

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.14 + phase: 0
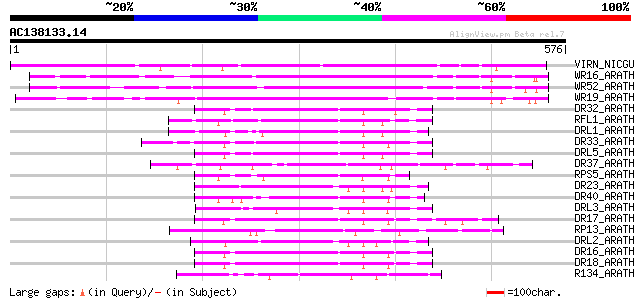
(576 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VIRN_NICGU (Q40392) TMV resistance protein N 319 1e-86
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 187 6e-47
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 166 2e-40
WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY ... 164 8e-40
DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730 83 2e-15
RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like p... 82 3e-15
DRL1_ARATH (P60838) Probable disease resistance protein At1g12280 81 6e-15
DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740 78 5e-14
DRL5_ARATH (Q9C8K0) Probable disease resistance protein At1g51480 78 7e-14
DR37_ARATH (Q9LVT4) Probable disease resistance protein At5g47250 75 5e-13
RPS5_ARATH (O64973) Disease resistance protein RPS5 (Resistance ... 73 2e-12
DR23_ARATH (O82484) Putative disease resistance protein At4g10780 72 3e-12
DR40_ARATH (Q8RXS5) Probable disease resistance protein At5g6302... 72 4e-12
DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890 72 5e-12
DR17_ARATH (O64790) Putative disease resistance protein At1g61300 71 7e-12
RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance... 70 2e-11
DRL2_ARATH (P60839) Probable disease resistance protein At1g12290 69 3e-11
DR16_ARATH (O22727) Putative disease resistance protein At1g61190 68 6e-11
DR18_ARATH (O64789) Putative disease resistance protein At1g61310 67 1e-10
R134_ARATH (Q38834) Putative disease resistance RPP13-like prote... 66 2e-10
>VIRN_NICGU (Q40392) TMV resistance protein N
Length = 1144
Score = 319 bits (817), Expect = 1e-86
Identities = 212/590 (35%), Positives = 330/590 (55%), Gaps = 46/590 (7%)
Query: 1 MATQSPSS-FTYQVFLSFRGADTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLK 59
MA+ S SS ++Y VFLSFRG DTR FT +LY+ L DKGI TF DD L+ G I L
Sbjct: 1 MASSSSSSRWSYDVFLSFRGEDTRKTFTSHLYEVLNDKGIKTFQDDKRLEYGATIPGELC 60
Query: 60 NAIEKSRIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRY 119
AIE+S+ I VFSENYA+S +CL+ELV I C V+P+F VDP+ VR+ +
Sbjct: 61 KAIEESQFAIVVFSENYATSRWCLNELVKIMECKTRFKQTVIPIFYDVDPSHVRNQKESF 120
Query: 120 GEALAVHKKKFQNDKDNTERLQQWKEALSQAANLSG-----------------QHYKHGI 162
+A H+ K+ KD+ E +Q+W+ AL++AANL G +
Sbjct: 121 AKAFEEHETKY---KDDVEGIQRWRIALNEAANLKGSCDNRDKTDADCIRQIVDQISSKL 177
Query: 163 SREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFI--- 219
+ L + VG+ + ++ ++ L E + V ++G++G GG+GK+T+A+AI++ +
Sbjct: 178 CKISLSYLQNIVGIDTHLEKIESLL-EIGINGVRIMGIWGMGGVGKTTIARAIFDTLLGR 236
Query: 220 ---ADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRK 276
+ QF+ CFL++++ N + LQ LL + +R G + RL K
Sbjct: 237 MDSSYQFDGACFLKDIKENKR--GMHSLQNALLSELLREKANYNNEEDGKHQMASRLRSK 294
Query: 277 KILLILDDVDKLDQ-LEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATE 335
K+L++LDD+D D LE LAG LDWFG GSR+IITTR+KHL++ + I + V L E
Sbjct: 295 KVLIVLDDIDNKDHYLEYLAGDLDWFGNGSRIIITTRDKHLIEKNDI--IYEVTALPDHE 352
Query: 336 ALELLRWMAFKENVPSSH-EDILNRALTYASGLPLAIVIIGSNLVGRSVQDSMSTLDGYE 394
+++L + AF + VP+ + E + + YA GLPLA+ + GS L + + S ++ +
Sbjct: 353 SIQLFKQHAFGKEVPNENFEKLSLEVVNYAKGLPLALKVWGSLLHNLRLTEWKSAIEHMK 412
Query: 395 EIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHA-HYGHCIVHHVAV 453
I LK+SYD LE ++Q +FLDIAC +G + + +IL + H G + + +
Sbjct: 413 NNSYSGIIDKLKISYDGLEPKQQEMFLDIACFLRGEEKDYILQILESCHIG--AEYGLRI 470
Query: 454 LAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDI------GTRK 507
L +KSL+ +Y+ V +HDLI+DMGK +V + +PGERSRLW +++ T
Sbjct: 471 LIDKSLVFISEYNQ-VQMHDLIQDMGKYIVNFQK--DPGERSRLWLAKEVEEVMSNNTGT 527
Query: 508 IKMINMKFPSMESDIDWNGNAFEKMTNLKTFITENGHHSKSLEYLPSSLR 557
+ M + S S + ++ A + M L+ F +++YLP++LR
Sbjct: 528 MAMEAIWVSSYSSTLRFSNQAVKNMKRLRVFNMGRSSTHYAIDYLPNNLR 577
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 187 bits (475), Expect = 6e-47
