

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138579.1 - phase: 0
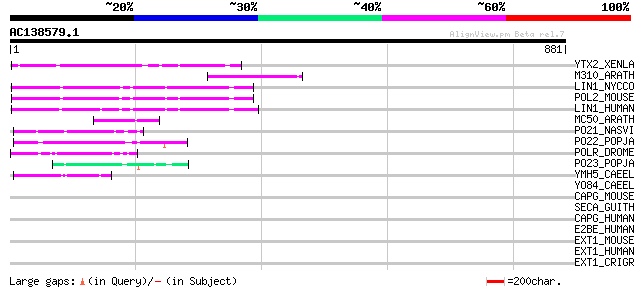
(881 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 123 2e-27
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 120 2e-26
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 110 1e-23
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 109 4e-23
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 104 1e-21
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 81 1e-14
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 57 2e-07
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 55 6e-07
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 49 4e-05
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 47 2e-04
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 46 5e-04
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 36 0.39
CAPG_MOUSE (P24452) Macrophage capping protein (Myc basic motif ... 33 2.5
SECA_GUITH (O78441) Preprotein translocase secA subunit 33 3.3
CAPG_HUMAN (P40121) Macrophage capping protein (Actin-regulatory... 33 3.3
E2BE_HUMAN (Q13144) Translation initiation factor eIF-2B epsilon... 32 5.7
EXT1_MOUSE (P97464) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225) (G... 32 7.4
EXT1_HUMAN (Q16394) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225) (G... 32 7.4
EXT1_CRIGR (Q9JK82) Exostosin-1 (EC 2.4.1.-) (Heparan sulfate co... 32 7.4
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 123 bits (309), Expect = 2e-27
Identities = 107/374 (28%), Positives = 178/374 (46%), Gaps = 33/374 (8%)
Query: 4 IALIPK-GDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSIL 62
++L+PK GD+++ +K+WRP++L + YKI AK ++ RLK VL + I +QS VPGR+I
Sbjct: 510 LSLLPKKGDLRL-IKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIF 568
Query: 63 DNAMAAIELVHYKKTKVKGTQGDVAF-KLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWI 121
DN +L+H+ + T +AF LD KA+DR+D YL + F Q+V ++
Sbjct: 569 DNVFLIRDLLHF----ARRTGLSLAFLSLDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYL 624
Query: 122 MMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTI-- 179
+ + V +N + P+ GRG+RQG PLS L+ + E L+RK +G +
Sbjct: 625 KTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRK-RLTGLVLK 683
Query: 180 -HGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFF 238
+++ +A +L A D RA+ + EV +AS IN+ KS
Sbjct: 684 EPDMRVVLSAYADDVILVAQDLVDLERAQECQ--------EVYAAASSARINWSKSSGLL 735
Query: 239 SRNGSQSKQLNIPNILQVQAVLGTGKYLGLPSMTGRSKKATFNFI--KDRIWKKINSWS- 295
GS P + KYLG+ ++ + NFI ++ + ++ W
Sbjct: 736 --EGSLKVDFLPPAFRDISWESKIIKYLGV-YLSAEEYPVSQNFIELEECVLTRLGKWKG 792
Query: 296 -SKCLSKAGREVLMKSMLQSIPTYFMSIFTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHW 354
+K LS GR +++ ++ S Y + + +I++ L F W G HW
Sbjct: 793 FAKVLSMRGRALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLW-------IGKHW 845
Query: 355 LSWDRLAAHKNDGG 368
+S + +GG
Sbjct: 846 VSAGVSSLPLKEGG 859
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 120 bits (301), Expect = 2e-26
Identities = 58/153 (37%), Positives = 89/153 (57%), Gaps = 3/153 (1%)
