

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.7 + phase: 0 /pseudo
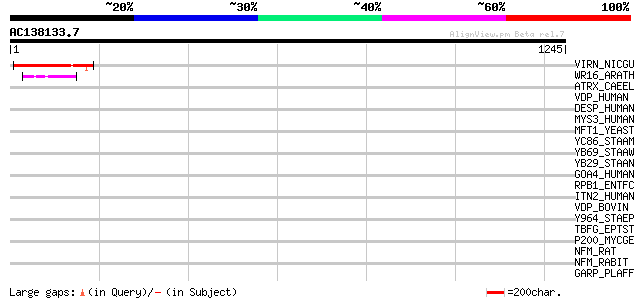
(1245 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 149 5e-35
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 49 1e-04
ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-li... 45 0.002
VDP_HUMAN (O60763) General vesicular transport factor p115 (Tran... 42 0.014
DESP_HUMAN (P15924) Desmoplakin (DP) (250/210 kDa paraneoplastic... 42 0.014
MYS3_HUMAN (Q92794) MYST histone acetyltransferase 3 (Runt-relat... 40 0.030
MFT1_YEAST (P33441) Mitochondrial fusion target protein 40 0.030
YC86_STAAM (P67277) Hypothetical UPF0144 protein SAV1286 39 0.067
YB69_STAAW (P67279) Hypothetical UPF0144 protein MW1169 39 0.067
YB29_STAAN (P67278) Hypothetical UPF0144 protein SA1129 39 0.067
GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member... 39 0.067
RPB1_ENTFC (Q8GCR6) DNA-directed RNA polymerase beta chain (EC 2... 39 0.088
ITN2_HUMAN (Q9NZM3) Intersectin 2 (SH3 domain-containing protein... 39 0.088
VDP_BOVIN (P41541) General vesicular transport factor p115 (Tran... 38 0.15
Y964_STAEP (Q8CSS7) Hypothetical UPF0144 protein SE0964 37 0.26
TBFG_EPTST (Q90502) Thread biopolymer filament gamma subunit 37 0.26
P200_MYCGE (Q49429) Protein P200 37 0.26
NFM_RAT (P12839) Neurofilament triplet M protein (160 kDa neurof... 37 0.26
NFM_RABIT (P54938) Neurofilament triplet M protein (160 kDa neur... 37 0.26
GARP_PLAFF (P13816) Glutamic acid-rich protein precursor 37 0.33
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 149 bits (376), Expect = 5e-35
Identities = 83/186 (44%), Positives = 117/186 (62%), Gaps = 8/186 (4%)
Query: 8 SSSSFSYAFTYQVFLSFRGTDTRYGFTGNLYKALTDKGIHTFIDENDLRRGDEITPALLK 67
+SSS S ++Y VFLSFRG DTR FT +LY+ L DKGI TF D+ L G I L K
Sbjct: 2 ASSSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELCK 61
Query: 68 AIDESRIFIPVFSIKYASSSFCLDELVHIIHCYTTKGRVVLPVFFGVEPSHVRHHKGSYG 127
AI+ES+ I VFS YA+S +CL+ELV I+ C T + V+P+F+ V+PSHVR+ K S+
Sbjct: 62 AIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESFA 121
Query: 128 QALAEHKKRFQNDEDNIKRLQRWKVALSQAANFSGYHDSPPGE-----RNLQQDQSSALT 182
+A EH+ ++ +D+++ +QRW++AL++AAN G D+ R + SS L
Sbjct: 122 KAFEEHETKY---KDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKLC 178
Query: 183 CCQLSY 188
LSY
Sbjct: 179 KISLSY 184
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 48.5 bits (114), Expect = 1e-04
Identities = 31/124 (25%), Positives = 58/124 (46%), Gaps = 9/124 (7%)
Query: 28 DTRYGFTGNLYKALTDKGIHTFIDENDLRRGDEITPALLKAIDESRIFIPVFSIKYASSS 87
+ RY F +L KAL KG++ ++D D ++ ++ +R+ + + + +
Sbjct: 15 EVRYSFVSHLSKALQRKGVNDVFIDSD----DSLSNESQSMVERARVSVMILP---GNRT 67
Query: 88 FCLDELVHIIHCYTTKGRVVLPVFFGVEPSHVRHHKG--SYGQALAEHKKRFQNDEDNIK 145
LD+LV ++ C K +VV+PV +GV S S G + H ++ +D +K