Identities = 159/577 (27%), Positives = 274/577 (46%), Gaps = 77/577 (13%)
Query: 21 DTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSRIFIPVFSENYASSS 80
+ R+ F +L KAL KG+ D+D D ++ ++ +E++R+ + + N S
Sbjct: 15 EVRYSFVSHLSKALQRKGVNDVFIDSD----DSLSNESQSMVERARVSVMILPGNRTVS- 69
Query: 81 FCLDELVHITHCYDTKGCLVLPVFIGV--DPTDVRHHTGRYGEALAVHKKKFQNDKDNTE 138
LD+LV + C K +V+PV GV T+ G + H +K +D
Sbjct: 70 --LDKLVKVLDCQKNKDQVVVPVLYGVRSSETEWLSALDSKGFSSVHHSRKECSDS---- 123
Query: 139 RLQQWKEALSQAANLSGQHYKHGISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMV 198
Q KE + + GI + L++ K + K ++ V
Sbjct: 124 --QLVKETVRDVYEKLFYMERIGIYSKLLEIEK---------------MINKQPLDIRCV 166
Query: 199 GLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIK 258
G++G GIGK+TLAKA+++ ++ +F+ CF+E+ + L E+ LK +
Sbjct: 167 GIWGMPGIGKTTLAKAVFDQMSGEFDAHCFIEDYTKAIQEKGVYCLLEEQFLKE---NAG 223
Query: 259 LGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLK 318
G + +++ RL K++L++LDDV +E+ GG DWFGP S +IIT+++K + +
Sbjct: 224 ASGTVTKLSLLRDRLNNKRVLVVLDDVRSPLVVESFLGGFDWFGPKSLIIITSKDKSVFR 283
Query: 319 IHGIESTHAVEGLNATEALELLRWMAFKENVPSSH-EDILNRALTYASGLPLAIVIIGSN 377
+ + + V+GLN EAL+L A +++ + ++ + + YA+G PLA+ + G
Sbjct: 284 LCRVNQIYEVQGLNEKEALQLFSLCASIDDMAEQNLHEVSMKVIKYANGHPLALNLYGRE 343
Query: 378 LVGRSVQDSMS-TLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVK 436
L+G+ M +E P +K SYD+L E+++FLDIAC F+G V
Sbjct: 344 LMGKKRPPEMEIAFLKLKECPPAIFVDAIKSSYDTLNDREKNIFLDIACFFQGENVDYVM 403
Query: 437 EILHAHYGHCIVHHVA--VLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGER 494
++L G HV VL EKSL+ ++ V +H+LI+D+G++++ +E+ + R
Sbjct: 404 QLLE---GCGFFPHVGIDVLVEKSLV--TISENRVRMHNLIQDVGRQIINRET-RQTKRR 457
Query: 495 SRLW-----------------------FERDIGTRKIKMINMKFPSMESDIDWNGNAFEK 531
SRLW FER +I+ + + ++ DI AF+
Sbjct: 458 SRLWEPCSIKYLLEDKEQNENEEQKTTFERAQVPEEIEGMFLDTSNLSFDI--KHVAFDN 515
Query: 532 MTNLKTFITENG----HH-----SKSLEYLPSSLRVM 559
M NL+ F + HH SL LP+ LR++
Sbjct: 516 MLNLRLFKIYSSNPEVHHVNNFLKGSLSSLPNVLRLL 552
Score = 37.7 bits (86), Expect = 0.082
Identities = 27/97 (27%), Positives = 50/97 (50%), Gaps = 9/97 (9%)
Query: 395 EIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFK----GCKWPEVKEILHAHYGHCIVHH 450
E+ E + +L+V Y L++ +++FL IA F G P + I+ + +
Sbjct: 1041 EVSGNEDEEVLRVRYAGLQEIYKALFLYIAGLFNDEDVGLVAPLIANIIDMD----VSYG 1096
Query: 451 VAVLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQES 487
+ VLA +SL+ + + + +H L+ MGKE++ ES
Sbjct: 1097 LKVLAYRSLI-RVSSNGEIVMHYLLRQMGKEILHTES 1132
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein
RRS1) (Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 166 bits (420), Expect = 2e-40
Identities = 157/579 (27%), Positives = 268/579 (46%), Gaps = 77/579 (13%)
Query: 21 DTRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKSRIFIPVFSENYASSS 80
+ R+ F +L +AL KGI + D D+ D + + IEK+ + + V N S
Sbjct: 18 EVRYSFVSHLSEALRRKGINNVVVDVDID--DLLFKESQAKIEKAGVSVMVLPGNCDPSE 75
Query: 81 FCLDELVHITHCY-DTKGCLVLPVFIGVDPTDVRHHTGRYGEALAVHKKKFQNDKDNTER 139
LD+ + C + K V+ V YG++L + + D R
Sbjct: 76 VWLDKFAKVLECQRNNKDQAVVSVL--------------YGDSLLRDQWLSELDFRGLSR 121
Query: 140 LQQWKEALSQAANLSGQHYKHGISREPLDVAKYP--VGLQSRVQHVKGHLDEKSDDEVHM 197
+ Q ++ S + + I R+ + Y +G+ S++ ++ ++ K +
Sbjct: 122 IHQSRKECSDSILVEE------IVRDVYETHFYVGRIGIYSKLLEIENMVN-KQPIGIRC 174
Query: 198 VGLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDI 257
VG++G GIGK+TLAKA+++ ++ F+ CF+E+ + L L E+ LL I
Sbjct: 175 VGIWGMPGIGKTTLAKAVFDQMSSAFDASCFIEDYDKSIHEKGLYCLLEEQLLPGNDATI 234
Query: 258 KLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLL 317
+ ++ RL K++L++LDDV E+ G DW GPGS +IIT+R+K +
Sbjct: 235 ------MKLSSLRDRLNSKRVLVVLDDVCNALVAESFLEGFDWLGPGSLIIITSRDKQVF 288
Query: 318 KIHGIESTHAVEGLNATEA--LELLRWMAFKENVPSSHEDILNRALTYASGLPLAIVIIG 375
++ GI + V+GLN EA L LL ++ + ++ R ++YA+G PLAI + G