Query: 314 SIPTYFMSIFTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHWLSWDRLAAHK-NDGGMSFK 372
++P Y MS F L LC ++ + FWW S R + W++W +L K +DGG+ F+
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWS-SCENKRKISWVAWQKLCKSKEDDGGLGFR 60
Query: 373 SLPVFNLAMLGKQGWRSMTNPDTLIARIYKARYFPRCDFLQSSLGHNPSYVWRSLCNSKF 432
L FN A+L KQ +R + P TL++R+ ++RYFP ++ S+G PSY WRS+ + +
Sbjct: 61 DLGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGRE 120
Query: 433 ILKAGSRWRIGDGDTISIWNNKWIAGDITLIPP 465
+L G IGDG +W ++WI D T +PP
Sbjct: 121 LLSRGLLRTIGDGIHTKVWLDRWIM-DETPLPP 152
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 110 bits (276), Expect = 1e-23
Identities = 102/391 (26%), Positives = 174/391 (44%), Gaps = 21/391 (5%)
Query: 3 NIALIPKGDMQVSMKD-WRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSI 61
NI LIPK + K+ +RPI+L N+ KI K+L NR+++ + K I +Q F+PG
Sbjct: 512 NITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQG 571
Query: 62 LDNAMAAIELV-HYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKW 120
N +I ++ H K K K + +D KA+D I ++ + K+G ++K
Sbjct: 572 WFNIRKSINVIQHINKLKNK---DHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKL 628
Query: 121 IMMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIH 180
I ++I+N G RQG PLS LF I E L+ IR+ +A I
Sbjct: 629 IEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIREEKA---IK 685
Query: 181 GVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSR 240
G+ I + +S LFADD ++ + +++ + SG IN KS F
Sbjct: 686 GIHIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYT 743
Query: 241 NGSQSKQLNIPNILQVQAVLGTGKYLG--LPSMTGRSKKATFNFIKDRIWKKINSWSSKC 298
N +Q+++ + + + V KYLG L K + ++ I + +N W +
Sbjct: 744 NNNQAEK-TVKDSIPFTVVPKKMKYLGVYLTKDVKDLYKENYETLRKEIAEDVNKWKNIP 802
Query: 299 LSKAGREVLMKSMLQSIPTYFMSIFTL--PTSLCDEIEKMLNSFWWGHSGTQGRGLHWLS 356
S GR ++K + Y + + P S ++EK++ F W Q ++
Sbjct: 803 CSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWNQKKPQ------IA 856
Query: 357 WDRLAAHKNDGGMSFKSLPVFNLAMLGKQGW 387
L+ GG++ L ++ +++ K W
Sbjct: 857 KTLLSNKNKAGGITLPDLRLYYKSIVIKTAW 887
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 109 bits (272), Expect = 4e-23
Identities = 105/391 (26%), Positives = 171/391 (42%), Gaps = 23/391 (5%)
Query: 4 IALIPKGDMQ-VSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSIL 62
I LIPK ++++RPI+L N+ KI K+LANR++ + I +Q F+PG
Sbjct: 540 ITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQGW 599
Query: 63 DNAMAAIELVHY-KKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWI 121
N +I ++HY K K K + LD KA+D+I ++ V+ + G ++ I
Sbjct: 600 FNIRKSINVIHYINKLKDK---NHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMI 656
Query: 122 MMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHG 181
++ VN E I G RQG PLS YLF I E L+ IR+ + I G
Sbjct: 657 KAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIRQQK---EIKG 713
Query: 182 VKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSRN 241
++I +S L ADD ++ + + N++ G IN KS F
Sbjct: 714 IQIGKEEVKIS--LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTK 771
