V + + + K ++ +P Y+ +
Sbjct: 260 GKGLSLLKPLGT----YILYGSSNMVTGETKSFFSFAKSWWQVEKVNPIKLYEENKVIAG 315
Query: 302 FSYLN 306
FS LN
Sbjct: 316 FSLLN 320
>ADH_SULSR (P50381) NAD-dependent alcohol dehydrogenase (EC 1.1.1.1)
Length = 347
Score = 87.4 bits (215), Expect = 4e-17
Identities = 88/334 (26%), Positives = 150/334 (44%), Gaps = 49/334 (14%)
Query: 31 NVPTPHPKEDQVLIKVVAAALNPVDIKRALGHF------KDIDSPLPTVPGYDVAGVVVS 84
++ P PK QVLIKV AA + D+ G F +D+ LP G+++AG +
Sbjct: 17 DIDIPKPKGAQVLIKVEAAGVCHSDVHMRQGRFGNLRIVEDLGVKLPVTLGHEIAGKIEE 76
Query: 85 VGEQVKKFKVGDEV------------YGDINEITL-HNPKTIG-----TLSEYTVAEEKV 126
+G++V + GD V Y I E L +P+ +G +EY +
Sbjct: 77 MGDEVVGYSKGDLVAVNPWQGEGNCYYCRIGEEHLCDSPRWLGINFDGAYAEYVLNPHYK 136
Query: 127 LAHKPSNLSFVEAASLPLAIITAYQGLERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFE 186
+K L+ VEA+ L + IT Y+ + + K+L+V+G GG+G++ +Q+AK V
Sbjct: 137 YMYKLRRLNAVEASPLTCSGITTYRAVRKASLDPTKTLVVVGAGGGLGTMAVQIAKAVGG 196
Query: 187 ASRIAATASTTKLDFLRKLGADLAIDYTKEN----YEELTE--KFDVVYDAVGDSERAV- 239
A+ I ++ ++ GAD I+ + ++ +TE D V D + +SE+ +
Sbjct: 197 ATIIGVDVREEAVETAKRAGADYVINASVQDPLAEIRRITEGKGVDAVID-LNNSEKTLS 255
Query: 240 ---KAANEGGKVVTI----------LPPGTPPAIPFL--LTSDGAVLEKLQPYLENGKVK 284
KA + GK + + P T I F+ L + + + E GKVK
Sbjct: 256 VYPKALAKQGKYIMVGLFGADLHFHAPLITLSEIQFVGSLVGNQSDFLGIMRLAEAGKVK 315
Query: 285 PILDPKSPFPFYQTLEAFSYLNTNRAIGKIVIHP 318
P++ + EA L +A+G+ V+ P
Sbjct: 316 PMI--TKTMKLEEANEAIDNLENFKAVGRQVLIP 347
>KF76_MOUSE (Q80TB8) Probable oxidoreductase KIAA1576 (EC 1.-.-.-)
Length = 417
Score = 87.0 bits (214), Expect = 6e-17
Identities = 84/305 (27%), Positives = 137/305 (44%), Gaps = 37/305 (12%)
Query: 12 KAWVYSQYGNIEEILKFDSNVPTPHPKEDQVLIKVVAAALNPVDIKRALGHFKDIDSP-- 69
+A V + +G + ++ S P P++ ++ I+V A LN +D+ G+ ID+P
Sbjct: 41 RAVVLAGFGGLNKLRL--SRKAMPEPQDGELKIRVKACGLNFIDLMVRQGN---IDNPPK 95
Query: 70 LPTVPGYDVAGVVVSVGEQVKKFKVGDEVYGDINEITLHNPKTIGTLSEYTVAEEKVLAH 129
P VPG++ +G+V ++G+ VK +++GD V +N +E + +
Sbjct: 96 TPLVPGFECSGIVEALGDSVKGYEIGDRVMAFVN---------YNAWAEVVCTPVEFVYK 146
Query: 130 KPSNLSFVEAASLPLAIITAYQGL-ERVEFSSGKSLLVLGGAGGVGSLVIQLAKHVFEAS 188
P ++SF EAA+ P+ +TAY L E G S+LV GGVG V QL V +
Sbjct: 147 IPDDMSFSEAAAFPMNFVTAYTMLFEIANLREGMSVLVHSAGGGVGQAVAQLCSTVPNVT 206
Query: 189 RIAATASTTKLDFLRKLGA---DLAIDYTKENYEELTEKFDVVYDAV-GDSERAVKAANE 244
+ TAST K + ++ D DY +E E D+V D + GD N
Sbjct: 207 -VFGTASTFKHEAIKDSVTHLFDRNADYVQEVKRISAEGVDIVLDCLCGD--------NT 257
Query: 245 GGKVVTILPPGTPPAIPFLLTSDGAVLEKLQPYLENGKVKPILDPKSPFPFYQ---TLEA 301
G + + P GT L S V + + + K ++ +P Y+ +
Sbjct: 258 GKGLSLLKPLGT----YILYGSSNMVTGETKSFFSFAKSWWQVEKVNPIKLYEENKVIAG 313
Query: 302 FSYLN 306
FS LN
Sbjct: 314 FSLLN 318
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.135 0.383
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 39,979,443
Number of Sequences: 164201
Number of extensions: 1779110
Number of successful extensions: 5022
Number of sequences better than 10.0: 296
Number of HSP's better than 10.0 without gapping: 196
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 4550
Number of HSP's gapped (non-prelim): 409
length of query: 320
length of database: 59,974,054
effective HSP length: 110
effective length of query: 210
effective length of database: 41,911,944
effective search space: 8801508240
effective search space used: 8801508240
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC139600.7


