

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138199.3 - phase: 0
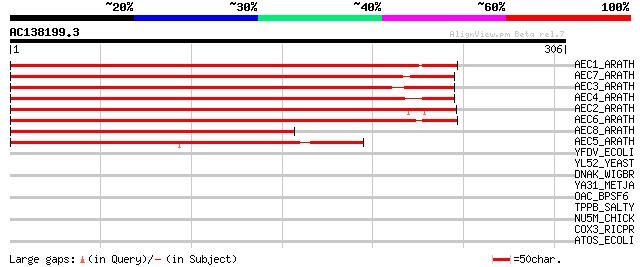
(306 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
AEC1_ARATH (Q9C6B8) Auxin efflux carrier component 1 (PIN-FORMED... 437 e-122
AEC7_ARATH (Q940Y5) Auxin efflux carrier component 7 (AtPIN7) 373 e-103
AEC3_ARATH (Q9S7Z8) Auxin efflux carrier component 3 (AtPIN3) 368 e-102
AEC4_ARATH (Q8RWZ6) Auxin efflux carrier component 4 (AtPIN4) 360 2e-99
AEC2_ARATH (Q9LU77) Auxin efflux carrier component 2 (AtPIN2) (A... 359 5e-99
AEC6_ARATH (Q9SQH6) Probable auxin efflux carrier component 6 (A... 294 2e-79
AEC8_ARATH (Q9FFD0) Putative auxin efflux carrier component 8 (A... 180 3e-45
AEC5_ARATH (Q9LFP6) Putative auxin efflux carrier component 5 (A... 173 4e-43
YFDV_ECOLI (P76519) Hypothetical protein yfdV 37 0.061
YL52_YEAST (P54072) Hypothetical 64.0 kDa protein in ACS2-MPT4 i... 35 0.23
DNAK_WIGBR (Q8D2Q5) Chaperone protein dnaK (Heat shock protein 7... 31 3.4
YA31_METJA (Q58437) Hypothetical protein MJ1031 30 5.7
OAC_BPSF6 (P23214) O-acetyl transferase (EC 2.3.1.-) (O-antigen ... 30 5.7
TPPB_SALTY (Q8ZPM6) Tripeptide permease tppB 30 7.5
NU5M_CHICK (P18940) NADH-ubiquinone oxidoreductase chain 5 (EC 1... 30 7.5
COX3_RICPR (Q9ZDX3) Probable cytochrome c oxidase polypeptide II... 30 7.5
ATOS_ECOLI (Q06067) Sensor protein atoS (EC 2.7.3.-) 30 7.5
>AEC1_ARATH (Q9C6B8) Auxin efflux carrier component 1 (PIN-FORMED
protein) (AtPIN1)
Length = 622
Score = 437 bits (1123), Expect = e-122
Identities = 216/247 (87%), Positives = 232/247 (93%), Gaps = 1/247 (0%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT DFYHVMTAMVPLYVAMILAYGSVKWWKIF+PDQCSGINRFVALFAVPLLSFHFIA
Sbjct: 1 MITAADFYHVMTAMVPLYVAMILAYGSVKWWKIFTPDQCSGINRFVALFAVPLLSFHFIA 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+NNPY MNLRFLAAD+LQK+I+L LL +W S+ G L+WTITLFSLSTLPNTLVMGIPL
Sbjct: 61 ANNPYAMNLRFLAADSLQKVIVLSLLFLWCKLSRNGSLDWTITLFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
LKGMYG FSG LMVQIVVLQCIIWYTLMLF+FE+RGA+LLI EQFPDTAGSIVSIHVDSD
Sbjct: 121 LKGMYGNFSGDLMVQIVVLQCIIWYTLMLFLFEYRGAKLLISEQFPDTAGSIVSIHVDSD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
+MSLDGR PLET+AEIKEDGKLH+TVR+SNASRSDIYSRRSQGLS+ TPRPSNLTNAEIY
Sbjct: 181 IMSLDGRQPLETEAEIKEDGKLHVTVRRSNASRSDIYSRRSQGLSA-TPRPSNLTNAEIY 239
Query: 241 SLQSSRN 247
SLQSSRN
Sbjct: 240 SLQSSRN 246
>AEC7_ARATH (Q940Y5) Auxin efflux carrier component 7 (AtPIN7)