Sbjct: 68 VSLDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLSALDSKGFSSVHHSRKECSDSQLVK 127
Query: 146 RLQR 149
R
Sbjct: 128 ETVR 131
>ATRX_CAEEL (Q9U7E0) Transcriptional regulator ATRX homolog (X-linked
nuclear protein-1)
Length = 1359
Score = 44.7 bits (104), Expect = 0.002
Identities = 66/314 (21%), Positives = 131/314 (41%), Gaps = 26/314 (8%)
Query: 898 KKNELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEEEIDTEEDDFSDDEI 957
KK + E +K SEDE R Q S + T+++ +E + S++E
Sbjct: 108 KKVDQKKKEKSKKKRTTSSSEDEDSDEEREQK----SKKKSKKTKKQTSSESSEESEEER 163
Query: 958 FTESSSQMVIHYQEKSNTEQIDTEE--DDFYRSAQMGIHVVKEKSNIKDHDLSEDEIFVP 1015
+ S + +EKS ++ +T E D+ + ++ +K+K+ + SEDE V
Sbjct: 164 KVKKSKKN----KEKSVKKRAETSEESDEDEKPSKKSKKGLKKKAKSESESESEDEKEVK 219
Query: 1016 DEIKKLSSAQIGIHEKSNTEDVI----FTNPNRTTTTTKPYYSQALFPNPPRKIRKFRPT 1071
KK + ++S +ED T + + T+ S++ + + ++ P
Sbjct: 220 KSKKKSKKV---VKKESESEDEAPEKKKTEKRKRSKTSSEESSESEKSDEEEEEKESSPK 276
Query: 1072 LKKQRFVEVSKTESLQRRS------LEETKSDGVKSLEVSETESLQQQYLAVVSGMRNLV 1125
KK++ + V K S + L + K G +L + Q+ + S + V
Sbjct: 277 PKKKKPLAVKKLSSDEESEESDVEVLPQKKKRGAVTLISDSEDEKDQKSESEASDVEEKV 336
Query: 1126 LTETKEKHRFVEVGVSETE---TAHRWSLEETKSDGVKRFEVSETESLQQQDLALLSSGM 1182
+ +K E G +E T +R S ++ K + K+ + ++ LQ++ + +
Sbjct: 337 SKKKAKKQESSESGSDSSEGSITVNRKSKKKEKPEKKKKGIIMDSSKLQKETIDAERAEK 396
Query: 1183 QNMVLTDKKEKENN 1196
+ +KK+KE N
Sbjct: 397 ERRKRLEKKQKEFN 410
Score = 35.0 bits (79), Expect = 1.3
Identities = 46/230 (20%), Positives = 87/230 (37%), Gaps = 27/230 (11%)
Query: 947 TEEDDFSDDEIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEKSNIKDHD 1006
+EEDD ++E +SS + + +S +++ D EED ++ + K++ + K
Sbjct: 64 SEEDDDDEEESPRKSSKKSRKRAKSESESDESDEEEDRKKSKSKKKVDQKKKEKSKKKRT 123
Query: 1007 LSEDEIFVPDEIKKLSSAQIGIHEKSNTEDVIFTNPNRTTTTTKPYYSQALFPNPPRKIR 1066
S E DE ++ S + K T + RK++
Sbjct: 124 TSSSEDEDSDEEREQKSKKKSKKTKKQTSS-----------------ESSEESEEERKVK 166
Query: 1067 KFRPTLKK--QRFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQYLAVVSGMRNL 1124
K + +K ++ E S+ + +++K G+K SE+ES + V +
Sbjct: 167 KSKKNKEKSVKKRAETSEESDEDEKPSKKSKK-GLKKKAKSESESESEDEKEVKKSKKKS 225
Query: 1125 VLTETKEKHRFVEVGVSETETAHRWSLEETKSDGVKRFEVSETESLQQQD 1174
KE SE E + E+ K E SE+E +++
Sbjct: 226 KKVVKKESE-------SEDEAPEKKKTEKRKRSKTSSEESSESEKSDEEE 268
>VDP_HUMAN (O60763) General vesicular transport factor p115
(Transcytosis associated protein) (TAP) (Vesicle docking
protein)
Length = 962
Score = 41.6 bits (96), Expect = 0.014
Identities = 73/332 (21%), Positives = 144/332 (42%), Gaps = 29/332 (8%)
Query: 884 YDNFNHMPEMIRTIKKNELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEE 943
Y F+H E + +K+ E + ++ + E+E+KK I V+ + E+
Sbjct: 606 YMIFDH--EFTKLVKELEGVITKAIYKSSEEDKKEEEVKKTLEQHDNI-VTHYKNMIREQ 662
Query: 944 EIDTEEDDFSDDEIFTESSSQMVIHYQEKSNTEQIDTEEDDF-YRSAQMGI---HVVKEK 999
++ EE ++ T + QI +D + Q+G H
Sbjct: 663 DLQLEE---LRQQVSTLKCQNEQLQTAVTQQVSQIQQHKDQYNLLKIQLGKDNQHQGSYS 719