Query: 242 GSQSKQLNIPNILQVQAVLGTGKYLGLPSMTGRSK---KATFNFIKDRIWKKINSWSSKC 298
Q+++ I V KYLG+ ++T K F +K I + + W
Sbjct: 772 NKQAEK-EIRETTPFSIVTNNIKYLGV-TLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLP 829
Query: 299 LSKAGREVLMKSMLQSIPTY-FMSI-FTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHWLS 356
S GR ++K + Y F +I +PT +E+E + F W + + ++
Sbjct: 830 CSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKKPR------IA 883
Query: 357 WDRLAAHKNDGGMSFKSLPVFNLAMLGKQGW 387
L + GG++ L ++ A++ K W
Sbjct: 884 KSLLKDKRTSGGITMPDLKLYYRAIVIKTAW 914
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 104 bits (259), Expect = 1e-21
Identities = 104/400 (26%), Positives = 179/400 (44%), Gaps = 25/400 (6%)
Query: 3 NIALIPKGDMQVSMKD-WRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSI 61
+I LIPK + K+ +RPI+L N+ KI K+LAN++++ + K I +Q F+P
Sbjct: 512 SIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQG 571
Query: 62 LDNAMAAIELV-HYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKW 120
N +I ++ H +TK + +D KA+D+I ++ + K+G ++K
Sbjct: 572 WFNIRKSINIIQHINRTK---DTNHMIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKI 628
Query: 121 IMMCVETVDYSVIVN-EEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTI 179
I + ++I+N +++ P + G RQG PLS L I E L+ IR+ + I
Sbjct: 629 IRAIYDKPTANIILNGQKLEAPPLK-TGTRQGCPLSPLLPNIVLEVLARAIRQEK---EI 684
Query: 180 HGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFS 239
G+++ +S LFADD ++ A + ++ SG IN QKS+ F
Sbjct: 685 KGIQLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLY 742
Query: 240 RNGSQSKQLNIPNILQVQAVLGTGKYLGLPSMTGRSK---KATFNFIKDRIWKKINSWSS 296
N Q++ I + L KYLG+ +T K K + + + I + N W +
Sbjct: 743 TNNRQTES-QIMSELPFTIASKRIKYLGI-QLTRDVKDLFKENYKPLLNEIKEDTNKWKN 800
Query: 297 KCLSKAGR-EVLMKSMLQSIPTYFMSI-FTLPTSLCDEIEKMLNSFWWGHSGTQGRGLHW 354
S GR ++ ++L + F +I LP + E+EK F W
Sbjct: 801 IPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIWNQKRAH------ 854
Query: 355 LSWDRLAAHKNDGGMSFKSLPVFNLAMLGKQGWRSMTNPD 394
++ L+ GG++ ++ A + K W N D
Sbjct: 855 IAKSTLSQKNKAGGITLPDFKLYYKATVTKTAWYWYQNRD 894
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 80.9 bits (198), Expect = 1e-14
Identities = 43/106 (40%), Positives = 60/106 (56%), Gaps = 1/106 (0%)
Query: 133 IVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKICNNAPIVS 192
I+N G + RGLRQGDPLS YLFI+C E LS L R+A+ G + G+++ NN+P ++
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRIN 72
Query: 193 HLLFADDCFLFFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFF 238
HLLFADD R P A + G +N+ S ++F
Sbjct: 73 HLLFADDT-SSARWIPLAAQIWPIFFLSMRLFQGNPVNHPMSNLYF 117
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 57.0 bits (136), Expect = 2e-07
Identities = 52/207 (25%), Positives = 91/207 (43%), Gaps = 15/207 (7%)
Query: 6 LIPKGDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSILDNA 65
LIPK + +RP+++ +V + ++LANR+ + Q F+ + +N
Sbjct: 376 LIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRAFIVADGVAENT 433
Query: 66 MAAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMMCV 125
