

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.8 + phase: 0 /pseudo
(751 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
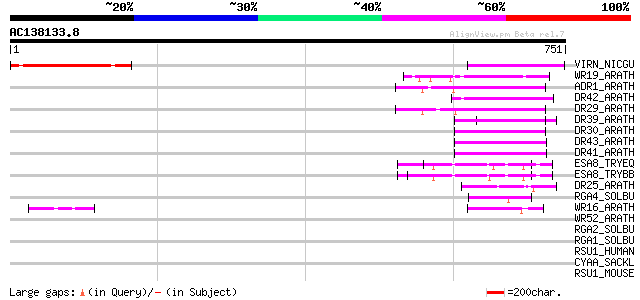
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 149 2e-35
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 86 4e-16
ADR1_ARATH (Q9FW44) Disease resistance protein ADR1 (Activated d... 64 1e-09
DR42_ARATH (Q9FKZ1) Probable disease resistance protein At5g66900 62 4e-09
DR29_ARATH (Q9SZA7) Probable disease resistance protein At4g33300 61 1e-08
DR39_ARATH (Q9LVT1) Putative disease resistance protein At5g47280 59 5e-08
DR30_ARATH (Q9LZ25) Probable disease resistance protein At5g04720 57 1e-07
DR43_ARATH (Q9FKZ0) Probable disease resistance protein At5g66910 53 3e-06
DR41_ARATH (Q9FKZ2) Probable disease resistance protein At5g66890 53 3e-06
ESA8_TRYEQ (P26337) Putative adenylate cyclase regulatory protein 51 1e-05
ESA8_TRYBB (P23799) Putative adenylate cyclase regulatory protei... 51 1e-05
DR25_ARATH (O50052) Putative disease resistance protein At4g19050 49 5e-05
RGA4_SOLBU (Q7XA39) Putative disease resistance protein RGA4 (RG... 47 1e-04
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 46 4e-04
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 45 0.001
RGA2_SOLBU (Q7XBQ9) Disease resistance protein RGA2 (RGA2-blb) (... 43 0.003
RGA1_SOLBU (Q7XA42) Putative disease resistance protein RGA1 (RG... 42 0.005
RSU1_HUMAN (Q15404) Ras suppressor protein 1 (Rsu-1) (RSP-1) 40 0.030
CYAA_SACKL (P23466) Adenylate cyclase (EC 4.6.1.1) (ATP pyrophos... 40 0.030
RSU1_MOUSE (Q01730) Ras suppressor protein 1 (Rsu-1) (RSP-1) 39 0.050
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 149 bits (377), Expect = 2e-35
Identities = 83/164 (50%), Positives = 109/164 (65%), Gaps = 7/164 (4%)
Query: 1 MQSPSSSTSISYDYKYQVFLSFRGSDTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEIT 60
M S SSS+ SYD VFLSFRG DTR FT +LY+ L DKGI TF DD L+ G I
Sbjct: 1 MASSSSSSRWSYD----VFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIP 56
Query: 61 PSLLKAIEESRIFIPVFSINYASSSFCLDELDHIIHCYKTKGRPVLPVFFGVDPSHVRHH 120
L KAIEES+ I VFS NYA+S +CL+EL I+ C + V+P+F+ VDPSHVR+
Sbjct: 57 GELCKAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQ 116
Query: 121 KGSYGEALAEHEKRFQNDPKNMERLQGWKDALSQAANLSGYHDS 164
K S+ +A EHE ++++D +E +Q W+ AL++AANL G D+
Sbjct: 117 KESFAKAFEEHETKYKDD---VEGIQRWRIALNEAANLKGSCDN 157
Score = 73.6 bits (179), Expect = 2e-12
Identities = 47/133 (35%), Positives = 75/133 (56%), Gaps = 2/133 (1%)
Query: 620 LTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLK 679
LT PD +G+ NLE ++ C NL +H+S+G +K+ L C++LKRFP + + SL+
Sbjct: 632 LTRTPDFTGMPNLEYVNLYQCSNLEEVHHSLGCCSKVIGLYLNDCKSLKRFPCVNVESLE 691
Query: 680 ELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSS-FQNLSELDELSVREF-GMLR 737
L L C SL+ P++ +M +I + IRELPSS FQ + + +L + ++
Sbjct: 692 YLGLRSCDSLEKLPEIYGRMKPEIQIHMQGSGIRELPSSIFQYKTHVTKLLLWNMKNLVA 751
Query: 738 FPKHNDRMYSIVS 750
P R+ S+VS
Sbjct: 752 LPSSICRLKSLVS 764
Score = 39.7 bits (91), Expect = 0.030
Identities = 38/152 (25%), Positives = 65/152 (42%), Gaps = 16/152 (10%)
Query: 603 AKKFQDMTILILDHCEYLTH-IPD-VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLS 660
A+ + L L +C + +P+ + LS+L+KL N + +SI L L+ L
Sbjct: 829 AEGLHSLEYLNLSYCNLIDGGLPEEIGSLSSLKKLDLSR-NNFEHLPSSIAQLGALQSLD 887
Query: 661 AFGCRTLKRFPPLGLASLKELKLSCCYSLKSFPKLLCKMTNIDKI------------WFW 708