Sbjct: 289 RLCGINQIYEVQGLNEKEARQLFLLSASIMEDMGEQNLHELSVRVISYANGNPLAISVYG 348
Query: 376 SNLVGRSVQDSMST-LDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPE 434
L G+ M T + P +I K SYD+L E+++FLDIAC F+G
Sbjct: 349 RELKGKKKLSEMETAFLKLKRRPPFKIVDAFKSSYDTLSDNEKNIFLDIACFFQG---EN 405
Query: 435 VKEILHAHYGHCIVHHVA--VLAEKSLMDHLKYDSYVTLHDLIEDMGKEVVRQESPDEPG 492
V ++ G HV VL +K L+ ++ V LH L +D+G+E++ E+ +
Sbjct: 406 VNYVIQLLEGCGFFPHVEIDVLVDKCLV--TISENRVWLHKLTQDIGREIINGETV-QIE 462
Query: 493 ERSRLW-----------------------FERDIGTRKIKMINMKFPSMESDIDWNGNAF 529
R RLW F+R G+ +I+ + + ++ D+ +AF
Sbjct: 463 RRRRLWEPWSIKYLLEYNEHKANGEPKTTFKRAQGSEEIEGLFLDTSNLRFDL--QPSAF 520
Query: 530 EKMTN---LKTFITENGHH------SKSLEYLPSSLRVM 559
+ M N LK + + H + SL LP+ LR++
Sbjct: 521 KNMLNLRLLKIYCSNPEVHPVINFPTGSLHSLPNELRLL 559
Score = 35.0 bits (79), Expect = 0.53
Identities = 25/84 (29%), Positives = 45/84 (52%), Gaps = 2/84 (2%)
Query: 404 ILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAVLAEKSLMDHL 463
+L+VSYD L++ ++ +FL IA F V ++ A + + VLA+ SL+ +
Sbjct: 1088 VLRVSYDDLQEMDKVLFLYIASLFNDEDVDFVAPLI-AGIDLDVSSGLKVLADVSLIS-V 1145
Query: 464 KYDSYVTLHDLIEDMGKEVVRQES 487
+ + +H L MGKE++ +S
Sbjct: 1146 SSNGEIVMHSLQRQMGKEILHGQS 1169
>WR19_ARATH (Q9SZ67) Probable WRKY transcription factor 19 (WRKY
DNA-binding protein 19)
Length = 1895
Score = 164 bits (414), Expect = 8e-40
Identities = 161/594 (27%), Positives = 273/594 (45%), Gaps = 88/594 (14%)
Query: 7 SSFTYQVFLSFRGAD-TRHGFTGNLYKALTDKGIYTFIDDNDLQRGDEITPSLKNAIEKS 65
SS Y V + + AD + F +L +L +GI + N++ +A+ K
Sbjct: 664 SSKDYDVVIRYGRADISNEDFISHLRASLCRRGISVYEKFNEV-----------DALPKC 712
Query: 66 RIFIPVFSENYASSSFCLDELVHITHCYDTKGCLVLPVFIGVDPTDVRHHTGRYGEALAV 125
R+ I V + Y S+ L++I T+ +V P+F + P D ++ Y
Sbjct: 713 RVLIIVLTSTYVPSN-----LLNILEHQHTEDRVVYPIFYRLSPYDFVCNSKNY------ 761
Query: 126 HKKKFQNDKDNTERLQQWKEALSQAANLSGQHYKHGISREPLDVAKYP------------ 173
++ + D+ ++W+ AL + + G E +D
Sbjct: 762 -ERFYLQDEP-----KKWQAALKEITQMPGYTLTDKSESELIDEIVRDALKVLCSADKVN 815
Query: 174 -VGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENV 232
+G+ +V+ + L +S D V +G++GT GIGK+T+A+ I+ I+ Q+E L+++
Sbjct: 816 MIGMDMQVEEILSLLCIESLD-VRSIGIWGTVGIGKTTIAEEIFRKISVQYETCVVLKDL 874
Query: 233 RVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQ-GIPIIKQRLCRKKILLILDDVDKLDQL 291
++E L + + ++ + +S ++ RL RK+IL+ILDDV+ +
Sbjct: 875 HKEVEVKGHDAVRENFLSEVLEVEPHVIRISDIKTSFLRSRLQRKRILVILDDVNDYRDV 934
Query: 292 EALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELL-RWMAFKENVP 350
+ G L++FGPGSR+I+T+RN+ + + I+ + V+ L+ ++L LL R P
Sbjct: 935 DTFLGTLNYFGPGSRIIMTSRNRRVFVLCKIDHVYEVKPLDIPKSLLLLDRGTCQIVLSP 994
Query: 351 SSHEDILNRALTYASGLPLAIVIIGS--NLVGRSVQDSMSTLDGYEEIPNKEIQRILKVS 408
++ + + +++G P + + S + Q+ +T Y I I + S
Sbjct: 995 EVYKTLSLELVKFSNGNPQVLQFLSSIDREWNKLSQEVKTTSPIY-------IPGIFEKS 1047
Query: 409 YDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIVHHVAV--LAEKSLMDHLKYD 466
L+ E+ +FLDIAC F V +L G HV L +KSL+ +
Sbjct: 1048 CCGLDDNERGIFLDIACFFNRIDKDNVAMLLD---GCGFSAHVGFRGLVDKSLLT-ISQH 1103
Query: 467 SYVTLHDLIEDMGKEVVRQESPDEPGERSRLW--------FERDIGTRKIK-----MINM 513
+ V + I+ G+E+VRQES D PG+RSRLW F D GT I+ M+N+
Sbjct: 1104 NLVDMLSFIQATGREIVRQESADRPGDRSRLWNADYIRHVFINDTGTSAIEGIFLDMLNL 1163
Query: 514 KFPSMESDIDWNGNAFEKMTNLKTF-----ITENGH---HSKSLEYLPSSLRVM 559
KF D N N FEKM NL+ E H + LEYLPS LR++
Sbjct: 1164 KF-------DANPNVFEKMCNLRLLKLYCSKAEEKHGVSFPQGLEYLPSKLRLL 1210
>DR32_ARATH (Q9FG91) Probable disease resistance protein At5g43730
Length = 848
Score = 83.2 bits (204), Expect = 2e-15
Identities = 84/263 (31%), Positives = 134/263 (50%), Gaps = 28/263 (10%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAIYNFIAD---QFEVLCFLENVRVNSTSDNLKHLQEKL 248
DDE+ +GLYG GGIGK+TL +++ N + +F+V+ ++ V S L+ +Q+++
Sbjct: 169 DDEIRTLGLYGMGGIGKTTLLESLNNKFVELESEFDVVIWV----VVSKDFQLEGIQDQI 224
Query: 249 LLKTVRLD--IKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSR 306