++ + K+KG + LD+ KA+D ++ + + + ++ +IM
Sbjct: 434 SLLSAMIKEARMKIKGLYIAI---LDVKKAFDSVEHRSILDALRRKKLPLEMRNYIMWVY 490
Query: 126 ETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKIC 185
+ V + I RG+RQGDPLS LF + + L R E +G + G
Sbjct: 491 RNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLFNCVMDAV--LRRLPENTGFLMG---- 544
Query: 186 NNAPIVSHLLFADDCFLFFRAEPSEAM 212
A + L+FADD L AE E +
Sbjct: 545 --AEKIGALVFADDLVLL--AETREGL 567
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 55.5 bits (132), Expect = 6e-07
Identities = 73/290 (25%), Positives = 131/290 (45%), Gaps = 31/290 (10%)
Query: 6 LIPK-GDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSILDN 64
LIPK GD++ + +WRPI + + + ++ ++LA RL+ ++ P G + +D
Sbjct: 63 LIPKDGDLE-NPSNWRPITIASALQRLLHRILAKRLEAAVE------LHPAQKGYARIDG 115
Query: 65 AMA-AIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMM 123
+ ++ L Y ++ + + LD+ KA+D + + + + ++G +I
Sbjct: 116 TLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITG 175
Query: 124 CVETVDYSVIVNE-EIAGPIIHGRGLRQGDPLSTYLF-IICAEGLSSLIRKAEASGTIHG 181
+ ++ V I RG++QGDPLS +LF + E L SL GTI
Sbjct: 176 SLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIGGTIGE 235
Query: 182 VKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNE--SASGQAINYQKS-EIFF 238
KI P+ L FADD L E ++ ++ + V G ++N +KS I
Sbjct: 236 EKI----PV---LAFADDLLLL---EDNDVLLPTTLATVANFFRLRGMSLNAKKSVSISV 285
Query: 239 SRNGS-----QSKQLNIPNI-LQVQAVLGTGKYLG-LPSMTGRSKKATFN 281
+ +G L + N+ L V + T +YLG ++G +K +N
Sbjct: 286 AASGGVCIPRTKPFLRVDNVLLPVTDRMQTFRYLGHFFGLSGAAKPTVYN 335
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 49.3 bits (116), Expect = 4e-05
Identities = 53/203 (26%), Positives = 89/203 (43%), Gaps = 13/203 (6%)
Query: 1 MTNIALIPKGDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRS 60
+ IPK +D+RPI++ +V+ + +LA RL ++ Q F+P
Sbjct: 397 LARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSINW--DPRQRGFLPTDG 454
Query: 61 ILDNAMAAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKW 120
DNA ++LV + K + LD+SKA+D + + + G +V +
Sbjct: 455 CADNA-TIVDLV--LRHSHKHFRSCYIANLDVSKAFDSLSHASIYDTLRAYGAPKGFVDY 511
Query: 121 IMMCVETVDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIH 180
+ E S+ + + + RG++QGDPLS LF + + L+R +
Sbjct: 512 VQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLV---MDRLLRTLPSE---I 565
Query: 181 GVKICNNAPIVSHLLFADDCFLF 203
G K+ N I + FADD LF
Sbjct: 566 GAKVGN--AITNAAAFADDLVLF 586
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 47.0 bits (110), Expect = 2e-04
Identities = 59/235 (25%), Positives = 93/235 (39%), Gaps = 37/235 (15%)
Query: 69 IELVHY-KKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMMCVET 127
I L HY K ++KG +V LDI KA+D + + M G +IM +
Sbjct: 7 IMLEHYIKLRRLKGKTYNVV-SLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITD 65
Query: 128 VDYSVIVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKICNN 187
+++V I G++QGDPLS LF I + L + + + ++
Sbjct: 66 AYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDEQPGASM------TP 119
Query: 188 APIVSHLLFADDCFL-----------------FFRAEPSEAMVMKNILEVNESASGQAIN 230