Query: 1000 SNIKDHDLSEDEIF-VPDEIKKLSSAQIGIHEKSNTEDVIFTNPNRTTTTTKPYYSQALF 1058
+ + + +EI + +EI++L Q + + +D + N + T+ S A+
Sbjct: 720 EGAQMNGIQPEEIGRLREEIEELKRNQELLQSQLTEKDSMIENMKSSQTSGTNEQSSAIV 779
Query: 1059 P----NPPRKIRKFRPTLKKQ---RFVEVSKTESLQRRSLEETKSDGVKSLEVS-ETESL 1110
++++ TLK Q + VE++K ++ ++ L++T++ KS+EV ETE++
Sbjct: 780 SARDSEQVAELKQELATLKSQLNSQSVEITKLQTEKQELLQKTEAFA-KSVEVQGETETI 838
Query: 1111 QQQYLAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKS------DGVKRFEV 1164
V G + +L ETKE ++ +SE TA + L+ + S + E+
Sbjct: 839 IATKTTDVEGRLSALLQETKELKNEIK-ALSEERTAIKEQLDSSNSTIAILQTEKDKLEL 897
Query: 1165 SETESLQQQD--LALLSSGMQNMVLTDKKEKE 1194
T+S ++QD L LL+ Q ++ K K+
Sbjct: 898 EITDSKKEQDDLLVLLADQDQKILSLKNKLKD 929
>DESP_HUMAN (P15924) Desmoplakin (DP) (250/210 kDa paraneoplastic
pemphigus antigen)
Length = 2871
Score = 41.6 bits (96), Expect = 0.014
Identities = 71/315 (22%), Positives = 132/315 (41%), Gaps = 61/315 (19%)
Query: 898 KKNELNHVE--IDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEEEIDTEEDDFSDD 955
K E+ ++ ID E +D EDE +L+R+Q + + + T+T ++ +E + +
Sbjct: 1479 KNKEIERLKQLIDKETNDRKCLEDENARLQRVQYDLQKANSSATETINKLKVQEQELTRL 1538
Query: 956 EIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEKSNIKDHDLSEDEIFVP 1015
I E S QE++ +Q T + + Q+ V+E+ N SED
Sbjct: 1539 RIDYERVS------QERTVKDQDITRFQNSLKELQLQKQKVEEELNRLKRTASEDSC--- 1589
Query: 1016 DEIKKLSSAQIGIHEKSNTEDVIFTNPNRTTTTTKPYYSQALFPNPPRKIRKFRPTL--- 1072
+ KKL G+ + + TN T + + + +R+ R L
Sbjct: 1590 -KRKKLEEELEGMRRSLKEQAIKITN-----LTQQLEQASIVKKRSEDDLRQQRDVLDGH 1643
Query: 1073 --KKQRFVE-----VSKTESLQRRSLEETKS----------------DGVKSLEVSE--- 1106
+KQR E S+ E+L+R+ L+E +S D +SL S+
Sbjct: 1644 LREKQRTQEELRRLSSEVEALRRQLLQEQESVKQAHLRNEHFQKAIEDKSRSLNESKIEI 1703
Query: 1107 ------TESLQQQYLAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKSDGVK 1160
TE+L +++L + +RNL L ++ + G SE ++ ++ E +S
Sbjct: 1704 ERLQSLTENLTKEHLMLEEELRNLRL-----EYDDLRRGRSEADSDKNATILELRS---- 1754
Query: 1161 RFEVSETESLQQQDL 1175
+ ++S +L+ Q L
Sbjct: 1755 QLQISNNRTLELQGL 1769
>MYS3_HUMAN (Q92794) MYST histone acetyltransferase 3 (Runt-related
transcription factor binding protein 2) (Monocytic
leukemia zinc finger protein) (Zinc finger protein 220)
Length = 2004
Score = 40.4 bits (93), Expect = 0.030
Identities = 72/285 (25%), Positives = 113/285 (39%), Gaps = 31/285 (10%)
Query: 936 TEETDTEEEIDTEEDDFSDDEIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHV 995
T E D EEE + +E++ E F SSQ V+ Q S + D EED+ A +
Sbjct: 1064 TFEIDEEEE-EEDENELFPREYFRRLSSQDVLRCQSSSKRKSKDEEEDEESDDAD-DTPI 1121
Query: 996 VKEKSNIKDHDLSEDEIFVPDEIKKLSSAQIGIHEKSNTEDVIFTNPNR-----TTTTTK 1050
+K S ++ D+ + PD L + KS P R +
Sbjct: 1122 LKPVSLLRKRDVKNSPL-EPDTSTPLKKKKGWPKGKSRKPIHWKKRPGRKPGFKLSREIM 1180
Query: 1051 PYYSQALFPNPPRKIRKFRPTLKKQRFVE-VSKTESL----QRRSLEETKSDGVKSLEVS 1105