L + +R D + S+ +I L RKK +L+LDD+ L + GS+
Sbjct: 225 LGR-LRPDKEWERETESKKASLINNNLKRKKFVLLLDDLWSEVDLIKIGVPPPSRENGSK 283
Query: 307 VIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYAS- 365
++ TTR+K + K + V+ L+ EA EL R + + + SH+DI A A+
Sbjct: 284 IVFTTRSKEVCKHMKADKQIKVDCLSPDEAWELFR-LTVGDIILRSHQDIPALARIVAAK 342
Query: 366 --GLPLAIVIIGSNLVGR-SVQDSMSTLDGYEEIPNK------EIQRILKVSYDSLEKEE 416
GLPLA+ +IG +V + +VQ+ ++ +K I ILK SYDSL+ E
Sbjct: 343 CHGLPLALNVIGKAMVCKETVQEWRHAINVLNSPGHKFPGMEERILPILKFSYDSLKNGE 402
Query: 417 QSVFLDIACCFKGCK-WPEVKEI 438
I CF C +PE EI
Sbjct: 403 ------IKLCFLYCSLFPEDFEI 419
>RFL1_ARATH (Q8L3R3) Disease resistance protein RFL1 (RPS5-like
protein 1) (pNd13/pNd14)
Length = 885
Score = 82.4 bits (202), Expect = 3e-15
Identities = 93/292 (31%), Positives = 137/292 (46%), Gaps = 35/292 (11%)
Query: 165 EPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAI---YNFIAD 221
E L + VG S + V L E D+V +VGLYG GG+GK+TL I ++ +
Sbjct: 149 EELPIQSTIVGQDSMLDKVWNCLME---DKVWIVGLYGMGGVGKTTLLTQINNKFSKLGG 205
Query: 222 QFEVLCFLENVRVNSTSDNL-KHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILL 280
F+V+ ++ V N+T + K + EKL L D K +Q I L RKK +L
Sbjct: 206 GFDVVIWVV-VSKNATVHKIQKSIGEKLGLVGKNWDEK--NKNQRALDIHNVLRRKKFVL 262
Query: 281 ILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELL 340
+LDD+ + +L+ + G +V TT +K + G+++ + L+ A +LL
Sbjct: 263 LLDDIWEKVELKVIGVPYPSGENGCKVAFTTHSKEVCGRMGVDNPMEISCLDTGNAWDLL 322
Query: 341 RWMAFKENVPSSHEDILNRALTYAS---GLPLAIVIIGSNL-VGRSVQD---------SM 387
+ EN SH DI A + GLPLA+ +IG + R++Q+ S
Sbjct: 323 K-KKVGENTLGSHPDIPQLARKVSEKCCGLPLALNVIGETMSFKRTIQEWRHATEVLTSA 381
Query: 388 STLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCK-WPEVKEI 438
+ G E+ EI ILK SYDSL E D CF C +PE EI
Sbjct: 382 TDFSGMED----EILPILKYSYDSLNGE------DAKSCFLYCSLFPEDFEI 423
>DRL1_ARATH (P60838) Probable disease resistance protein At1g12280
Length = 894
Score = 81.3 bits (199), Expect = 6e-15
Identities = 92/290 (31%), Positives = 137/290 (46%), Gaps = 39/290 (13%)
Query: 165 EPLDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQ-- 222
E + + VG ++ ++ V L E D+ +VGLYG GG+GK+TL I N +++
Sbjct: 149 EEMPIQPTIVGQETMLERVWTRLTEDGDE---IVGLYGMGGVGKTTLLTRINNKFSEKCS 205
Query: 223 -FEVLCFLENVRVNSTSDNLKHLQEKLLLKTVRLDIKLGG-------VSQGIPIIKQRLC 274
F V+ ++ V S S ++ +Q + RLD LGG +Q I L
Sbjct: 206 GFGVVIWV----VVSKSPDIHRIQGDI---GKRLD--LGGEEWDNVNENQRALDIYNVLG 256
Query: 275 RKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGIESTHAVEGLNAT 334
++K +L+LDD+ + LE L G +V+ TTR++ + ++ V L
Sbjct: 257 KQKFVLLLDDIWEKVNLEVLGVPYPSRQNGCKVVFTTRSRDVCGRMRVDDPMEVSCLEPN 316
Query: 335 EALELLRWMAFKENVPSSHEDILNRALTYAS---GLPLAIVIIGSNLV-GRSVQD---SM 387
EA EL + M EN H DI A A GLPLA+ +IG + R VQ+ ++
Sbjct: 317 EAWELFQ-MKVGENTLKGHPDIPELARKVAGKCCGLPLALNVIGETMACKRMVQEWRNAI 375
Query: 388 STLDGY-EEIPNKE-IQRILKVSYDSLEKEEQSVFLDIACCFKGCK-WPE 434
L Y E P E I ILK SYD+L KE+ + CF C +PE
Sbjct: 376 DVLSSYAAEFPGMEQILPILKYSYDNLNKEQ------VKPCFLYCSLFPE 419
>DR33_ARATH (Q9FG90) Probable disease resistance protein At5g43740
Length = 862
Score = 78.2 bits (191), Expect = 5e-14
Identities = 90/319 (28%), Positives = 154/319 (48%), Gaps = 35/319 (10%)
Query: 137 TERLQQWKEALSQAA-NLSGQHYKHGISREPLDVAKYPVGLQSRVQHVKGHLDEKSDDEV 195
++ L++ KE LS+ + Q H + ++ + VGL V+ L +DE+
Sbjct: 118 SKMLEEVKELLSKKDFRMVAQEIIHKVEKKLIQTT---VGLDKLVEMAWSSL---MNDEI 171
Query: 196 HMVGLYGTGGIGKSTLAKAIYNFIAD---QFEVLCFLENVRVNSTSDNLKHLQEKLL--L 250
+GLYG GG+GK+TL +++ N + +F+V+ ++ V S + +Q+++L L
Sbjct: 172 GTLGLYGMGGVGKTTLLESLNNKFVELESEFDVVIWV----VVSKDFQFEGIQDQILGRL 227
Query: 251 KTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIIT 310
++ + + + S+ +I L RKK +L+LDD+ + + GS+++ T
Sbjct: 228 RSDK-EWERETESKKASLIYNNLERKKFVLLLDDLWSEVDMTKIGVPPPTRENGSKIVFT 286
Query: 311 TRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYAS---GL 367