C+ L + P L L EL + C +LK L+ K + ++ F
Sbjct: 888 LKDCQRLTQLPELP-PELNELHVDCHMALKFIHYLVTKRKKLHRVKLDDAHNDTMYNLFA 946
Query: 709 YTSIRELPSSFQNLSELDELSVREFGMLRFPK 740
YT + + S ++S D LS+ F +P+
Sbjct: 947 YTMFQNISSMRHDISASDSLSLTVFTGQPYPE 978
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 85.9 bits (211), Expect = 4e-16
Identities = 71/218 (32%), Positives = 112/218 (50%), Gaps = 31/218 (14%)
Query: 534 FKKMTRLKTLIIENGHCSK-----------GLKYLRSSLKALKWE----GCLSKSLSSSI 578
F+KM L+ L + +CSK GL+YL S L+ L WE L KS +
Sbjct: 1172 FEKMCNLRLLKL---YCSKAEEKHGVSFPQGLEYLPSKLRLLHWEYYPLSSLPKSFNPEN 1228
Query: 579 LSKASEIKSFSNCIYL*-----CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLE 633
L + + S + ++ CT S S +K + M L + + LT IP +S +NLE
Sbjct: 1229 LVELNLPSSCAKKLWKGKKARFCTTNS-SLEKLKKMR---LSYSDQLTKIPRLSSATNLE 1284
Query: 634 KLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPL-GLASLKELKLSCCYSLKSF 692
+ E C +L+++ SI +L KL L+ GC L+ P + L SL+ L LS C L +F
Sbjct: 1285 HIDLEGCNSLLSLSQSISYLKKLVFLNLKGCSKLENIPSMVDLESLEVLNLSGCSKLGNF 1344
Query: 693 PKLLCKMTNIDKIWFWYTSIRELPSSFQNLSELDELSV 730
P++ N+ +++ T I+E+PSS +NL L++L +
Sbjct: 1345 PEI---SPNVKELYMGGTMIQEIPSSIKNLVLLEKLDL 1379
>ADR1_ARATH (Q9FW44) Disease resistance protein ADR1 (Activated
disease resistance protein 1)
Length = 787
Score = 64.3 bits (155), Expect = 1e-09
Identities = 57/217 (26%), Positives = 97/217 (44%), Gaps = 17/217 (7%)
Query: 523 FKRKHYKKGKAFKKMTRLKTLIIENGHCSKGLKY---LRSSLKALK--WEGCLSKSLSSS 577
F +Y KM+RL+ L+I N S + + ++L L+ W L +
Sbjct: 535 FSSDNYVLPPFIGKMSRLRVLVIINNGMSPARLHGFSIFANLAKLRSLW---LKRVHVPE 591
Query: 578 ILSKASEIKSFSNCIYL*CTM------ISFSAKK-FQDMTILILDHCEYLTHIPDVSGLS 630
+ S +K+ + C + SF K F ++ L +DHC+ L + + G++
Sbjct: 592 LTSCTIPLKNLHKIHLIFCKVKNSFVQTSFDISKIFPSLSDLTIDHCDDLLELKSIFGIT 651
Query: 631 NLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFP--PLGLASLKELKLSCCYS 688
+L LS C ++ + ++ ++ LERL + C L P L LK + +S C S
Sbjct: 652 SLNSLSITNCPRILELPKNLSNVQSLERLRLYACPELISLPVEVCELPCLKYVDISQCVS 711
Query: 689 LKSFPKLLCKMTNIDKIWFWYTSIRELPSSFQNLSEL 725
L S P+ K+ +++KI S+ LPSS L L
Sbjct: 712 LVSLPEKFGKLGSLEKIDMRECSLLGLPSSVAALVSL 748
>DR42_ARATH (Q9FKZ1) Probable disease resistance protein At5g66900
Length = 809
Score = 62.4 bits (150), Expect = 4e-09
Identities = 45/142 (31%), Positives = 72/142 (50%), Gaps = 6/142 (4%)
Query: 598 MISFSAKKFQDMTILILDHCEYLTHIPD-VSGLSNLEKLSFEYCKNLITIHNSIGHLNKL 656
++S + K Q++ I D+C L +P +S + +L+ LS C L + +IG+L++L
Sbjct: 643 VVSNALSKLQEIDI---DYCYDLDELPYWISEIVSLKTLSITNCNKLSQLPEAIGNLSRL 699
Query: 657 ERLSAFGCRTLKRFPPL--GLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRE 714
E L L P GL++L+ L +S C L+ P+ + K+ N+ KI S E
Sbjct: 700 EVLRLCSSMNLSELPEATEGLSNLRFLDISHCLGLRKLPQEIGKLQNLKKISMRKCSGCE 759
Query: 715 LPSSFQNLSELDELSVREFGML 736
LP S NL L+ E G+L
Sbjct: 760 LPESVTNLENLEVKCDEETGLL 781
>DR29_ARATH (Q9SZA7) Probable disease resistance protein At4g33300
Length = 855
Score = 60.8 bits (146), Expect = 1e-08
Identities = 62/219 (28%), Positives = 93/219 (42%), Gaps = 20/219 (9%)
Query: 523 FKRKHYKKGKAFKKMTRLKTLIIENGHCSKGLKY------LRSSLKALKWEGCLSKSLSS 576
F Y KM+RLK L+I N S + + S L++L E LS+
Sbjct: 602 FSSDKYVLPPFISKMSRLKVLVIINNGMSPAVLHDFSIFAHLSKLRSLWLERVHVPQLSN 661
Query: 577 SILSKASEIKSFSNCIYL*CTMI-SFS------AKKFQDMTILILDHCEYLTHIPD-VSG 628
S + +K+ + C + SF A F + L +DHC+ L +P + G
Sbjct: 662 S----TTPLKNLHKMSLILCKINKSFDQTGLDVADIFPKLGDLTIDHCDDLVALPSSICG 717