P +QA P I K K Q E V E + +R+ EE +++ ++ E
Sbjct: 1181 PVSTQACVIEPIVSIPKAGRKPKIQESEETVEPKEDMPLPEERKEEEEMQAEAEEAEEGE 1240
Query: 1106 ETESLQQQY-LAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKSDGVKRFEV 1164
E ++ + A + N TETKE V E E R S E+ +S+ ++ E+
Sbjct: 1241 EEDAASSEVPAASPADSSNSPETETKEPE------VEEEEEKPRVSEEQRQSEEEQQ-EL 1293
Query: 1165 SETESLQQQDLALLSSGMQ----------NMVLTDKKEKENNMTR 1199
E E +++D A ++ ++ T KKE E TR
Sbjct: 1294 EEPEPEEEEDAAAETAQNDDHDADDEDDGHLESTKKKELEEQPTR 1338
>MFT1_YEAST (P33441) Mitochondrial fusion target protein
Length = 392
Score = 40.4 bits (93), Expect = 0.030
Identities = 38/142 (26%), Positives = 59/142 (40%), Gaps = 8/142 (5%)
Query: 902 LNHVEIDWEKDDLHLSEDEIKKLR--RLQMGIHVSWTEETDTEEEIDTEEDDFSDDEIFT 959
LN E D +D EDE +K R R + + V+ E D +EE D E DD D+E
Sbjct: 252 LNEAEGDNIDEDYESDEDEERKERFKRQRSMVEVNTIENVDEKEESDHEYDDQEDEE--N 309
Query: 960 ESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIH--VVKEKSNIKDHDLSED--EIFVP 1015
E M + ++ ++D E +++ G + KE I++ D D E
Sbjct: 310 EEEDDMEVDVEDIKEDNEVDGESSQQEDNSRQGNNEETDKETGVIEEPDAVNDAEEADSD 369
Query: 1016 DEIKKLSSAQIGIHEKSNTEDV 1037
+KL S+ E+V
Sbjct: 370 HSSRKLGGTTSDFSASSSVEEV 391
>YC86_STAAM (P67277) Hypothetical UPF0144 protein SAV1286
Length = 519
Score = 39.3 bits (90), Expect = 0.067
Identities = 35/143 (24%), Positives = 61/143 (42%), Gaps = 4/143 (2%)
Query: 1054 SQALFPNPPRKIRKFRPTLKKQRFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQ 1113
+Q L ++R+ R L++Q + K E+L+R+S K D + + S+ E QQQ
Sbjct: 65 NQILREQTEAELRERRSELQRQETRLLQKEENLERKSDLLDKKDEILEQKESKIEEKQQQ 124
Query: 1114 YLAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKSDG----VKRFEVSETES 1169
A S ++ L++ +E R + E +EE S VK E E
Sbjct: 125 VDAKESSVQTLIMKHEQELERISGLTQEEAINEQLQRVEEELSQDIAVLVKEKEKEAKEK 184
Query: 1170 LQQQDLALLSSGMQNMVLTDKKE 1192
+ + LL++ +Q + E
Sbjct: 185 VDKTAKELLATAVQRLAADHTSE 207
>YB69_STAAW (P67279) Hypothetical UPF0144 protein MW1169
Length = 519
Score = 39.3 bits (90), Expect = 0.067
Identities = 35/143 (24%), Positives = 61/143 (42%), Gaps = 4/143 (2%)
Query: 1054 SQALFPNPPRKIRKFRPTLKKQRFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQ 1113
+Q L ++R+ R L++Q + K E+L+R+S K D + + S+ E QQQ
Sbjct: 65 NQILREQTEAELRERRSELQRQETRLLQKEENLERKSDLLDKKDEILEQKESKIEEKQQQ 124
Query: 1114 YLAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKSDG----VKRFEVSETES 1169
A S ++ L++ +E R + E +EE S VK E E
Sbjct: 125 VDAKESSVQTLIMKHEQELERISGLTQEEAINEQLQRVEEELSQDIAVLVKEKEKEAKEK 184
Query: 1170 LQQQDLALLSSGMQNMVLTDKKE 1192
+ + LL++ +Q + E
Sbjct: 185 VDKTAKELLATAVQRLAADHTSE 207
>YB29_STAAN (P67278) Hypothetical UPF0144 protein SA1129
Length = 519
Score = 39.3 bits (90), Expect = 0.067
Identities = 35/143 (24%), Positives = 61/143 (42%), Gaps = 4/143 (2%)
Query: 1054 SQALFPNPPRKIRKFRPTLKKQRFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQ 1113