TR+ + K + V L+ EA EL R + + + SH+DI A A+ GL
Sbjct: 287 TRSTEVCKHMKADKQIKVACLSPDEAWELFR-LTVGDIILRSHQDIPALARIVAAKCHGL 345
Query: 368 PLAIVIIGSNL-VGRSVQDSMSTLD----GYEEIPNKE--IQRILKVSYDSLEKEEQSVF 420
PLA+ +IG + ++Q+ ++ E P E I ILK SYDSL+ E
Sbjct: 346 PLALNVIGKAMSCKETIQEWSHAINVLNSAGHEFPGMEERILPILKFSYDSLKNGE---- 401
Query: 421 LDIACCFKGCK-WPEVKEI 438
I CF C +PE EI
Sbjct: 402 --IKLCFLYCSLFPEDSEI 418
>DRL5_ARATH (Q9C8K0) Probable disease resistance protein At1g51480
Length = 854
Score = 77.8 bits (190), Expect = 7e-14
Identities = 79/264 (29%), Positives = 132/264 (49%), Gaps = 29/264 (10%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAIYNFIAD---QFEVLCFLENVRVNSTSDNLKHLQEKL 248
+DE+ + L+G GG+GK+TL I N + +F+V+ ++ V S L+ +Q+++
Sbjct: 170 NDEIRTLCLHGMGGVGKTTLLACINNKFVELESEFDVVIWV----VVSKDFQLEGIQDQI 225
Query: 249 LLKTVRLDIKLGGVSQG--IPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSR 306
L + +RLD + ++ +I L RKK +L+LDD+ L + G++
Sbjct: 226 LGR-LRLDKEWERETENKKASLINNNLKRKKFVLLLDDLWSEVDLNKIGVPPPTRENGAK 284
Query: 307 VIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYAS- 365
++ T R+K + K + V L+ EA EL R + + + SSHEDI A A+
Sbjct: 285 IVFTKRSKEVSKYMKADMQIKVSCLSPDEAWELFR-ITVDDVILSSHEDIPALARIVAAK 343
Query: 366 --GLPLAIVIIGSNLVGR-SVQD-----SMSTLDGYEEIPNKE--IQRILKVSYDSLEKE 415
GLPLA+++IG + + ++Q+ ++ + P E I +LK SYDSL+
Sbjct: 344 CHGLPLALIVIGEAMACKETIQEWHHAINVLNSPAGHKFPGMEERILLVLKFSYDSLKNG 403
Query: 416 EQSVFLDIACCFKGCK-WPEVKEI 438
E I CF C +PE EI
Sbjct: 404 E------IKLCFLYCSLFPEDFEI 421
>DR37_ARATH (Q9LVT4) Probable disease resistance protein At5g47250
Length = 843
Score = 75.1 bits (183), Expect = 5e-13
Identities = 102/440 (23%), Positives = 189/440 (42%), Gaps = 66/440 (15%)
Query: 147 LSQAANLSGQHYKHGISREPLDVAKY-----PVGLQSRVQHVKGHLDEKSDDEVHMVGLY 201
L++ +LSG+ ++ + P V + VGL + ++ L + DE M+G++
Sbjct: 126 LTEVKSLSGKDFQEVTEQPPPPVVEVRLCQQTVGLDTTLEKTWESLRK---DENRMLGIF 182
Query: 202 GTGGIGKSTLAKAIYN---FIADQFEVLCFLENVRVNSTSDNLKHLQEKLLL-----KTV 253
G GG+GK+TL I N ++D ++V+ ++E+ + + E+L + T
Sbjct: 183 GMGGVGKTTLLTLINNKFVEVSDDYDVVIWVESSKDADVGKIQDAIGERLHICDNNWSTY 242
Query: 254 RLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRN 313
K +S+ + +K R +L+LDD+ + + A G+ G +V+ TTR+
Sbjct: 243 SRGKKASEISRVLRDMKPR-----FVLLLDDL--WEDVSLTAIGIPVLGKKYKVVFTTRS 295
Query: 314 KHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYASGLPLAIVI 373
K + + V+ L+ +A +L + + + DI + + GLPLA+ +
Sbjct: 296 KDVCSVMRANEDIEVQCLSENDAWDLFDMKVHCDGL-NEISDIAKKIVAKCCGLPLALEV 354
Query: 374 IGSNLVGRSV----QDSMSTLDGYE---EIPNKEIQRILKVSYDSLEKEEQSVFLDIACC 426
I + +S + ++ TL+ Y + K I ++LK+SYD L+ + FL A
Sbjct: 355 IRKTMASKSTVIQWRRALDTLESYRSEMKGTEKGIFQVLKLSYDYLKTKNAKCFLYCA-L 413
Query: 427 FKGCKWPEVKEILHAHYGHCIVHH--------------VAVLAEKSLMDHLKYDSYVTLH 472
F + + E++ G + + L L+ L+ + V +H
Sbjct: 414 FPKAYYIKQDELVEYWIGEGFIDEKDGRERAKDRGYEIIDNLVGAGLL--LESNKKVYMH 471
Query: 473 DLIEDMGKEVVRQESPDEPGER---------SRLWFERDIGT-RKIKMINMKFPSMESDI 522
D+I DM +V S GER S+L D T K+ + N + ++ D
Sbjct: 472 DMIRDMALWIV---SEFRDGERYVVKTDAGLSQLPDVTDWTTVTKMSLFNNEIKNIPDDP 528
Query: 523 DWNGNAFEKMTNLKTFITEN 542
+ F TNL T +N
Sbjct: 529 E-----FPDQTNLVTLFLQN 543
>RPS5_ARATH (O64973) Disease resistance protein RPS5 (Resistance to
Pseudomonas syringae protein 5) (pNd3/pNd10)
Length = 889
Score = 73.2 bits (178), Expect = 2e-12
Identities = 74/248 (29%), Positives = 119/248 (47%), Gaps = 38/248 (15%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAI---YNFIADQFEVLCFLENVRVNSTSDNLKHLQEKL 248
+D ++GLYG GG+GK+TL I ++ I D+F+V+ ++ V S S ++ +Q +
Sbjct: 173 EDGSGILGLYGMGGVGKTTLLTKINNKFSKIDDRFDVVIWV----VVSRSSTVRKIQRDI 228
Query: 249 LLKTVRLDIKLGGV-------SQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWF 301
K + LGG+ +Q I L R+K +L+LDD+ + L+A+
Sbjct: 229 AEK-----VGLGGMEWSEKNDNQIAVDIHNVLRRRKFVLLLDDIWEKVNLKAVGVPYPSK 283
Query: 302 GPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRAL 361