Length = 619
Score = 373 bits (957), Expect = e-103
Identities = 184/245 (75%), Positives = 213/245 (86%), Gaps = 3/245 (1%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+TA++PLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITWHDLYTVLTAVIPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
SNNPY MNLRF+AADTLQKLI+L LL IW+NF++ G LEW+IT+FSLSTLPNTLVMGIPL
Sbjct: 61 SNNPYAMNLRFIAADTLQKLIMLTLLIIWANFTRSGSLEWSITIFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYGE+SGSLMVQIVVLQCIIWYTL+LF+FE+RGA++LI EQFP+T SIVS V+SD
Sbjct: 121 LIAMYGEYSGSLMVQIVVLQCIIWYTLLLFLFEYRGAKILIMEQFPETGASIVSFKVESD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDG LETDA+I +DGKLH+TVRKSNASR Y G ++ TPRPSNLT AEIY
Sbjct: 181 VVSLDGHDFLETDAQIGDDGKLHVTVRKSNASRRSFY---GGGGTNMTPRPSNLTGAEIY 237
Query: 241 SLQSS 245
SL ++
Sbjct: 238 SLNTT 242
>AEC3_ARATH (Q9S7Z8) Auxin efflux carrier component 3 (AtPIN3)
Length = 640
Score = 368 bits (945), Expect = e-102
Identities = 183/245 (74%), Positives = 213/245 (86%), Gaps = 6/245 (2%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MI+ D Y V+TA++PLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MISWHDLYTVLTAVIPLYVAMILAYGSVRWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+NNPY MNLRF+AADTLQK+I+L LL +W+NF++ G LEW+IT+FSLSTLPNTLVMGIPL
Sbjct: 61 TNNPYAMNLRFIAADTLQKIIMLSLLVLWANFTRSGSLEWSITIFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYGE+SGSLMVQIVVLQCIIWYTL+LF+FEFRGA++LI EQFP+TA SIVS V+SD
Sbjct: 121 LIAMYGEYSGSLMVQIVVLQCIIWYTLLLFLFEFRGAKMLIMEQFPETAASIVSFKVESD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDG LETDAEI +DGKLH+TVRKSNA SRRS + TPRPSNLT AEIY
Sbjct: 181 VVSLDGHDFLETDAEIGDDGKLHVTVRKSNA------SRRSFCGPNMTPRPSNLTGAEIY 234
Query: 241 SLQSS 245
SL ++
Sbjct: 235 SLSTT 239
>AEC4_ARATH (Q8RWZ6) Auxin efflux carrier component 4 (AtPIN4)
Length = 616
Score = 360 bits (924), Expect = 2e-99
Identities = 179/245 (73%), Positives = 206/245 (84%), Gaps = 9/245 (3%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+TA+VPLYVAMILAYGSV+WWKIFSPDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITWHDLYTVLTAVVPLYVAMILAYGSVQWWKIFSPDQCSGINRFVAIFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+N+PY MN RF+AADTLQK+I+L LLA+W+N +K G LEW IT+FSLSTLPNTLVMGIPL
Sbjct: 61 TNDPYAMNFRFVAADTLQKIIMLVLLALWANLTKNGSLEWMITIFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L MYG ++GSLMVQ+VVLQCIIWYTL+LF+FE+RGA+LLI EQFP+T SIVS V+SD
