

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138464.5 + phase: 0
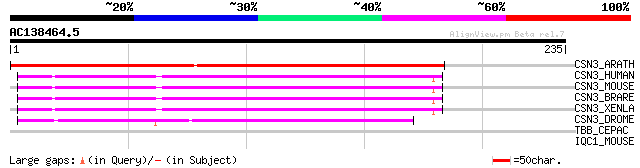
(235 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CSN3_ARATH (Q8W575) COP9 signalosome complex subunit 3 (Signalos... 182 5e-46
CSN3_HUMAN (Q9UNS2) COP9 signalosome complex subunit 3 (Signalos... 93 5e-19
CSN3_MOUSE (O88543) COP9 signalosome complex subunit 3 (Signalos... 93 6e-19
CSN3_BRARE (Q6P2U9) COP9 signalosome complex subunit 3 (Signalos... 92 1e-18
CSN3_XENLA (Q7ZTN8) COP9 signalosome complex subunit 3 (Signalos... 91 2e-18
CSN3_DROME (Q8SYG2) COP9 signalosome complex subunit 3 (Signalos... 64 2e-10
TBB_CEPAC (P41741) Tubulin beta chain (Beta tubulin) 30 3.8
IQC1_MOUSE (Q8BP00) IQ calmodulin-binding motif containing prote... 29 8.4
>CSN3_ARATH (Q8W575) COP9 signalosome complex subunit 3 (Signalosome
subunit 3) (FUSCA protein 11) (FUSCA11)
Length = 429
Score = 182 bits (463), Expect = 5e-46
Identities = 92/184 (50%), Positives = 126/184 (68%), Gaps = 1/184 (0%)
Query: 1 MDPLEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY 60
++ +E +I IQGLS S D++ LH +L+ + ++LR++ + L +D S HSLG+LY
Sbjct: 5 VNSVEAVITSIQGLSGSPEDLSALHDLLRGAQDSLRAEPGVNFSTLDQLDASKHSLGYLY 64
Query: 61 LLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPI 120
L+ + VS ++ A E +PIIARFIN+C QIRLA KFVS+CK LKD V L P+
Sbjct: 65 FLEVLTCGPVSKEKAAYE-IPIIARFINSCDAGQIRLASYKFVSLCKILKDHVIALGDPL 123
Query: 121 RGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLYLY 180
RGV P+ A++KLQVS++ LT LH + L LCL AK YK+G SIL DD+ E+D PRD +LY
Sbjct: 124 RGVGPLLNAVQKLQVSSKRLTALHPDVLQLCLQAKSYKSGFSILSDDIVEIDQPRDFFLY 183
Query: 181 CYYG 184
YYG
Sbjct: 184 SYYG 187
>CSN3_HUMAN (Q9UNS2) COP9 signalosome complex subunit 3 (Signalosome
subunit 3) (SGN3) (JAB1-containing signalosome subunit
3)
Length = 423
Score = 93.2 bits (230), Expect = 5e-19
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSIHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>CSN3_MOUSE (O88543) COP9 signalosome complex subunit 3 (Signalosome
subunit 3) (SGN3) (JAB1-containing signalosome subunit
3)
Length = 423
Score = 92.8 bits (229), Expect = 6e-19
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSVPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++ LT +HA+ LCLLAKC+K L LD D+ ++ Y
Sbjct: 122 GILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPALPYLDVDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>CSN3_BRARE (Q6P2U9) COP9 signalosome complex subunit 3 (Signalosome
subunit 3)
Length = 423
Score = 91.7 bits (226), Expect = 1e-18
Identities = 56/184 (30%), Positives = 94/184 (50%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LS+ + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNNVRQLSAQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDIQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ N E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPNIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ + A+ K+Q++ LT +HA+ LCLLAKC+K + L+ D+ ++ Y
Sbjct: 122 SILKQAIDKMQMNTNQLTSVHADLCQLCLLAKCFKPAVPFLELDMMDICKENGAYDAKHF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>CSN3_XENLA (Q7ZTN8) COP9 signalosome complex subunit 3 (Signalosome
subunit 3)
Length = 423