Query: 629 LSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPP--LGLASLKELKLSCC 686
L++L LS C L + ++ L LE L + C LK P L LK L +S C
Sbjct: 718 LTSLSCLSITNCPRLGELPKNLSKLQALEILRLYACPELKTLPGEICELPGLKYLDISQC 777
Query: 687 YSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSFQNLSEL 725
SL P+ + K+ ++KI + PSS +L L
Sbjct: 778 VSLSCLPEEIGKLKKLEKIDMRECCFSDRPSSAVSLKSL 816
>DR39_ARATH (Q9LVT1) Putative disease resistance protein At5g47280
Length = 623
Score = 58.9 bits (141), Expect = 5e-08
Identities = 36/126 (28%), Positives = 64/126 (50%), Gaps = 3/126 (2%)
Query: 603 AKKFQDMTILILDHCEYLTHIPD-VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSA 661
A+ F +T + +D+C+ L +P + G+++L +S C N+ + +I L L+ L
Sbjct: 459 AQIFPKLTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISKLQALQLLRL 518
Query: 662 FGCRTLKRFPP--LGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSF 719
+ C LK P L L + +S C SL S P+ + + ++KI S+ +PSS
Sbjct: 519 YACPELKSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMRECSLSSIPSSA 578
Query: 720 QNLSEL 725
+L+ L
Sbjct: 579 VSLTSL 584
Score = 47.0 bits (110), Expect = 2e-04
Identities = 28/111 (25%), Positives = 54/111 (48%), Gaps = 3/111 (2%)
Query: 632 LEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPP--LGLASLKELKLSCCYSL 689
L ++ +YC +L + ++I + L +S C +K P L +L+ L+L C L
Sbjct: 465 LTDITIDYCDDLAELPSTICGITSLNSISITNCPNIKELPKNISKLQALQLLRLYACPEL 524
Query: 690 KSFPKLLCKMTNIDKIWFWYT-SIRELPSSFQNLSELDELSVREFGMLRFP 739
KS P +C++ + + + S+ LP N+ L+++ +RE + P
Sbjct: 525 KSLPVEICELPRLVYVDISHCLSLSSLPEKIGNVRTLEKIDMRECSLSSIP 575
>DR30_ARATH (Q9LZ25) Probable disease resistance protein At5g04720
Length = 811
Score = 57.4 bits (137), Expect = 1e-07
Identities = 35/126 (27%), Positives = 63/126 (49%), Gaps = 3/126 (2%)
Query: 603 AKKFQDMTILILDHCEYLTHIPD-VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSA 661
A+ F ++ L +DHC+ L +P + G+++L +S C + + ++ L L+ L
Sbjct: 647 AQIFPKLSDLTIDHCDDLLELPSTICGITSLNSISITNCPRIKELPKNLSKLKALQLLRL 706
Query: 662 FGCRTLKRFPP--LGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSF 719
+ C L P L LK + +S C SL S P+ + K+ ++KI S+ +P+S
Sbjct: 707 YACHELNSLPVEICELPRLKYVDISQCVSLSSLPEKIGKVKTLEKIDTRECSLSSIPNSV 766
Query: 720 QNLSEL 725
L+ L
Sbjct: 767 VLLTSL 772
Score = 41.6 bits (96), Expect = 0.008
Identities = 34/153 (22%), Positives = 61/153 (39%), Gaps = 35/153 (22%)
Query: 622 HIPDVSG----LSNLEKLSFEYCK----------------------------NLITIHNS 649
H+P++S L NL KLS +CK +L+ + ++
Sbjct: 611 HVPELSSSTVPLQNLHKLSLIFCKINTSLDQTELDIAQIFPKLSDLTIDHCDDLLELPST 670
Query: 650 IGHLNKLERLSAFGCRTLKRFPP--LGLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWF 707
I + L +S C +K P L +L+ L+L C+ L S P +C++ + +
Sbjct: 671 ICGITSLNSISITNCPRIKELPKNLSKLKALQLLRLYACHELNSLPVEICELPRLKYVDI 730
Query: 708 -WYTSIRELPSSFQNLSELDELSVREFGMLRFP 739
S+ LP + L+++ RE + P
Sbjct: 731 SQCVSLSSLPEKIGKVKTLEKIDTRECSLSSIP 763
>DR43_ARATH (Q9FKZ0) Probable disease resistance protein At5g66910
Length = 815
Score = 53.1 bits (126), Expect = 3e-06
Identities = 35/127 (27%), Positives = 63/127 (49%), Gaps = 3/127 (2%)
Query: 603 AKKFQDMTILILDHCEYLTHIPD-VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSA 661
+K ++ + +D+C L +P + + +L+ LS C L + +IG+L++LE L
Sbjct: 651 SKALSNLQEIDIDYCYDLDELPYWIPEVVSLKTLSITNCNKLSQLPEAIGNLSRLEVLRM 710
Query: 662 FGCRTLKRFPPLG--LASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSF 719