+Q L ++R+ R L++Q + K E+L+R+S K D + + S+ E QQQ
Sbjct: 65 NQILREQTEAELRERRSELQRQETRLLQKEENLERKSDLLDKKDEILEQKESKIEEKQQQ 124
Query: 1114 YLAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKSDG----VKRFEVSETES 1169
A S ++ L++ +E R + E +EE S VK E E
Sbjct: 125 VDAKESSVQTLIMKHEQELERISGLTQEEAINEQLQRVEEELSQDIAVLVKEKEKEAKEK 184
Query: 1170 LQQQDLALLSSGMQNMVLTDKKE 1192
+ + LL++ +Q + E
Sbjct: 185 VDKTAKELLATAVQRLAADHTSE 207
>GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member 4
(Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (72.1
protein)
Length = 2230
Score = 39.3 bits (90), Expect = 0.067
Identities = 70/350 (20%), Positives = 141/350 (40%), Gaps = 62/350 (17%)
Query: 890 MPEMIRTIKKN----------ELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEET 939
M E I+TI+K EL+ V+ + SE++I KL++L +E
Sbjct: 437 MDEQIKTIEKTSEEERISLQQELSRVKQEVVDVMKKSSEEQIAKLQKLHEKELAR--KEQ 494
Query: 940 DTEEEIDTEEDDFSDDE--IFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVK 997
+ +++ T E +F + +S S+ + QEK E + EE + + A ++
Sbjct: 495 ELTKKLQTREREFQEQMKVALEKSQSEYLKISQEKEQQESLALEELELQKKA-----ILT 549
Query: 998 EKSNIKDHDLSEDEIFVPDEIKKLSSA-------------QIGIH-----EKSNTEDVIF 1039
E N K DL ++ I +L S+ + +H K N E +
Sbjct: 550 ESEN-KLRDLQQEAETYRTRILELESSLEKSLQENKNQSKDLAVHLEAEKNKHNKEITVM 608
Query: 1040 TNPNRTTTTTKPYYSQALFPNPPRKIR--------KFRPTLKKQRFVEVSKTESLQRRSL 1091
++T + + AL+ + ++ K R ++++ + E + + +
Sbjct: 609 VEKHKTELESLKHQQDALWTEKLQVLKQQYQTEMEKLREKCEQEKETLLKDKEIIFQAHI 668
Query: 1092 EETKSDGVKSLEVSET--ESLQQQYLAVVSGMRNL------VLTETKEKHRFVEVGVSET 1143
EE ++ L+V +T ESL + V+ L + +T + + +E + E
Sbjct: 669 EEMNEKTLEKLDVKQTELESLSSELSEVLKARHKLEEELSVLKDQTDKMKQELEAKMDEQ 728
Query: 1144 ETAHRWSLEETKSDGVKRFEVSETESLQQQDLALLSSGMQNMVLTDKKEK 1193
+ H+ ++ +K EV S+Q+ + AL Q +L +++K
Sbjct: 729 KNHHQQQVDSI----IKEHEV----SIQRTEKALKDQINQLELLLKERDK 770
>RPB1_ENTFC (Q8GCR6) DNA-directed RNA polymerase beta chain (EC
2.7.7.6) (RNAP beta subunit) (Transcriptase beta chain)
(RNA polymerase beta subunit)
Length = 1208
Score = 38.9 bits (89), Expect = 0.088
Identities = 21/65 (32%), Positives = 36/65 (55%), Gaps = 1/65 (1%)
Query: 921 IKKLRRLQMGIHVSWTEETDTE-EEIDTEEDDFSDDEIFTESSSQMVIHYQEKSNTEQID 979
+K+L+ L + + V EET+ E ++D E+DD + T+ + Q EK EQ++
Sbjct: 1131 VKELQSLGLDMRVLDIEETEIELRDMDDEDDDLITVDALTKFAEQQTAKELEKKAAEQVE 1190
Query: 980 TEEDD 984
E+DD
Sbjct: 1191 DEKDD 1195
>ITN2_HUMAN (Q9NZM3) Intersectin 2 (SH3 domain-containing protein 1B)
(SH3P18) (SH3P18-like WASP associated protein)
Length = 1696
Score = 38.9 bits (89), Expect = 0.088
Identities = 65/324 (20%), Positives = 129/324 (39%), Gaps = 40/324 (12%)
Query: 908 DWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEEE--IDTEEDDFSDDEIFTESSSQM 965
+WE+ L E E KK L+ + E EEE D E + + E+ + +
Sbjct: 402 EWERKQRELQEQEWKKQLELEKRLEKQRELERQREEERRKDIERREAAKQELERQRRLEW 461