Score = 90.9 bits (224), Expect = 2e-18
Identities = 57/184 (30%), Positives = 93/184 (49%), Gaps = 7/184 (3%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLYLLD 63
LE + ++ LSS + +L ++ S E L + S L +L +D HSLG L +L
Sbjct: 5 LEQFVNSVRQLSSQG-QMTQLCELINKSGELLAKNLSHLDTVLGALDVQEHSLGVLAVL- 62
Query: 64 AFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRGV 123
F S+ + E + FI+ C+ E IR A + F +C +L + + + P+RG+
Sbjct: 63 -FVKFSMPSIPDFETLFSQVQLFISTCNGEHIRYATDTFAGLCHQLTNALVERKQPLRGI 121
Query: 124 APMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEVDHPRDLY----L 179
+ A+ K+Q++A LT +H + L LLAKC+K L+ LD D+ ++ Y
Sbjct: 122 CVIRQAIDKMQMNANQLTSIHGDLCQLSLLAKCFKPALAYLDVDMMDICKENGAYDAKPF 181
Query: 180 YCYY 183
CYY
Sbjct: 182 LCYY 185
>CSN3_DROME (Q8SYG2) COP9 signalosome complex subunit 3 (Signalosome
subunit 3) (Dch3)
Length = 445
Score = 64.3 bits (155), Expect = 2e-10
Identities = 49/169 (28%), Positives = 82/169 (47%), Gaps = 3/169 (1%)
Query: 4 LEPLIAQIQGLSSSSADIARLHTILKHSDETLRSDSSRLYPILQLIDPSIHSLGFLY-LL 62
LE + Q++ LS+S + L L S L + S L +L+ +D HSLG LY LL
Sbjct: 5 LENYVNQVRTLSASGS-YRELAEELPESLSLLARNWSILDNVLETLDMQQHSLGVLYVLL 63
Query: 63 DAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSVCKRLKDLVTLLEAPIRG 122
+AS +N + + + ++ F+ + EQ+R A F C + V I G
Sbjct: 64 AKLHSASTANPEPV-QLIQLMRDFVQRNNNEQLRYAVCAFYETCHLFTEFVVQKNLSILG 122
Query: 123 VAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILDDDVFEV 171
+ + A+ +++ LTP+HA+ +L L AK + L LD D+ ++
Sbjct: 123 IRIISRAIDQIRQLETQLTPIHADLCLLSLKAKNFSVVLPYLDADITDI 171
>TBB_CEPAC (P41741) Tubulin beta chain (Beta tubulin)
Length = 447
Score = 30.4 bits (67), Expect = 3.8
Identities = 16/40 (40%), Positives = 24/40 (60%), Gaps = 4/40 (10%)
Query: 24 LHTILKHSDETLRSDSSRLYPI----LQLIDPSIHSLGFL 59
+H +++HSDET D+ LY I L+L +PS L +L
Sbjct: 189 VHQLVEHSDETFCIDNEALYDICMRTLKLSNPSYGDLNYL 228
>IQC1_MOUSE (Q8BP00) IQ calmodulin-binding motif containing protein
1
Length = 598
Score = 29.3 bits (64), Expect = 8.4
Identities = 34/132 (25%), Positives = 55/132 (40%), Gaps = 14/132 (10%)
Query: 46 LQLIDPSIHSLGFLYLLDAFSAASVSNQQQAEEAVPIIARFINACSVEQIRLAPEKFVSV 105
+QLI + S FL+LL N Q + ++ + S +++ + S+
Sbjct: 157 VQLIQNVLQSDHFLHLLQT------DNVQIGASVMTLLQNILQINSGNLLKIEGKALHSI 210
Query: 106 CKRLKDLVTLLEAPIRGVAPMFTALRKLQVSAEHLTPLHAEFLMLCLLAKCYKTGLSILD 165
+ L LL P + TA + L V AE H E L+L L+ CYK S+L+
Sbjct: 211 LDEI--LFKLLSTPSPVIRS--TATKLLLVLAES----HQEILILLRLSACYKGLRSLLN 262
Query: 166 DDVFEVDHPRDL 177
+ R+L
Sbjct: 263 KQETLTEFSREL 274
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.329 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 25,667,111
Number of Sequences: 164201
Number of extensions: 1001189
Number of successful extensions: 2955
Number of sequences better than 10.0: 8
Number of HSP's better than 10.0 without gapping: 6
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 2947
Number of HSP's gapped (non-prelim): 9
length of query: 235
length of database: 59,974,054
effective HSP length: 107
effective length of query: 128
effective length of database: 42,404,547
effective search space: 5427782016
effective search space used: 5427782016
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC138464.5