C L P L++L+ L +S C L+ P+ + K+ ++ I S ELP S
Sbjct: 711 CSCMNLSELPEATERLSNLRSLDISHCLGLRKLPQEIGKLQKLENISMRKCSGCELPDSV 770
Query: 720 QNLSELD 726
+ L L+
Sbjct: 771 RYLENLE 777
Score = 36.2 bits (82), Expect = 0.33
Identities = 27/100 (27%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query: 587 SFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSG-LSNLEKLSFEYCKNLIT 645
S +NC L + + + + +L + C L+ +P+ + LSNL L +C L
Sbjct: 685 SITNCNKL--SQLPEAIGNLSRLEVLRMCSCMNLSELPEATERLSNLRSLDISHCLGLRK 742
Query: 646 IHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSC 685
+ IG L KLE +S C + P + L+ L++ C
Sbjct: 743 LPQEIGKLQKLENISMRKCSGCEL--PDSVRYLENLEVKC 780
>DR41_ARATH (Q9FKZ2) Probable disease resistance protein At5g66890
Length = 415
Score = 53.1 bits (126), Expect = 3e-06
Identities = 38/127 (29%), Positives = 63/127 (48%), Gaps = 3/127 (2%)
Query: 603 AKKFQDMTILILDHCEYLTHIPD-VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSA 661
++ Q + + +D+C L +P +S + +L+KLS C L + +IG L LE L
Sbjct: 251 SETLQSLQEIEIDYCYNLDELPYWISQVVSLKKLSVTNCNKLCRVIEAIGDLRDLETLRL 310
Query: 662 FGCRTLKRFPPL--GLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSF 719
C +L P L +L+ L +S + LK+ P + K+ ++KI ELP S
Sbjct: 311 SSCASLLELPETIDRLDNLRFLDVSGGFQLKNLPLEIGKLKKLEKISMKDCYRCELPDSV 370
Query: 720 QNLSELD 726
+NL L+
Sbjct: 371 KNLENLE 377
Score = 39.3 bits (90), Expect = 0.039
Identities = 52/226 (23%), Positives = 94/226 (41%), Gaps = 54/226 (23%)
Query: 540 LKTLIIENGHCSKGLKYLRSSLKALKWEGCLSKSLSSSILSKASEIKSFSNCIYL*CTMI 599
++ L++ + L +++K LK ++ L + L+ S + S N +
Sbjct: 156 VEALVLNISSSNYALPNFIATMKELKVVIIINHGLEPAKLTNLSCLSSLPNLKRI----- 210
Query: 600 SFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLS----------------------- 636
+F+ ++I +LD IP + GL +LEKLS
Sbjct: 211 -----RFEKVSISLLD-------IPKL-GLKSLEKLSLWFCHVVDALNELEDVSETLQSL 257
Query: 637 ----FEYCKNLITIHNSIGHLNKLERLSAFGCRTLKR-FPPLG-LASLKELKLSCCYSLK 690
+YC NL + I + L++LS C L R +G L L+ L+LS C SL
Sbjct: 258 QEIEIDYCYNLDELPYWISQVVSLKKLSVTNCNKLCRVIEAIGDLRDLETLRLSSCASLL 317
Query: 691 SFPKLLCKMTNIDKIWFWYTS----IRELPSSFQNLSELDELSVRE 732
P+ + +D + F S ++ LP L +L+++S+++
Sbjct: 318 ELPETI---DRLDNLRFLDVSGGFQLKNLPLEIGKLKKLEKISMKD 360
>ESA8_TRYEQ (P26337) Putative adenylate cyclase regulatory protein
Length = 630
Score = 50.8 bits (120), Expect = 1e-05
Identities = 52/213 (24%), Positives = 90/213 (41%), Gaps = 42/213 (19%)
Query: 526 KHYKKGKAFKKMTRLKTLIIENGHCSKGLKYLR--SSLKALKWEGCLS-------KSLSS 576
K++K +++ L L + H L ++ S+LK L GC S + L++
Sbjct: 335 KNFKDLNGLERLVNLDKLNLSGCHGVSSLGFVANLSNLKELDISGCESLVCFDGLQDLNN 394
Query: 577 SILSKASEIKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLS 636
+ ++KSF+N + K M L L CE +T + + L LE+LS
Sbjct: 395 LEVLYLRDVKSFTNV---------GAIKNLSKMRELDLSGCERITSLSGLETLKGLEELS 445
Query: 637 FEYCKNLITIHN--SIGHL--------------------NKLERLSAFGCRTLKRFPPL- 673
E C +++ S+ HL LE L GCR F P+
Sbjct: 446 LEGCGEIMSFDPIWSLHHLRVLYVSECGNLEDLSGLEGITGLEELYLHGCRKCTNFGPIW 505
Query: 674 GLASLKELKLSCCYSLKSFPKLLCKMTNIDKIW 706
L ++ ++LSCC +L+ L C +T +++++
Sbjct: 506 NLRNVCVVELSCCENLEDLSGLQC-LTGLEELY 537
Score = 46.2 bits (108), Expect = 3e-04
Identities = 48/183 (26%), Positives = 82/183 (44%), Gaps = 18/183 (9%)
Query: 560 SLKALKWEGCLSKSLSSSILSKASEIKSF--SNCIYL*CTMISFSAKKFQDMTILILDHC 617
SL+ L GC + + L K S ++ S C+ L ++ K ++ +L + +C
Sbjct: 278 SLEKLSLSGCWNVTKGLEELCKFSNLRELDISGCLVLGSAVV---LKNLINLKVLSVSNC 334