Sbjct: 121 LIAMYGTYAGSLMVQVVVLQCIIWYTLLLFLFEYRGAKLLIMEQFPETGASIVSFKVESD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
V+SLDG LETDAEI DGKLH+TVRKSNASR + TPRPSNLT AEIY
Sbjct: 181 VVSLDGHDFLETDAEIGNDGKLHVTVRKSNASRRSLMM---------TPRPSNLTGAEIY 231
Query: 241 SLQSS 245
SL S+
Sbjct: 232 SLSST 236
>AEC2_ARATH (Q9LU77) Auxin efflux carrier component 2 (AtPIN2)
(Auxin efflux carrier AGR) (Polar-auxin-transport efflux
component AGRAVITROPIC 1) (AtAGR1) (Ethylene insensitive
root 1) (AtEIR1) (WAVY6)
Length = 647
Score = 359 bits (921), Expect = 5e-99
Identities = 183/251 (72%), Positives = 211/251 (83%), Gaps = 5/251 (1%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT D Y V+ AMVPLYVAMILAYGSV+WW IF+PDQCSGINRFVA+FAVPLLSFHFI+
Sbjct: 1 MITGKDMYDVLAAMVPLYVAMILAYGSVRWWGIFTPDQCSGINRFVAVFAVPLLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
SN+PY MN FLAAD+LQK++IL L +W FS+RG LEW ITLFSLSTLPNTLVMGIPL
Sbjct: 61 SNDPYAMNYHFLAADSLQKVVILAALFLWQAFSRRGSLEWMITLFSLSTLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSD 180
L+ MYG+FSG+LMVQIVVLQ IIWYTLMLF+FEFRGA+LLI EQFP+TAGSI S VDSD
Sbjct: 121 LRAMYGDFSGNLMVQIVVLQSIIWYTLMLFLFEFRGAKLLISEQFPETAGSITSFRVDSD 180
Query: 181 VMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYS---RRSQGLSSN--TPRPSNLT 235
V+SL+GR PL+TDAEI +DGKLH+ VR+S+A+ S I S GL+S+ TPR SNLT
Sbjct: 181 VISLNGREPLQTDAEIGDDGKLHVVVRRSSAASSMISSFNKSHGGGLNSSMITPRASNLT 240
Query: 236 NAEIYSLQSSR 246
EIYS+QSSR
Sbjct: 241 GVEIYSVQSSR 251
>AEC6_ARATH (Q9SQH6) Probable auxin efflux carrier component 6
(AtPIN6)
Length = 570
Score = 294 bits (753), Expect = 2e-79
Identities = 148/248 (59%), Positives = 187/248 (74%), Gaps = 4/248 (1%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MIT +FY VM AM PLY AM +AYGSVKW KIF+P QCSGINRFV++FAVP+LSFHFI+
Sbjct: 1 MITGNEFYTVMCAMAPLYFAMFVAYGSVKWCKIFTPAQCSGINRFVSVFAVPVLSFHFIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
NNPYKM+ F+ ADTL K+ + LL++W+ F K G L+W ITLFS++TLPNTLVMGIPL
Sbjct: 61 QNNPYKMDTMFILADTLSKIFVFVLLSLWAVFFKAGGLDWLITLFSIATLPNTLVMGIPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFP-DTAGSIVSIHVDS 179
L+ MYG+++ +LMVQ+VVLQCIIWYTL+LF+FE R ARLLI +FP AGSI I VD
Sbjct: 121 LQAMYGDYTQTLMVQLVVLQCIIWYTLLLFLFELRAARLLIRAEFPGQAAGSIAKIQVDD 180
Query: 180 DVMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEI 239
DV+SLDG PL T+ E +G++ + +R+S +S D S L TPR SNL+NAEI
Sbjct: 181 DVISLDGMDPLRTETETDVNGRIRLRIRRSVSSVPDSVMSSSLCL---TPRASNLSNAEI 237
Query: 240 YSLQSSRN 247
+S+ + N