A ++ L FADD L +FR K + SG+++
Sbjct: 120 ACKIASLAFADDLLLLEDRDIDVPNSLATTCAYFRTRGMTLNPEKCASISAATVSGRSV- 178
Query: 231 YQKSEIFFSRNGSQSKQLNIPNILQVQAVLGTGKYLGLP-SMTGRSKKATFNFIK 284
+S+ F+ +G K L + T KYLGL S TG +K +N +
Sbjct: 179 -PRSKPSFTIDGRYIKPL---------GGINTFKYLGLTFSSTGVAKPTVYNLTR 223
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 45.8 bits (107), Expect = 5e-04
Identities = 40/158 (25%), Positives = 70/158 (43%), Gaps = 8/158 (5%)
Query: 7 IPKGDMQVSMKDWRPIALCNVVYKIAAKVLANRLKRVLDKCISDNQSPFVPGRSILDNAM 66
IPK S ++RPI+L + +I +++ +R++ +S +Q F+ RS + +
Sbjct: 673 IPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPSSLV 732
Query: 67 AAIELVHYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMMCVE 126
+I L H K D+ F D +KA+D++ L + G W +
Sbjct: 733 RSISLYHSILKNEKSL--DILF-FDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLH 789
Query: 127 TVDYSVIVNEEI---AGPIIHGRGLRQGDPLSTYLFII 161
+SV +N+ + A PI G+ QG LFI+
Sbjct: 790 LRTFSVKINKFVSSNAYPI--SSGVPQGSVSGPLLFIL 825
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 36.2 bits (82), Expect = 0.39
Identities = 24/96 (25%), Positives = 47/96 (48%), Gaps = 1/96 (1%)
Query: 73 HYKKTKVKGTQGDVAFKLDISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMMCVETVDYSV 132
++ K +G Q DV + LD SKA+D++ + L + + + ++W+ + + + V
Sbjct: 7 NWTKNIDEGAQTDVKY-LDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTNRSFKV 65
Query: 133 IVNEEIAGPIIHGRGLRQGDPLSTYLFIICAEGLSS 168
V ++ P G+ QG +S LF I +S+
Sbjct: 66 KVGNTLSEPKKTVCGVPQGSVISPVLFGIFVNEISA 101
>CAPG_MOUSE (P24452) Macrophage capping protein (Myc basic motif
homolog-1) (Actin-capping protein GCAP39)
Length = 352
Score = 33.5 bits (75), Expect = 2.5
Identities = 20/74 (27%), Positives = 40/74 (54%), Gaps = 3/74 (4%)
Query: 167 SSLIRKAEASGTIHGVKICNNAPIVSHLLFADDCFLFFRAEPSEAMVMKNILEVNESASG 226
++L + ++A+G ++ K+ +++P S LL DDCF+ ++ + K + NE
Sbjct: 253 AALYKVSDATGQMNLTKVADSSPFASELLIPDDCFVLDNGLCAQIYIWKG-RKANEKERQ 311
Query: 227 QAINYQKSEIFFSR 240
A+ Q ++ F SR
Sbjct: 312 AAL--QVADGFISR 323
>SECA_GUITH (O78441) Preprotein translocase secA subunit
Length = 877
Score = 33.1 bits (74), Expect = 3.3
Identities = 25/98 (25%), Positives = 44/98 (44%), Gaps = 5/98 (5%)
Query: 143 IHGRGLRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKICNNAPIVSHLLFADDCFL 202
+ GR RQGDP S+ F+ + L + + + + ++I + PI S LL +
Sbjct: 616 LRGRAGRQGDPGSSRFFLSLDDNLLRIFNGDKIAKIMDQLQIDEDLPIESKLLNSSLESA 675
Query: 203 FFRAEPSEAMVMKNILEVNESASGQAINYQKSEIFFSR 240
+ E K + + +E +NYQ+ I+F R
Sbjct: 676 QKKVEAYYYDARKQLFDYDE-----VLNYQRQAIYFER 708
>CAPG_HUMAN (P40121) Macrophage capping protein (Actin-regulatory
protein CAP-G)
Length = 348
Score = 33.1 bits (74), Expect = 3.3
Identities = 23/93 (24%), Positives = 45/93 (47%), Gaps = 3/93 (3%)
Query: 148 LRQGDPLSTYLFIICAEGLSSLIRKAEASGTIHGVKICNNAPIVSHLLFADDCFLFFRAE 207
L++G+P ++L + ++A+G ++ K+ +++P LL +DDCF+
Sbjct: 230 LKEGNPEEDLTADKANAQAAALYKVSDATGQMNLTKVADSSPFALELLISDDCFVLDNGL 289
Query: 208 PSEAMVMKNILEVNESASGQAINYQKSEIFFSR 240
+ + K + NE A+ Q +E F SR
Sbjct: 290 CGKIYIWKG-RKANEKERQAAL--QVAEGFISR 319