Query: 618 EYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLGLAS 677
+ + + L NL+KL+ C + ++ + +L+ L+ L GC +L F GL
Sbjct: 335 KNFKDLNGLERLVNLDKLNLSGCHGVSSL-GFVANLSNLKELDISGCESLVCFD--GLQD 391
Query: 678 LKELKLSCCYSLKSFP-----KLLCKMTNIDKIWF-WYTSIRELPSSFQNLSELDELSVR 731
L L++ +KSF K L KM +D TS+ S + L L+ELS+
Sbjct: 392 LNNLEVLYLRDVKSFTNVGAIKNLSKMRELDLSGCERITSL----SGLETLKGLEELSLE 447
Query: 732 EFG 734
G
Sbjct: 448 GCG 450
Score = 40.8 bits (94), Expect = 0.013
Identities = 34/123 (27%), Positives = 52/123 (41%), Gaps = 2/123 (1%)
Query: 609 MTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLK 668
+ +L C +T + + G+ +LEKLS C N+ + + L L GC L
Sbjct: 256 LKVLRYSSCHEITDLTAIGGMRSLEKLSLSGCWNVTKGLEELCKFSNLRELDISGCLVLG 315
Query: 669 RFPPL-GLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWFWYTSIRELPSSFQNLSELDE 727
L L +LK L +S C + K L ++ N+DK+ NLS L E
Sbjct: 316 SAVVLKNLINLKVLSVSNCKNFKDLNGLE-RLVNLDKLNLSGCHGVSSLGFVANLSNLKE 374
Query: 728 LSV 730
L +
Sbjct: 375 LDI 377
Score = 33.9 bits (76), Expect = 1.6
Identities = 26/71 (36%), Positives = 35/71 (48%), Gaps = 1/71 (1%)
Query: 612 LILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFP 671
L L CE +T I V L NL+ LS +C NL + + L LE+L GC L
Sbjct: 536 LYLIGCEEITPIGVVGNLRNLKCLSTCWCANLKEL-GGLDRLVNLEKLDLSGCCGLSSSV 594
Query: 672 PLGLASLKELK 682
+ L SL +L+
Sbjct: 595 FMELMSLPKLQ 605
>ESA8_TRYBB (P23799) Putative adenylate cyclase regulatory protein
(Leucine repeat protein) (VSG expression site-associated
protein F14.9)
Length = 630
Score = 50.8 bits (120), Expect = 1e-05
Identities = 50/213 (23%), Positives = 89/213 (41%), Gaps = 42/213 (19%)
Query: 526 KHYKKGKAFKKMTRLKTLIIENGHCSKGLKYLR--SSLKALKWEGCLS-------KSLSS 576
K++K +++ L+ L + H L ++ S+LK L GC S + L++
Sbjct: 335 KNFKDLNGLERLVNLEKLNLSGCHGVSSLGFVANLSNLKELDISGCESLVCFDGLQDLNN 394
Query: 577 SILSKASEIKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLS 636
+ ++KSF+N + K M L L CE +T + + L LE+LS
Sbjct: 395 LEVLYLRDVKSFTNV---------GAIKNLSKMRELDLSGCERITSLSGLETLKGLEELS 445
Query: 637 FEYCKNLITIH----------------------NSIGHLNKLERLSAFGCRTLKRFPPL- 673
E C +++ + + L LE + GCR F P+
Sbjct: 446 LEGCGEIMSFDPIWSLYHLRVLYVSECGNLEDLSGLQCLTGLEEMYLHGCRKCTNFGPIW 505
Query: 674 GLASLKELKLSCCYSLKSFPKLLCKMTNIDKIW 706
L ++ L+LSCC +L L C +T +++++
Sbjct: 506 NLRNVCVLELSCCENLDDLSGLQC-LTGLEELY 537
Score = 47.8 bits (112), Expect = 1e-04
Identities = 55/206 (26%), Positives = 91/206 (43%), Gaps = 20/206 (9%)
Query: 539 RLKTLIIENGHCSKGLKYLRS--SLKALKWEGCLSKSLSSSILSKASEIKSF--SNCIYL 594
+LK L I + H L + SL+ L GC + + L K S ++ S C+ L
Sbjct: 255 KLKMLDISSCHEITDLTAIGGVRSLEKLSLSGCWNVTKGLEELCKFSNLRELDISGCLVL 314
Query: 595 *CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLN 654
++ K ++ +L + +C+ + + L NLEKL+ C + ++ + +L+
Sbjct: 315 GSAVV---LKNLINLKVLSVSNCKNFKDLNGLERLVNLEKLNLSGCHGVSSL-GFVANLS 370
Query: 655 KLERLSAFGCRTLKRFPPLGLASLKELKLSCCYSLKSFP-----KLLCKMTNIDKIWF-W 708
L+ L GC +L F GL L L++ +KSF K L KM +D
Sbjct: 371 NLKELDISGCESLVCFD--GLQDLNNLEVLYLRDVKSFTNVGAIKNLSKMRELDLSGCER 428
Query: 709 YTSIRELPSSFQNLSELDELSVREFG 734
TS+ S + L L+ELS+ G
Sbjct: 429 ITSL----SGLETLKGLEELSLEGCG 450
Score = 33.5 bits (75), Expect = 2.1
Identities = 26/71 (36%), Positives = 35/71 (48%), Gaps = 1/71 (1%)
Query: 612 LILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFP 671
L L CE +T I V L NL+ LS +C NL + + L LE+L GC L