G +V TTR++ + G++ V L E+ +L + M +N SH DI A
Sbjct: 284 DNGCKVAFTTRSRDVCGRMGVDDPMEVSCLQPEESWDLFQ-MKVGKNTLGSHPDIPGLAR 342
Query: 362 TYA---SGLPLAIVIIGSNLV-GRSVQDSMSTLD----------GYEEIPNKEIQRILKV 407
A GLPLA+ +IG + R+V + +D G E+ EI +LK
Sbjct: 343 KVARKCRGLPLALNVIGEAMACKRTVHEWCHAIDVLTSSAIDFSGMED----EILHVLKY 398
Query: 408 SYDSLEKE 415
SYD+L E
Sbjct: 399 SYDNLNGE 406
>DR23_ARATH (O82484) Putative disease resistance protein At4g10780
Length = 892
Score = 72.4 bits (176), Expect = 3e-12
Identities = 78/260 (30%), Positives = 119/260 (45%), Gaps = 29/260 (11%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKL--L 249
DD V +GLYG GG+GK+TL I+N + D + + V V+S + +QE +
Sbjct: 170 DDGVGTMGLYGMGGVGKTTLLTQIHNTLHDTKNGVDIVIWVVVSSDL-QIHKIQEDIGEK 228
Query: 250 LKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVII 309
L + + SQ I L +K+ +L+LDD+ K L + +V+
Sbjct: 229 LGFIGKEWNKKQESQKAVDILNCLSKKRFVLLLDDIWKKVDLTKIGIPSQTRENKCKVVF 288
Query: 310 TTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVP----SSHEDILNRALTYAS 365
TTR+ + G+ V+ L+ +A W F+E V SH DIL A A
Sbjct: 289 TTRSLDVCARMGVHDPMEVQCLSTNDA-----WELFQEKVGQISLGSHPDILELAKKVAG 343
Query: 366 ---GLPLAIVIIGSNLVG-RSVQD---SMSTLDGYE---EIPNKEIQRILKVSYDSLEKE 415
GLPLA+ +IG + G R+VQ+ ++ L Y + I ILK SYD+L +
Sbjct: 344 KCRGLPLALNVIGETMAGKRAVQEWHHAVDVLTSYAAEFSGMDDHILLILKYSYDNLNDK 403
Query: 416 EQSVFLDIACCFKGCK-WPE 434
+ CF+ C +PE
Sbjct: 404 H------VRSCFQYCALYPE 417
>DR40_ARATH (Q8RXS5) Probable disease resistance protein At5g63020
(pNd11)
Length = 888
Score = 72.0 bits (175), Expect = 4e-12
Identities = 75/263 (28%), Positives = 122/263 (45%), Gaps = 44/263 (16%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAI---YNFIADQFEVLCFL---ENVRVNSTSD----NL 241
+DE+ ++GL+G GG+GK+TL I ++ + +F+++ ++ + +++ D L
Sbjct: 171 EDEIGILGLHGMGGVGKTTLLSHINNRFSRVGGEFDIVIWIVVSKELQIQRIQDEIWEKL 230
Query: 242 KHLQEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWF 301
+ EK KT DIK I L K+ +L+LDD+ L +
Sbjct: 231 RSDNEKWKQKTE--DIKASN-------IYNVLKHKRFVLLLDDIWSKVDLTEVGVPFPSR 281
Query: 302 GPGSRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRAL 361
G +++ TTR K + G++S V L +A +L E SH +I A
Sbjct: 282 ENGCKIVFTTRLKEICGRMGVDSDMEVRCLAPDDAWDLFT-KKVGEITLGSHPEIPTVAR 340
Query: 362 TYAS---GLPLAIVIIGSNLV-GRSVQDSMSTLD----------GYEEIPNKEIQRILKV 407
T A GLPLA+ +IG + R+VQ+ S +D G E+ EI ILK
Sbjct: 341 TVAKKCRGLPLALNVIGETMAYKRTVQEWRSAIDVLTSSAAEFSGMED----EILPILKY 396
Query: 408 SYDSLEKEEQSVFLDIACCFKGC 430
SYD+L+ E+ + CF+ C
Sbjct: 397 SYDNLKSEQ------LKLCFQYC 413
>DRL3_ARATH (Q9LMP6) Probable disease resistance protein At1g15890
Length = 921
Score = 71.6 bits (174), Expect = 5e-12
Identities = 79/265 (29%), Positives = 120/265 (44%), Gaps = 34/265 (12%)
Query: 193 DEVHMVGLYGTGGIGKSTLAKAIYNFIADQFEVLCFLENVRVNSTSDNLKHLQEKLLLKT 252
DE +GLYG GG+GK+TL +I N + + V V+ N + +QE++L
Sbjct: 242 DERRTLGLYGMGGVGKTTLLASINNKFLEGMNGFDLVIWVVVSKDLQN-EGIQEQIL--- 297
Query: 253 VRLDIKLGGVSQGIPIIKQRLCR----KKILLILDDVDKLDQLEALAGGLDWFGPGSRVI 308
RL + G +C KK +L+LDD+ LE + GS+++
Sbjct: 298 GRLGLHRGWKQVTEKEKASYICNILNVKKFVLLLDDLWSEVDLEKIGVPPLTRENGSKIV 357
Query: 309 ITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVP----SSHEDILNRALTYA 364
TTR+K + + ++ V+ L EA W F++ V SHEDI A A
Sbjct: 358 FTTRSKDVCRDMEVDGEMKVDCLPPDEA-----WELFQKKVGPIPLQSHEDIPTLARKVA 412
Query: 365 S---GLPLAIVIIGSNLVGR-SVQDSMSTL----DGYEEIPNKE--IQRILKVSYDSLEK 414
GLPLA+ +IG + R +VQ+ + E P+ E I +LK SYD L+
Sbjct: 413 EKCCGLPLALSVIGKAMASRETVQEWQHVIHVLNSSSHEFPSMEEKILPVLKFSYDDLKD 472
Query: 415 EEQSVFLDIACCFKGCK-WPEVKEI 438
E+ + CF C +PE E+
Sbjct: 473 EK------VKLCFLYCSLFPEDYEV 491
>DR17_ARATH (O64790) Putative disease resistance protein At1g61300
Length = 766
Score = 71.2 bits (173), Expect = 7e-12
Identities = 95/341 (27%), Positives = 148/341 (42%), Gaps = 44/341 (12%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAIYNFIA---DQFEVLCFLENVRVNSTSDNLKHLQEKL 248