Sbjct: 238 FSVNTPNN 245
>AEC8_ARATH (Q9FFD0) Putative auxin efflux carrier component 8
(AtPIN8)
Length = 351
Score = 180 bits (457), Expect = 3e-45
Identities = 86/157 (54%), Positives = 111/157 (69%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MI D Y V+ AMVPLYVA+IL YGSVKWW IF+ DQC INR V F +PL + F A
Sbjct: 1 MINCGDVYKVIEAMVPLYVALILGYGSVKWWHIFTRDQCDAINRLVCYFTLPLFTIEFTA 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+P+ MN RF+AAD L K+II+ +LA+W+ +S +G W+IT FSL TL N+LV+G+PL
Sbjct: 61 HVDPFNMNYRFIAADVLSKVIIVTVLALWAKYSNKGSYCWSITSFSLCTLTNSLVVGVPL 120
Query: 121 LKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGA 157
K MYG+ + L+VQ V Q I+W TL+LF+ EFR A
Sbjct: 121 AKAMYGQQAVDLVVQSSVFQAIVWLTLLLFVLEFRKA 157
>AEC5_ARATH (Q9LFP6) Putative auxin efflux carrier component 5
(AtPIN5)
Length = 367
Score = 173 bits (439), Expect = 4e-43
Identities = 87/200 (43%), Positives = 131/200 (65%), Gaps = 10/200 (5%)
Query: 1 MITLTDFYHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIA 60
MI+ D YHV++A VPLYV+M L + S + K+FSP+QC+GIN+FVA F++PLLSF I+
Sbjct: 1 MISWLDIYHVVSATVPLYVSMTLGFLSARHLKLFSPEQCAGINKFVAKFSIPLLSFQIIS 60
Query: 61 SNNPYKMNLRFLAADTLQKLIILCLLAIWSNF-----SKRGCLEWTITLFSLSTLPNTLV 115
NNP+KM+ + + +D LQK +++ +LA+ F + G L W IT S+S LPNTL+
Sbjct: 61 ENNPFKMSPKLILSDILQKFLVVVVLAMVLRFWHPTGGRGGKLGWVITGLSISVLPNTLI 120
Query: 116 MGIPLLKGMYGEFSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSI 175
+G+P+L +YG+ + S++ QIVVLQ +IWYT++LF+FE AR L P + S+
Sbjct: 121 LGMPILSAIYGDEAASILEQIVVLQSLIWYTILLFLFELNAARAL-----PSSGASLEHT 175
Query: 176 HVDSDVMSLDGRTPLETDAE 195
D + +++ E D E
Sbjct: 176 GNDQEEANIEDEPKEEEDEE 195
>YFDV_ECOLI (P76519) Hypothetical protein yfdV
Length = 314
Score = 37.0 bits (84), Expect = 0.061
Identities = 35/132 (26%), Positives = 59/132 (44%), Gaps = 14/132 (10%)
Query: 14 MVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIASNNPYKMNLRFLA 73
++P+ V M+L Y S + + FS DQ N+ V +A+P F I + N +
Sbjct: 9 LLPIIVIMLLGYFSGRR-ETFSEDQARAFNKLVLNYALPAALFVSIT-----RANREMIF 62
Query: 74 ADTLQKLIILCLLAIWSNFSKRGCLEW-------TITLFSLSTLPNTLVMGIPLLKGMYG 126
ADT L+ L ++ FS GC ++ ++ P +G +L +YG
Sbjct: 63 ADTRLTLVSLVVIVGCFFFSWFGCYKFFKRTHAEAAVCALIAGSPTIGFLGFAVLDPIYG 122
Query: 127 E-FSGSLMVQIV 137
+ S L+V I+
Sbjct: 123 DSVSTGLVVAII 134
>YL52_YEAST (P54072) Hypothetical 64.0 kDa protein in ACS2-MPT4
intergenic region
Length = 576
Score = 35.0 bits (79), Expect = 0.23