Sbjct: 536 LYLIGCEEITTIGVVGNLRNLKCLSTCWCANLKEL-GGLERLVNLEKLDLSGCCGLSSSV 594
Query: 672 PLGLASLKELK 682
+ L SL +L+
Sbjct: 595 FMELMSLPKLQ 605
>DR25_ARATH (O50052) Putative disease resistance protein At4g19050
Length = 1181
Score = 48.9 bits (115), Expect = 5e-05
Identities = 37/131 (28%), Positives = 64/131 (48%), Gaps = 5/131 (3%)
Query: 612 LILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFP 671
L+L +C + +P + L++LE C L I+ S G ++ L ++ L P
Sbjct: 707 LLLRNCSLIEELPSIEKLTHLEVFDVSGCIKLKNINGSFGEMSYLHEVN-LSETNLSELP 765
Query: 672 PL--GLASLKELKLSCCYSLKSFPKLLCKMTNIDKI-WFWYTSIRELPSSFQNLSELDEL 728
L++LKEL + C LK+ P L K+TN++ T + + SF+NLS L ++
Sbjct: 766 DKISELSNLKELIIRKCSKLKTLPNLE-KLTNLEIFDVSGCTELETIEGSFENLSCLHKV 824
Query: 729 SVREFGMLRFP 739
++ E + P
Sbjct: 825 NLSETNLGELP 835
Score = 45.8 bits (107), Expect = 4e-04
Identities = 40/134 (29%), Positives = 69/134 (50%), Gaps = 9/134 (6%)
Query: 612 LILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFP 671
LI+ C L +P++ L+NLE C L TI S +L+ L +++ L P
Sbjct: 777 LIIRKCSKLKTLPNLEKLTNLEIFDVSGCTELETIEGSFENLSCLHKVN-LSETNLGELP 835
Query: 672 PL--GLASLKELKLSCCYSLKSFPKLLCKMTNIDKIWF---WYTSIRELPSSFQNLSELD 726
L++LKEL L C LK+ P L K+T++ + F T++ ++ SF+++S L
Sbjct: 836 NKISELSNLKELILRNCSKLKALPNLE-KLTHL--VIFDVSGCTNLDKIEESFESMSYLC 892
Query: 727 ELSVREFGMLRFPK 740
E+++ + FP+
Sbjct: 893 EVNLSGTNLKTFPE 906
>RGA4_SOLBU (Q7XA39) Putative disease resistance protein RGA4
(RGA4-blb)
Length = 988
Score = 47.4 bits (111), Expect = 1e-04
Identities = 33/90 (36%), Positives = 48/90 (52%), Gaps = 5/90 (5%)
Query: 621 THIPD--VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPL---GL 675
T +P+ + L+NLE LSF KNL + S+ LN L+RL C +L+ FP GL
Sbjct: 867 TSLPEEMFTSLTNLEFLSFFDFKNLKDLPTSLTSLNALKRLQIESCDSLESFPEQGLEGL 926
Query: 676 ASLKELKLSCCYSLKSFPKLLCKMTNIDKI 705
SL +L + C LK P+ L +T + +
Sbjct: 927 TSLTQLFVKYCKMLKCLPEGLQHLTALTNL 956
Score = 35.8 bits (81), Expect = 0.43
Identities = 18/60 (30%), Positives = 32/60 (53%), Gaps = 2/60 (3%)
Query: 612 LILDHCEYLTHIPD--VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKR 669
L ++ C+ L P+ + GL++L +L +YCK L + + HL L L GC +++
Sbjct: 907 LQIESCDSLESFPEQGLEGLTSLTQLFVKYCKMLKCLPEGLQHLTALTNLGVSGCPEVEK 966
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 45.8 bits (107), Expect = 4e-04
Identities = 29/90 (32%), Positives = 44/90 (48%), Gaps = 7/90 (7%)
Query: 26 DTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIPVFSINYASSS 85
+ RY F +L KAL KG+N D+ NE +E +R+ + + N S
Sbjct: 15 EVRYSFVSHLSKALQRKGVNDVFIDSDDSLSNESQ----SMVERARVSVMILPGNRTVS- 69
Query: 86 FCLDELDHIIHCYKTKGRPVLPVFFGVDPS 115
LD+L ++ C K K + V+PV +GV S
Sbjct: 70 --LDKLVKVLDCQKNKDQVVVPVLYGVRSS 97
Score = 45.8 bits (107), Expect = 4e-04
Identities = 34/107 (31%), Positives = 55/107 (50%), Gaps = 10/107 (9%)
Query: 620 LTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPL-GLASL 678
L IP +SG+SNLE+ + +L+ I S + KL L C L+ P + L L
Sbjct: 690 LAEIPGLSGVSNLEQSDLKPLTSLMKISTSYQNPGKLSCLELNDCSRLRSLPNMVNLELL 749
Query: 679 KELKLSCCYSLKS---FPKLLCKMTNIDKIWFWYTSIRELPSSFQNL 722
K L LS C L++ FP+ N+ +++ T++R++P Q+L
Sbjct: 750 KALDLSGCSELETIQGFPR------NLKELYLVGTAVRQVPQLPQSL 790
Score = 37.4 bits (85), Expect = 0.15
Identities = 30/91 (32%), Positives = 46/91 (49%), Gaps = 5/91 (5%)
Query: 629 LSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLG-LASLKELKLSCCY 687
L L+ + + + L+ I + + N LE + GC L+ FP G L L+ + LS C