Query: 966 V-IHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEKSNIKDHDLSEDEIFVPDEIKKLSSA 1024
I QE N + + EE S + +H+ E N K +S + D K +
Sbjct: 462 ERIRRQELLNQKNREQEEIVRLNSKKKNLHLELEALNGKHQQISGR---LQDVRLKKQTQ 518
Query: 1025 QIGIHEKSNTEDVIFTNPNRTTTTTKPYYSQALFPNPPRKIRKFRPTLKKQRF------- 1077
+ + D+ + + Y ++ ++ P +++ R +K +F
Sbjct: 519 KTELEVLDKQCDLEIMEIKQLQQELQEYQNKLIYLVPEKQLLNER--IKNMQFSNTPDSG 576
Query: 1078 VEVSKTESLQRRSLEETKSDGVKSLE------VSETESLQQQ-------------YLAVV 1118
V + +SL++ L + + + +LE +SE +S Q L+++
Sbjct: 577 VSLLHKKSLEKEELCQRLKEQLDALEKETASKLSEMDSFNNQLKCGNMDDSVLQCLLSLL 636
Query: 1119 SGMRN--LVLTETKEKHRFVEVGVSETETAHRWSLEETKSDGVKRFEVSETESLQQQDLA 1176
S + N L+L E +E + ++ + + R L+E + KR E+ + + L+ +
Sbjct: 637 SCLNNLFLLLKELRETYNTQQLALEQLYKIKRDKLKEIER---KRLELMQKKKLEDEAAR 693
Query: 1177 LLSSGMQNMVLTD-KKEKENNMTR 1199
G +N+ + +KE+E R
Sbjct: 694 KAKQGKENLWKENLRKEEEEKQKR 717
>VDP_BOVIN (P41541) General vesicular transport factor p115
(Transcytosis associated protein) (TAP) (Vesicle docking
protein)
Length = 961
Score = 38.1 bits (87), Expect = 0.15
Identities = 37/135 (27%), Positives = 67/135 (49%), Gaps = 12/135 (8%)
Query: 1071 TLKKQ---RFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQYLAVVSGMRNLVLT 1127
TLK Q + VE++K ++ ++ L++T++ + E+E++ V G + +L
Sbjct: 795 TLKSQLNSQSVEITKLQTEKQELLQKTEAFAKSAPVPGESETVIATKTTDVEGRLSALLQ 854
Query: 1128 ETKEKHRFVEVGVSETETAHRWSLEETKS------DGVKRFEVSETESLQQQD--LALLS 1179
ETKE ++ +SE TA + L+ + S + + EV T+S ++QD L LL+
Sbjct: 855 ETKELKNEIK-ALSEERTAIKEQLDSSNSTIAILQNEKNKLEVDITDSKKEQDDLLVLLA 913
Query: 1180 SGMQNMVLTDKKEKE 1194
Q + K KE
Sbjct: 914 DQDQKIFSLKNKLKE 928
>Y964_STAEP (Q8CSS7) Hypothetical UPF0144 protein SE0964
Length = 519
Score = 37.4 bits (85), Expect = 0.26
Identities = 34/136 (25%), Positives = 59/136 (43%), Gaps = 4/136 (2%)
Query: 1054 SQALFPNPPRKIRKFRPTLKKQRFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQ 1113
+Q L ++R+ R L++Q + K E+L R+S K D + + S+ E QQQ
Sbjct: 65 NQILKEQAENELRERRGELQRQETRLLQKEENLDRKSDLLDKKDEILEQKESKLEERQQQ 124
Query: 1114 YLAVVSGMRNLVLTETKEKHRFVEVGVSETETAHRWSLEETKSDG----VKRFEVSETES 1169
A S ++ L++ +E R + E +EE S VK E E
Sbjct: 125 VDAKESSVQTLIMKHEQELERISGLTQEEAVKEQLQRVEEELSQDIAILVKEKEKEAKEK 184
Query: 1170 LQQQDLALLSSGMQNM 1185
+ + LL++ +Q +
Sbjct: 185 VDKTAKELLATTVQRL 200
>TBFG_EPTST (Q90502) Thread biopolymer filament gamma subunit
Length = 603
Score = 37.4 bits (85), Expect = 0.26
Identities = 27/120 (22%), Positives = 58/120 (47%), Gaps = 5/120 (4%)
Query: 892 EMIRTIKKNELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSW-TEETDTEEEIDTEED 950
EM+ + L V ++ + D L + DEIK ++G+ + T+ + + +++ D
Sbjct: 216 EMLTEVSNLTLERVRLEIDVDHLRATADEIKSKYEFELGVRMQLETDIANMKRDLEAAND 275
Query: 951 DFSD-DEIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEKSNIKDHDLSE 1009
D D F + ++ +Q K+ E+++T + F R + V+ E N+K +L++