Identities = 53/238 (22%), Positives = 99/238 (41%), Gaps = 16/238 (6%)
Query: 8 YHVMTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIASNNPYKM 67
Y + + +Y M++ Y K+ I S + GI+ V +P L+F+ I SN ++
Sbjct: 10 YIALKPIFKIYTIMLVGYLVAKF-DIVSMENAKGISNMVVNAILPCLTFNKIVSNISWR- 67
Query: 68 NLRFLAADTLQKLIILCL---LAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPLLKGM 124
+++ + L I+ L A+++ F+ ++ L PN + I ++ M
Sbjct: 68 DIKEIGVIILSAFILFVLGATGALFTTFATTVPKKFFWGLIFAGFFPNISDLPIAYIQSM 127
Query: 125 YGE--FSGSLMVQIVVLQCIIWYTLMLFMFEFRGARLLIFEQFPDTAGSIVSIHVDSDVM 182
F+ + V CI + M F G ++ F DT DS+ +
Sbjct: 128 GNGSIFTAEEADKGVAYSCIFLFIQSFLMMNF-GMWRVVGLDFRDTK------EPDSENI 180
Query: 183 SLDGRTPLETDAEIKEDGKLHITVRKSNASRSDIYSRRSQGLSSNTPRPSNLTNAEIY 240
+ +P D ++ E KL R +NA +S+ +R + LS N+ + +T Y
Sbjct: 181 T-PSVSPAIDDRKLTEITKLPNITRPTNAYQSE-DARSNSDLSCNSITTNEMTPQAFY 236
>DNAK_WIGBR (Q8D2Q5) Chaperone protein dnaK (Heat shock protein 70)
(Heat shock 70 kDa protein) (HSP70)
Length = 645
Score = 31.2 bits (69), Expect = 3.4
Identities = 17/58 (29%), Positives = 29/58 (49%), Gaps = 1/58 (1%)
Query: 171 SIVSIHVDSDVMSLDGRTPLETDAEIKEDGKLHITVRKSNASRSD-IYSRRSQGLSSN 227
S+ ++D ++ G +E +I DG LH++ + N+ R I + S GLS N
Sbjct: 457 SLGQFNLDGIAPAMRGMPQIEVTFDIDADGILHVSAKDKNSGREQKITIKASSGLSEN 514
>YA31_METJA (Q58437) Hypothetical protein MJ1031
Length = 308
Score = 30.4 bits (67), Expect = 5.7
Identities = 33/130 (25%), Positives = 53/130 (40%), Gaps = 24/130 (18%)
Query: 11 MTAMVPLYVAMILAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIASNNPYKMNLR 70
M ++ + + +++ Y S K + I + +N V A+P F I+ N L
Sbjct: 4 MDVVLIVLILVLVGYFS-KIFGILKEEHAKILNNIVIYIAMPSTIFLTISKNVSSSQILE 62
Query: 71 FLAADTLQKLIILCLLAIWSNFSKRGCLEWTIT-------------LFSLSTLPNTLVMG 117
FL L +I LC L + G L + + L +S L NT +G
Sbjct: 63 FLK---LPVVIFLCCLFV-------GILAYLLGKHIFKLKDEKLGGLILVSMLGNTGFLG 112
Query: 118 IPLLKGMYGE 127
P+ GM+GE
Sbjct: 113 YPVALGMFGE 122
>OAC_BPSF6 (P23214) O-acetyl transferase (EC 2.3.1.-) (O-antigen
acetylase)
Length = 333
Score = 30.4 bits (67), Expect = 5.7
Identities = 30/109 (27%), Positives = 54/109 (49%), Gaps = 18/109 (16%)
Query: 48 LFAVPL-------LSFHFIASNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEW 100
+F++PL L+F F A+ Y+ + K+ ++ LLA+++ S +++
Sbjct: 188 MFSIPLWLYPLRGLAFFFGATMAMYEKSWNVSNV----KITVVSLLAMYAYASYGKGIDY 243
Query: 101 TIT---LFSLSTLPNTLVMGIPLLKG----MYGEFSGSLMVQIVVLQCI 142
T+T L S ST+ +G PL+KG YG + + VQ VV+ +
Sbjct: 244 TMTCYILVSFSTIAICTSVGDPLVKGRFDYSYGVYIYAFPVQQVVINTL 292