Sbjct: 591 LEMLKTIRLCHSQQLVDIDDLLKAQN-LEVVDLQGCTRLQSFPATGQLLHLRVVNLSGCT 649
Query: 688 SLKSFPKLLCKMTNIDKIWFWYTSIRELPSS 718
+KSFP++ NI+ + T I ELP S
Sbjct: 650 EIKSFPEI---PPNIETLNLQGTGIIELPLS 677
Score = 32.0 bits (71), Expect = 6.2
Identities = 36/151 (23%), Positives = 64/151 (41%), Gaps = 18/151 (11%)
Query: 604 KKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFG 663
K + + + L H + L I D+ NLE + + C L + + G L L ++ G
Sbjct: 589 KDLEMLKTIRLCHSQQLVDIDDLLKAQNLEVVDLQGCTRLQSFP-ATGQLLHLRVVNLSG 647
Query: 664 CRTLKRFP--PLGLASLK-------ELKLSCCY-SLKSFPKLLCKMTNIDKIWFW----- 708
C +K FP P + +L EL LS + + LL ++ + +
Sbjct: 648 CTEIKSFPEIPPNIETLNLQGTGIIELPLSIVKPNYRELLNLLAEIPGLSGVSNLEQSDL 707
Query: 709 --YTSIRELPSSFQNLSELDELSVREFGMLR 737
TS+ ++ +S+QN +L L + + LR
Sbjct: 708 KPLTSLMKISTSYQNPGKLSCLELNDCSRLR 738
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 44.7 bits (104), Expect = 0.001
Identities = 37/111 (33%), Positives = 55/111 (49%), Gaps = 14/111 (12%)
Query: 619 YLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASL 678
+LT IP +S S LE+L+ +L+ ++S L KL L C L+ P + L
Sbjct: 696 FLTEIPGLSEASKLERLT-----SLLESNSSCQDLGKLICLELKDCSCLQSLPNMANLDL 750
Query: 679 KELKLSCCYSLKS---FPKLLCKMTNIDKIWFWYTSIRELPSSFQNLSELD 726
L LS C SL S FP+ L +++ T+IRE+P Q+L L+
Sbjct: 751 NVLDLSGCSSLNSIQGFPRFL------KQLYLGGTAIREVPQLPQSLEILN 795
Score = 37.4 bits (85), Expect = 0.15
Identities = 52/200 (26%), Positives = 75/200 (37%), Gaps = 48/200 (24%)
Query: 533 AFKKMTRLKTLIIENGHCSK------------GLKYLRSSLKALKWEGCLSKSLSSSILS 580
AFK M L+ L I +CS L L + L+ L WE KSL +
Sbjct: 519 AFKNMLNLRLLKI---YCSNPEVHPVINFPTGSLHSLPNELRLLHWENYPLKSLPQNFDP 575
Query: 581 K-ASEIKSFSNCIYL*CTMISFSAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEY 639
+ EI N Y + K + + + L H ++L I D+ NLE + +
Sbjct: 576 RHLVEI----NMPYSQLQKLWGGTKNLEMLRTIRLCHSQHLVDIDDLLKAENLEVIDLQ- 630
Query: 640 CKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLG-LASLKELKLSCCYSLKSFPKLLCK 698
GC L+ FP G L L+ + LS C +KS ++
Sbjct: 631 -----------------------GCTRLQNFPAAGRLLRLRVVNLSGCIKIKSVLEI--- 664
Query: 699 MTNIDKIWFWYTSIRELPSS 718
NI+K+ T I LP S
Sbjct: 665 PPNIEKLHLQGTGILALPVS 684
Score = 33.5 bits (75), Expect = 2.1
Identities = 23/87 (26%), Positives = 41/87 (46%), Gaps = 3/87 (3%)
Query: 26 DTRYGFTGNLYKALTDKGINTFIDDNGLQRGNEITPSLLKAIEESRIFIPVFSINYASSS 85
+ RY F +L +AL KGIN + D + + + IE++ + + V N S
Sbjct: 18 EVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPSE 75
Query: 86 FCLDELDHIIHCYK-TKGRPVLPVFFG 111
LD+ ++ C + K + V+ V +G
Sbjct: 76 VWLDKFAKVLECQRNNKDQAVVSVLYG 102
>RGA2_SOLBU (Q7XBQ9) Disease resistance protein RGA2 (RGA2-blb)
(Blight resistance protein RPI)
Length = 970
Score = 43.1 bits (100), Expect = 0.003
Identities = 33/115 (28%), Positives = 55/115 (47%), Gaps = 6/115 (5%)
Query: 596 CTMISFSAKKFQDMTILILDHCEYLTHIPD--VSGLSNLEKLSFEYCKNLITIHNSIGHL 653
C ++ S+ + +T L + + + T P+ L+NL+ L+ C NL + S+ L
Sbjct: 824 CPFLTLSSN-LRALTSLRICYNKVATSFPEEMFKNLANLKYLTISRCNNLKELPTSLASL 882
Query: 654 NKLERLSAFGCRTLKRFPP---LGLASLKELKLSCCYSLKSFPKLLCKMTNIDKI 705
N L+ L C L+ P GL+SL EL + C LK P+ L +T + +
Sbjct: 883 NALKSLKIQLCCALESLPEEGLEGLSSLTELFVEHCNMLKCLPEGLQHLTTLTSL 937
>RGA1_SOLBU (Q7XA42) Putative disease resistance protein RGA1
(RGA3-blb)
Length = 1025
Score = 42.4 bits (98), Expect = 0.005
Identities = 32/104 (30%), Positives = 48/104 (45%), Gaps = 5/104 (4%)
Query: 602 SAKKFQDMTILILDHCEYLTHIPD--VSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERL 659