>E2BE_HUMAN (Q13144) Translation initiation factor eIF-2B epsilon
subunit (eIF-2B GDP-GTP exchange factor)
Length = 721
Score = 32.3 bits (72), Expect = 5.7
Identities = 32/121 (26%), Positives = 50/121 (40%), Gaps = 14/121 (11%)
Query: 38 NRLKRVLDKCIS------DNQSPFVPGRSILDNAMAAIELVHYKKTKVKGTQGDVAFKLD 91
+RL+R L+K +S SP P R DN + A++ + + TQG F
Sbjct: 168 HRLRRKLEKNVSVMTMIFKESSPSHPTRCHEDNVVVAVDSTTNRVLHFQKTQGLRRFAFP 227
Query: 92 ISKAYDRIDWNYLKCVMLKMGFSIQWVKWIMMCVETVDY--------SVIVNEEIAGPII 143
+S D ++ +L SI + + + DY ++VNEEI G I
Sbjct: 228 LSLFQGSSDGVEVRYDLLDCHISICSPQVAQLFTDNFDYQTRDDFVRGLLVNEEILGNQI 287
Query: 144 H 144
H
Sbjct: 288 H 288
>EXT1_MOUSE (P97464) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Multiple
exostoses protein 1 homolog)
Length = 746
Score = 32.0 bits (71), Expect = 7.4
Identities = 22/91 (24%), Positives = 35/91 (38%), Gaps = 6/91 (6%)
Query: 440 WRIGDGDTISIWNNKWIAGDITL---IPPTVGDFHLSHLRVSHCMLQHHKMWDVPFLETI 496
W + + I+ WN + GD L IP T+ H + Q +W+ F
Sbjct: 364 WELPFSEVIN-WNQAAVIGDERLLLQIPSTIRSIHQDKILALRQQTQF--LWEAYFSSVE 420
Query: 497 FDQQTVAAIIKTSLYTSIKEDRFVWNKENNG 527
T II+ ++ I + +WNK G
Sbjct: 421 KIVLTTLEIIQDRIFKHISRNSLIWNKHPGG 451
>EXT1_HUMAN (Q16394) Exostosin-1 (EC 2.4.1.224) (EC 2.4.1.225)
(Glucuronosyl-N-acetylglucosaminyl-proteoglycan/N-
acetylglucosaminyl-proteoglycan
4-alpha-N-acetylglucosaminyltransferase) (Putative tumor
suppressor protein EXT1) (Multiple exostoses protein
Length = 746
Score = 32.0 bits (71), Expect = 7.4
Identities = 22/91 (24%), Positives = 35/91 (38%), Gaps = 6/91 (6%)
Query: 440 WRIGDGDTISIWNNKWIAGDITL---IPPTVGDFHLSHLRVSHCMLQHHKMWDVPFLETI 496
W + + I+ WN + GD L IP T+ H + Q +W+ F
Sbjct: 364 WELPFSEVIN-WNQAAVIGDERLLLQIPSTIRSIHQDKILALRQQTQF--LWEAYFSSVE 420
Query: 497 FDQQTVAAIIKTSLYTSIKEDRFVWNKENNG 527
T II+ ++ I + +WNK G
Sbjct: 421 KIVLTTLEIIQDRIFKHISRNSLIWNKHPGG 451
>EXT1_CRIGR (Q9JK82) Exostosin-1 (EC 2.4.1.-) (Heparan sulfate
copolymerase) (Multiple exostoses protein 1 homolog)
Length = 746
Score = 32.0 bits (71), Expect = 7.4
Identities = 22/91 (24%), Positives = 35/91 (38%), Gaps = 6/91 (6%)
Query: 440 WRIGDGDTISIWNNKWIAGDITL---IPPTVGDFHLSHLRVSHCMLQHHKMWDVPFLETI 496
W + + I+ WN + GD L IP T+ H + Q +W+ F
Sbjct: 364 WELPFSEVIN-WNQAAVIGDERLLLQIPSTIRSIHQDKILALRQQTQF--LWEAYFSSVE 420
Query: 497 FDQQTVAAIIKTSLYTSIKEDRFVWNKENNG 527
T II+ ++ I + +WNK G
Sbjct: 421 KIVLTTLEIIQDRIFKHISRNSLIWNKHPGG 451
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.136 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 104,817,210
Number of Sequences: 164201
Number of extensions: 4350333
Number of successful extensions: 8741
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 8715
Number of HSP's gapped (non-prelim): 21
length of query: 881
length of database: 59,974,054
effective HSP length: 119
effective length of query: 762
effective length of database: 40,434,135
effective search space: 30810810870
effective search space used: 30810810870
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 70 (31.6 bits)
Medicago: description of AC138579.1