>TPPB_SALTY (Q8ZPM6) Tripeptide permease tppB
Length = 501
Score = 30.0 bits (66), Expect = 7.5
Identities = 20/89 (22%), Positives = 40/89 (44%), Gaps = 2/89 (2%)
Query: 34 FSPDQCSGINRFVALFAVPLLSFHFIASNNPYKMNLRFLAADTL--QKLIILCLLAIWSN 91
F P+Q +N F + P+L+ + + M ++F L +IL L A ++N
Sbjct: 316 FEPEQYQALNPFWIIIGSPILAAIYNRMGDTLPMPMKFAIGMVLCSGAFLILPLGAKFAN 375
Query: 92 FSKRGCLEWTITLFSLSTLPNTLVMGIPL 120
+ + W I + L ++ ++ G+ L
Sbjct: 376 DAGIVSVNWLIASYGLQSIGELMISGLGL 404
>NU5M_CHICK (P18940) NADH-ubiquinone oxidoreductase chain 5 (EC
1.6.5.3)
Length = 605
Score = 30.0 bits (66), Expect = 7.5
Identities = 29/106 (27%), Positives = 47/106 (43%), Gaps = 7/106 (6%)
Query: 23 LAYGSVKWWKIFSPDQCSGINRFVALFAVPLLSFHFIASNNPYKMNLRFLAADTLQKLII 82
L +K FS G+ +P L+F I+++ +K L FL + +I
Sbjct: 293 LTQNDIKKIIAFSTSSQLGLMMVTIGLDLPQLAFLHISTHAFFKAML-FLCSG-----LI 346
Query: 83 LCLLAIWSNFSKRGCLEWTITLF-SLSTLPNTLVMGIPLLKGMYGE 127
+ L + K GCL+ T+ + S T+ N +MG P L G Y +
Sbjct: 347 IHSLNGEQDIRKMGCLQKTLPMTTSCLTIGNLALMGTPFLAGFYSK 392
>COX3_RICPR (Q9ZDX3) Probable cytochrome c oxidase polypeptide III
(EC 1.9.3.1) (Cytochrome AA3 subunit 3)
Length = 278
Score = 30.0 bits (66), Expect = 7.5
Identities = 17/51 (33%), Positives = 28/51 (54%), Gaps = 2/51 (3%)
Query: 49 FAVPLLSFHFIASNNPYKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLE 99
FA+ LL I+S + YK N+ L+A + +I CL + W + + G +E
Sbjct: 28 FALLLLVIGGISSMHGYKFNMYILSAGVIS--VIYCLYSWWRDVVQEGIVE 76
>ATOS_ECOLI (Q06067) Sensor protein atoS (EC 2.7.3.-)
Length = 608
Score = 30.0 bits (66), Expect = 7.5
Identities = 22/68 (32%), Positives = 33/68 (48%), Gaps = 3/68 (4%)
Query: 65 YKMNLRFLAADTLQKLIILCLLAIWSNFSKRGCLEWTITLFSLSTLPNTLVMGIPLLKGM 124
+KM++R + T LI L L+ + FS+R I LSTL + +P L G
Sbjct: 186 WKMDVRIIIVLTAGLLISLLLIVL---FSRRLSANIDIITDGLSTLAQNIPTRLPQLPGE 242
Query: 125 YGEFSGSL 132
G+ S S+
Sbjct: 243 MGQISQSV 250
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 29,765,056
Number of Sequences: 164201
Number of extensions: 1054974
Number of successful extensions: 3322
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 3298
Number of HSP's gapped (non-prelim): 22
length of query: 306
length of database: 59,974,054
effective HSP length: 110
effective length of query: 196
effective length of database: 41,911,944
effective search space: 8214741024
effective search space used: 8214741024
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC138199.3