Sbjct: 276 MRVDLDSKFNFLTEELT--FQRKTQMEELNTLKQQFGRLGPVQTSVI-ELDNVKSVNLTD 332
>P200_MYCGE (Q49429) Protein P200
Length = 1616
Score = 37.4 bits (85), Expect = 0.26
Identities = 36/136 (26%), Positives = 67/136 (48%), Gaps = 22/136 (16%)
Query: 885 DNFNHMPEMIRTIKKN-ELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEE 943
D+ N + + + T + ELNH EI E +L +SE E+++ Q+ ETD+E
Sbjct: 675 DSVNDVDKSLETKTTSVELNHEEIGNEFINLDVSEKEVQEQPTTQL--------ETDSEF 726
Query: 944 EIDT---EEDDFSDD-EIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEK 999
+ T ED F++ E E SS+ + S T++++T E + + V+++
Sbjct: 727 VLPTYQIVEDSFTESAETPNEFSSEQKDTLEFISQTQEVETSESN--------VPTVEQE 778
Query: 1000 SNIKDHDLSEDEIFVP 1015
+ + +H E+ +F P
Sbjct: 779 TKLFEHQ-DENNLFTP 793
>NFM_RAT (P12839) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium polypeptide)
(NF-M)
Length = 845
Score = 37.4 bits (85), Expect = 0.26
Identities = 66/362 (18%), Positives = 142/362 (38%), Gaps = 49/362 (13%)
Query: 867 LPSKHTFLFDLGLKSRIYDNFNHMPEMIRTIKKNELNHVEIDWEKDDLHLSEDEIKKLRR 926
L +H F+ ++ ++++ D + M + + T+ EL EK++ E+E + +
Sbjct: 445 LKVQHKFVEEIIEETKVEDEKSEMEDAL-TVIAEELA-ASAKEEKEEAEEKEEEPEVEKS 502
Query: 927 LQMGIHVSWTEETDTEEEIDTEEDDFSDDEIFTESSSQMVIHYQEKSNTEQIDTEEDDFY 986
EE + EEE + +E++ +DE S Q EK + + D E +
Sbjct: 503 PVKSPEAKEEEEGEKEEEEEGQEEEEEEDE--GVKSDQAEEGGSEKEGSSEKDEGEQE-- 558
Query: 987 RSAQMGIHVVKEKSNIKDHDLSEDEIFVPDEIKKLSSAQIGIHEKSNTEDVIFTNPNRTT 1046
E+ + E+ E K+ + + E + E++ P +
Sbjct: 559 -----------EEGETEAEGEGEEA-----EAKEEKKTEGKVEEMAIKEEIKVEKPEKAK 602
Query: 1047 TTTKPYYSQALFPNPPRKIRK------FRPTLKKQRFVEVSKTESLQRRSLEETKSDGVK 1100
+ + + P P K K + KK+ E K E ++++ + K
Sbjct: 603 SPVPKSPVEEVKPKPEAKAGKDEQKEEEKVEEKKEVAKESPKEEKVEKKEEKPKDVPDKK 662
Query: 1101 SLEVSETESLQQQYLAVVSGMRNLVLTETKE---------KHRFVEVGVSETETAHRWSL 1151
E E ++ + + ++ + +TKE K + E G SE E + S
Sbjct: 663 KAESPVKEKAVEEMITITKSVKVSLEKDTKEEKPQQQEKVKEKAEEEGGSEEEVGDK-SP 721
Query: 1152 EETKSDGV--------KRFEVSETE---SLQQQDLALLSSGMQNMVLTDKKEKENNMTRY 1200
+E+K + + K E ET+ S Q+++ ++++G+ +KK ++ + +
Sbjct: 722 QESKKEDIAINGEVEGKEEEEQETQEKGSGQEEEKGVVTNGLDVSPAEEKKGEDRSDDKV 781
Query: 1201 II 1202
++
Sbjct: 782 VV 783
>NFM_RABIT (P54938) Neurofilament triplet M protein (160 kDa
neurofilament protein) (Neurofilament medium polypeptide)
(NF-M) (Fragment)
Length = 644
Score = 37.4 bits (85), Expect = 0.26
Identities = 63/343 (18%), Positives = 129/343 (37%), Gaps = 31/343 (9%)
Query: 880 KSRIYDNFNHMPEMIRTIKKNELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEET 939
KS + D + E + K E E + ++++ E+ + EE
Sbjct: 252 KSEMEDALTAIAEELAVSVKEEEKEEEAEGKEEEQEAEEEVAAAKKSPVKATTPEIKEEE 311
Query: 940 DTEEEIDTEEDDFSDDEIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEK 999
+EE EE++ +DE S Q EK + + + E+++ A+ + + K
Sbjct: 312 GEKEEEGQEEEEEEEDE--GVKSDQAEEGGSEKEGSSKNEGEQEEGETEAEGEVEEAEAK 369