+D V ++GL+G GG+GK+TL K I+N A +F+++ ++ V S L LQE +
Sbjct: 58 EDRVGIMGLHGMGGVGKTTLFKKIHNKFAKMSSRFDIVIWI----VVSKGAKLSKLQEDI 113
Query: 249 LLKTVRLD--IKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSR 306
K D K S I + L K+ +L+LDD+ + LEA+ +
Sbjct: 114 AEKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGVPYPSEVNKCK 173
Query: 307 VIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYAS- 365
V TTR++ + G V+ L +A EL + +N S I+ A A
Sbjct: 174 VAFTTRDQKVCGEMGDHKPMQVKCLEPEDAWELFK-NKVGDNTLRSDPVIVELAREVAQK 232
Query: 366 --GLPLAIVIIGSNLVGRS-VQDSMSTLD-------GYEEIPNKEIQRILKVSYDSLEKE 415
GLPLA+ +IG + ++ VQ+ +D + + NK I ILK SYDSL E
Sbjct: 233 CRGLPLALSVIGETMASKTMVQEWEHAIDVLTRSAAEFSNMGNK-ILPILKYSYDSLGDE 291
Query: 416 EQSVFLDIACCFKGCK-WPEVKEILHAHYGHCIVHH---VAVLAEKSLMDHLKYDSY--- 468
I CF C +PE EI Y ++ + + E ++ + Y
Sbjct: 292 H------IKSCFLYCALFPEDDEI----YNEKLIDYWICEGFIGEDQVIKRARNKGYEML 341
Query: 469 --VTLHDLIEDMGKEVVRQESPDEPGERSRLWFERDIGTRK 507
+TL +L+ +G E V + LW D G +K
Sbjct: 342 GTLTLANLLTKVGTEHVVMH---DVVREMALWIASDFGKQK 379
>RP13_ARATH (Q9M667) Disease resistance protein RPP13 (Resistance to
Peronospora parasitica protein 13)
Length = 835
Score = 69.7 bits (169), Expect = 2e-11
Identities = 92/377 (24%), Positives = 162/377 (42%), Gaps = 60/377 (15%)
Query: 167 LDVAKYPVGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYNF--IADQFE 224
+D + VGL+ + + L + + ++ ++G GG+GK+ LA+ +YN + ++FE
Sbjct: 157 VDQEEVVVGLEDDAKILLEKLLDYEEKNRFIISIFGMGGLGKTALARKLYNSRDVKERFE 216
Query: 225 VLCFLENVRVNSTSDNLKHLQEKL------LLKTVR------LDIKLGGVSQGIPIIKQR 272
+ + T D L + L L+ +R L++ L G+ +G
Sbjct: 217 YRAWTYVSQEYKTGDILMRIIRSLGMTSGEELEKIRKFAEEELEVYLYGLLEG------- 269
Query: 273 LCRKKILLILDDVDKLDQLEALAGGLDWFGPGSRVIITTRNKHLLK-IHGIESTHAVEGL 331
KK L+++DD+ + + ++L L GSRVIITTR K + + + G H + L
Sbjct: 270 ---KKYLVVVDDIWEREAWDSLKRALPCNHEGSRVIITTRIKAVAEGVDGRFYAHKLRFL 326
Query: 332 NATEALELLRWMAFKENVPSSHEDIL---NRALTYASGLPLAIVIIGSNLVGRSVQDSMS 388
E+ EL AF+ N+ ED+L + GLPL IV++ L S
Sbjct: 327 TFEESWELFEQRAFR-NIQRKDEDLLKTGKEMVQKCRGLPLCIVVLAGLL-------SRK 378
Query: 389 TLDGYEEIPNKEIQR-----------ILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKE 437
T + ++ N +R + +S+ L E + FL ++ +PE E
Sbjct: 379 TPSEWNDVCNSLWRRLKDDSIHVAPIVFDLSFKELRHESKLCFLYLSI------FPEDYE 432
Query: 438 ILHAHYGHCIVHHVAVLAEKSLM--DHLKYDSYVTLHDLIEDMGKEVVRQESPDEPGERS 495
I H +V + ++ +M D +Y + +LI+ E VR+E R
Sbjct: 433 IDLEKLIHLLVAEGFIQGDEEMMMEDVARY----YIEELIDRSLLEAVRRERGKVMSCRI 488
Query: 496 RLWFERDIGTRKIKMIN 512
RD+ +K K +N
Sbjct: 489 HDLL-RDVAIKKSKELN 504
>DRL2_ARATH (P60839) Probable disease resistance protein At1g12290
Length = 884
Score = 68.9 bits (167), Expect = 3e-11
Identities = 77/266 (28%), Positives = 114/266 (41%), Gaps = 33/266 (12%)
Query: 188 DEKSDDEVHMVGLYGTGGIGKSTLAKAIYNFIADQ---FEVLCFLENVRVNSTSDNLKHL 244
D DD ++GLYG GG+GK+TL I N D E++ ++ K +
Sbjct: 168 DHLMDDGTKIMGLYGMGGVGKTTLLTQINNRFCDTDDGVEIVIWVVVSGDLQIHKIQKEI 227
Query: 245 QEKLLLKTVRLDIKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPG 304
EK+ V + K +Q I L +K+ +L+LDD+ K +L + G
Sbjct: 228 GEKIGFIGVEWNQK--SENQKAVDILNFLSKKRFVLLLDDIWKRVELTEIGIPNPTSENG 285
Query: 305 SRVIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVP----SSHEDILNRA 360
++ TTR + + G+ V L A +A W FK+ V SSH DI A
Sbjct: 286 CKIAFTTRCQSVCASMGVHDPMEVRCLGADDA-----WDLFKKKVGDITLSSHPDIPEIA 340
Query: 361 LTYAS---GLPLAIVIIGSNLV--------GRSVQDSMSTLDGYEEIPNKEIQRILKVSY 409
A GLPLA+ +IG + R+V S + + + + I ILK SY
Sbjct: 341 RKVAQACCGLPLALNVIGETMACKKTTQEWDRAVDVSTTYAANFGAV-KERILPILKYSY 399
Query: 410 DSLEKEEQSVFLDIACCFKGCK-WPE 434
D+LE E + CF C +PE
Sbjct: 400 DNLESE------SVKTCFLYCSLFPE 419
>DR16_ARATH (O22727) Putative disease resistance protein At1g61190
Length = 967
Score = 68.2 bits (165), Expect = 6e-11