S + +T L + T +P+ L+NL+ L + +NL + S+ LN L+ L
Sbjct: 884 SISNLRALTSLDISDNVEATSLPEEMFKSLANLKYLKISFFRNLKELPTSLASLNALKSL 943
Query: 660 SAFGCRTLKRFPP---LGLASLKELKLSCCYSLKSFPKLLCKMT 700
C L+ P GL SL EL +S C LK P+ L +T
Sbjct: 944 KFEFCDALESLPEEGVKGLTSLTELSVSNCMMLKCLPEGLQHLT 987
Score = 31.6 bits (70), Expect = 8.0
Identities = 20/64 (31%), Positives = 25/64 (38%), Gaps = 7/64 (10%)
Query: 629 LSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLKRFPPLGLASLKELKLSCCYS 688
L NL+ L YC +L + L L L GC P +GL L+C S
Sbjct: 617 LQNLQTLDLHYCDSLSCLPKQTSKLGSLRNLLLDGCSLTSTPPRIGL-------LTCLKS 669
Query: 689 LKSF 692
L F
Sbjct: 670 LSCF 673
>RSU1_HUMAN (Q15404) Ras suppressor protein 1 (Rsu-1) (RSP-1)
Length = 276
Score = 39.7 bits (91), Expect = 0.030
Identities = 38/136 (27%), Positives = 61/136 (43%), Gaps = 7/136 (5%)
Query: 609 MTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLK 668
+T L+L H + P+++ L NLE L+F + + + I L KL+ L+ G L
Sbjct: 41 ITQLVLSHNKLTMVPPNIAELKNLEVLNF-FNNQIEELPTQISSLQKLKHLN-LGMNRLN 98
Query: 669 RFPPLGLASLKELK-LSCCY---SLKSFPKLLCKMTNIDKIWFWYTSIRELPSSFQNLSE 724
P G SL L+ L Y S S P +T + ++ LP L++
Sbjct: 99 TL-PRGFGSLPALEVLDLTYNNLSENSLPGNFFYLTTLRALYLSDNDFEILPPDIGKLTK 157
Query: 725 LDELSVREFGMLRFPK 740
L LS+R+ ++ PK
Sbjct: 158 LQILSLRDNDLISLPK 173
>CYAA_SACKL (P23466) Adenylate cyclase (EC 4.6.1.1) (ATP
pyrophosphate-lyase) (Adenylyl cyclase)
Length = 1839
Score = 39.7 bits (91), Expect = 0.030
Identities = 36/131 (27%), Positives = 66/131 (49%), Gaps = 8/131 (6%)
Query: 602 SAKKFQDMTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSA 661
SA K + ++ + ++ ++ D L +L+ L + K + +SI LN L ++
Sbjct: 655 SAIKLSSLRMVNIRASKFPANVTDAYKLVSLD-LERNFIKK---VPDSIFKLNNLTIVN- 709
Query: 662 FGCRTLKRFPPLGLASLKELKLSCCYSLK--SFPKLLCKMTNIDKIWFWYTSIRELPSSF 719
C L+R PP G + LK L+L S K ++P+++ TN+ +I Y I LP S
Sbjct: 710 LQCNNLERLPP-GFSKLKNLQLLDISSNKFVNYPEVINSCTNLLQIDLSYNKIHSLPVSI 768
Query: 720 QNLSELDELSV 730
L +L ++++
Sbjct: 769 NQLVKLAKMNL 779
>RSU1_MOUSE (Q01730) Ras suppressor protein 1 (Rsu-1) (RSP-1)
Length = 276
Score = 38.9 bits (89), Expect = 0.050
Identities = 38/136 (27%), Positives = 62/136 (44%), Gaps = 7/136 (5%)
Query: 609 MTILILDHCEYLTHIPDVSGLSNLEKLSFEYCKNLITIHNSIGHLNKLERLSAFGCRTLK 668
+T L+L H + T P+V+ L NLE L+F + + + I L KL+ L+ G L
Sbjct: 41 ITQLVLSHNKLTTVPPNVAELKNLEVLNF-FNNQIEELPTQISSLQKLKHLN-LGMNRLN 98
Query: 669 RFP-PLGLASLKELKLSCCY---SLKSFPKLLCKMTNIDKIWFWYTSIRELPSSFQNLSE 724
P G + L E+ L Y + S P +T + ++ LP L++
Sbjct: 99 TLPRGFGSSRLLEV-LELTYNNLNEHSLPGNFFYLTTLRALYLSDNDFEILPPDIGKLTK 157
Query: 725 LDELSVREFGMLRFPK 740
L LS+R+ ++ PK
Sbjct: 158 LQILSLRDNDLISLPK 173
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.347 0.151 0.545
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 79,411,437
Number of Sequences: 164201
Number of extensions: 2992869
Number of successful extensions: 11859
Number of sequences better than 10.0: 44
Number of HSP's better than 10.0 without gapping: 12
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 11699
Number of HSP's gapped (non-prelim): 148
length of query: 751
length of database: 59,974,054
effective HSP length: 118
effective length of query: 633
effective length of database: 40,598,336
effective search space: 25698746688
effective search space used: 25698746688
T: 11
A: 40
X1: 14 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC138133.8