Query: 1000 SNIKDHDLSEDEIFVPDEIKKLSSAQIGIHEKSNTEDVIFTNPNRTTTTTKPYYSQALFP 1059
K + SE+ V + + ++ A++G EK+ + P KP
Sbjct: 370 EEKKTEEKSEE---VAAKEEPVTEAKVGKPEKAKSPV-----PKSPVEEVKPKAEATAGK 421
Query: 1060 NPPRKIRKFRPTLKKQRFVEVSKTESLQRRSLEETKSDGVKSLEVSETESLQQQYLAVVS 1119
++ + KK+ E K E ++++ E+ K K E E ++ +
Sbjct: 422 GEQKEEEEKVEEEKKKAAKESPKEEKVEKKE-EKPKDVPKKKAESPVKEEAAEEAATITK 480
Query: 1120 GMRNLVLTETKE----------KHRFVEVGVSETETAHRWSLEETKSD----------GV 1159
+ + ETKE K + E G SE E + + S K D
Sbjct: 481 PTKVGLEKETKEGEKPLQQEKEKEKAGEEGGSEEEGSDQGSKRAKKEDIAVNGEGEGKEE 540
Query: 1160 KRFEVSETESLQQQDLALLSSGMQNMVLTDKKEKENNMTRYII 1202
+ E E S ++++ ++++G+ +KK + + + ++
Sbjct: 541 EEPETKEKGSGREEEKGVVTNGLDLSPADEKKGGDRSEEKVVV 583
>GARP_PLAFF (P13816) Glutamic acid-rich protein precursor
Length = 678
Score = 37.0 bits (84), Expect = 0.33
Identities = 34/177 (19%), Positives = 75/177 (42%), Gaps = 5/177 (2%)
Query: 835 IESESVEIPQVNLFLNGDECPLHWELNYENVLLPSKHTFLFDLGLKSRIYDNFNHMPEMI 894
+ES + PQ L ++ L + E L + + +G + + NH +M
Sbjct: 473 VESRPLSQPQCKLIDEPEQLTLMDKSKVEEKNLSIQEQLIGTIGRVNVVPRRDNHKKKMA 532
Query: 895 RTIKKNELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEEEIDTEEDDFSD 954
+ + +D E+D S++ ++ + +Q +E + EEE + EE++ +
Sbjct: 533 KIEEAELQKQKHVDKEEDKKEESKEVQEESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEE 592
Query: 955 DEIFTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEKSNIKDHDLSEDE 1011
+E E + +++ ++ D EED+ A+ +E + +D D +D+
Sbjct: 593 EE---EEEEEEEEEEEDEDEEDEDDAEEDE--DDAEEDEDDAEEDDDEEDDDEEDDD 644
Score = 33.9 bits (76), Expect = 2.8
Identities = 30/138 (21%), Positives = 61/138 (43%), Gaps = 21/138 (15%)
Query: 898 KKNELNHVEIDWEKDDLHLSEDEIKKLRRLQMGIHVSWTEETDTEEEIDTEEDDFSDDEI 957
+ E+ E + E+D+ E+E ++ + EE + E+E + +EDD +DE
Sbjct: 561 ESKEVQEDEEEVEEDEEEEEEEEEEEEEEEEEEEEEEEEEEEEEEDEDEEDEDDAEEDED 620
Query: 958 FTESSSQMVIHYQEKSNTEQIDTEEDDFYRSAQMGIHVVKEKSNIKDHDLSEDEIFVPDE 1017
E +++ + E+ D EEDD E+ + +D D E++ +E
Sbjct: 621 DAE---------EDEDDAEEDDDEEDD------------DEEDDDEDEDEDEEDEEEEEE 659
Query: 1018 IKKLSSAQIGIHEKSNTE 1035
++ S +I + + N +
Sbjct: 660 EEEESEKKIKRNLRKNAK 677
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.346 0.151 0.503
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 125,202,436
Number of Sequences: 164201
Number of extensions: 4802507
Number of successful extensions: 28168
Number of sequences better than 10.0: 105
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 106
Number of HSP's that attempted gapping in prelim test: 27788
Number of HSP's gapped (non-prelim): 309
length of query: 1245
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1123
effective length of database: 39,941,532
effective search space: 44854340436
effective search space used: 44854340436
T: 11
A: 40
X1: 15 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.7 bits)
S2: 72 (32.3 bits)
Medicago: description of AC138133.7