Identities = 81/264 (30%), Positives = 122/264 (45%), Gaps = 29/264 (10%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAIYNFIAD---QFEVLCFLENVRVNSTSDNLKHLQEKL 248
+D V ++GL+G GG+GK+TL K I+N A+ F+++ ++ V S L LQE +
Sbjct: 170 EDGVGIMGLHGMGGVGKTTLFKKIHNKFAETGGTFDIVIWI----VVSQGAKLSKLQEDI 225
Query: 249 LLKTVRLD--IKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSR 306
K D K S I + L K+ +L+LDD+ + LEA+ +
Sbjct: 226 AEKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCK 285
Query: 307 VIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYAS- 365
V TTR++ + G V+ L +A EL + +N S I+ A A
Sbjct: 286 VAFTTRDQKVCGQMGDHKPMQVKCLEPEDAWELFK-NKVGDNTLRSDPVIVGLAREVAQK 344
Query: 366 --GLPLAIVIIGSNLVGRS-VQDSMSTLD-------GYEEIPNKEIQRILKVSYDSLEKE 415
GLPLA+ IG + ++ VQ+ +D + ++ NK I ILK SYDSLE E
Sbjct: 345 CRGLPLALSCIGETMASKTMVQEWEHAIDVLTRSAAEFSDMQNK-ILPILKYSYDSLEDE 403
Query: 416 EQSVFLDIACCFKGCK-WPEVKEI 438
I CF C +PE +I
Sbjct: 404 H------IKSCFLYCALFPEDDKI 421
>DR18_ARATH (O64789) Putative disease resistance protein At1g61310
Length = 925
Score = 67.0 bits (162), Expect = 1e-10
Identities = 81/264 (30%), Positives = 121/264 (45%), Gaps = 29/264 (10%)
Query: 192 DDEVHMVGLYGTGGIGKSTLAKAIYNFIAD---QFEVLCFLENVRVNSTSDNLKHLQEKL 248
+D V ++GL+G GG+GK+TL K I+N A+ F+++ ++ V S L LQE +
Sbjct: 171 EDGVGIMGLHGMGGVGKTTLFKKIHNKFAEIGGTFDIVIWI----VVSQGAKLSKLQEDI 226
Query: 249 LLKTVRLD--IKLGGVSQGIPIIKQRLCRKKILLILDDVDKLDQLEALAGGLDWFGPGSR 306
K D K S I + L K+ +L+LDD+ + LEA+ +
Sbjct: 227 AEKLHLCDDLWKNKNESDKATDIHRVLKGKRFVLMLDDIWEKVDLEAIGIPYPSEVNKCK 286
Query: 307 VIITTRNKHLLKIHGIESTHAVEGLNATEALELLRWMAFKENVPSSHEDILNRALTYAS- 365
V TTR++ + G V L +A EL + +N SS I+ A A
Sbjct: 287 VAFTTRSREVCGEMGDHKPMQVNCLEPEDAWELFK-NKVGDNTLSSDPVIVGLAREVAQK 345
Query: 366 --GLPLAIVIIGSNLVGRS-VQDSMSTLD-------GYEEIPNKEIQRILKVSYDSLEKE 415
GLPLA+ +IG + ++ VQ+ +D + + NK I ILK SYDSL E
Sbjct: 346 CRGLPLALNVIGETMASKTMVQEWEYAIDVLTRSAAEFSGMENK-ILPILKYSYDSLGDE 404
Query: 416 EQSVFLDIACCFKGCK-WPEVKEI 438
I CF C +PE +I
Sbjct: 405 H------IKSCFLYCALFPEDGQI 422
>R134_ARATH (Q38834) Putative disease resistance RPP13-like protein
4
Length = 852
Score = 66.2 bits (160), Expect = 2e-10
Identities = 81/300 (27%), Positives = 136/300 (45%), Gaps = 39/300 (13%)
Query: 174 VGLQSRVQHVKGHLDEKSDDEVHMVGLYGTGGIGKSTLAKAIYN--FIADQFEVLCFLEN 231
VGL+ + +K L +D ++ ++ G GG+GK+T+A+ ++N I +FE +
Sbjct: 161 VGLEGDKRKIKEWLFRSNDSQLLIMAFVGMGGLGKTTIAQEVFNDKEIEHRFERRIW--- 217
Query: 232 VRVNSTSDNLKHLQEKLLLKTVRLDIKLGGVSQGIPI------IKQRLCRKKILLILDDV 285
V V+ T +E+++ +R LG S G I I+Q L K+ L+++DDV
Sbjct: 218 VSVSQT-----FTEEQIMRSILR---NLGDASVGDDIGTLLRKIQQYLLGKRYLIVMDDV 269
Query: 286 -DK-LDQLEALAGGLDWFGPGSRVIITTRNKHLLKIHGI--ESTHAVEGLNATEALELLR 341
DK L + + GL G G VI+TTR++ + K + TH E L+ + L
Sbjct: 270 WDKNLSWWDKIYQGLP-RGQGGSVIVTTRSESVAKRVQARDDKTHRPELLSPDNSWLLFC 328
Query: 342 WMAFKENVPSSH----EDILNRALTYASGLPLAIVIIGSNLV---------GRSVQDSMS 388
+AF N + ED+ +T GLPL I +G L+ R +
Sbjct: 329 NVAFAANDGTCERPELEDVGKEIVTKCKGLPLTIKAVGGLLLCKDHVYHEWRRIAEHFQD 388
Query: 389 TLDGYEEIPNKEIQRILKVSYDSLEKEEQSVFLDIACCFKGCKWPEVKEILHAHYGHCIV 448
L G + L++SYD L +S L ++ + C P+ ++++H G V
Sbjct: 389 ELRG-NTSETDNVMSSLQLSYDELPSHLKSCILTLSLYPEDCVIPK-QQLVHGWIGEGFV 446
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.400
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 69,201,755
Number of Sequences: 164201
Number of extensions: 3038126
Number of successful extensions: 9888
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 42
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 9717
Number of HSP's gapped (non-prelim): 116
length of query: 576
length of database: 59,974,054
effective HSP length: 116
effective length of query: 460
effective length of database: 40,926,738
effective search space: 18826299480
effective search space used: 18826299480
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 69 (31.2 bits)
Medicago: description of AC138133.14


