

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138133.11 - phase: 0
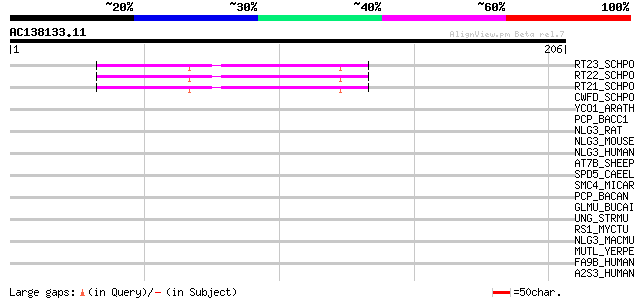
(206 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protei... 59 6e-09
RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protei... 59 6e-09
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 59 6e-09
CWFD_SCHPO (Q09882) Cell cycle control protein cwf13 (Transcript... 32 1.4
YCO1_ARATH (P61430) Hypothetical protein At1g22275 31 2.3
PCP_BACC1 (Q735N6) Pyrrolidone-carboxylate peptidase (EC 3.4.19.... 31 2.3
NLG3_RAT (Q62889) Neuroligin 3 precursor (Gliotactin homolog) 31 2.3
NLG3_MOUSE (Q8BYM5) Neuroligin 3 precursor (Gliotactin homolog) 31 2.3
NLG3_HUMAN (Q9NZ94) Neuroligin 3 precursor (Gliotactin homolog) 31 2.3
AT7B_SHEEP (Q9XT50) Copper-transporting ATPase 2 (EC 3.6.3.4) (C... 30 3.9
SPD5_CAEEL (P91349) Spindle-defective protein 5 29 6.7
SMC4_MICAR (Q9ERA5) Structural maintenance of chromosomes 4-like... 29 6.7
PCP_BACAN (Q81NT5) Pyrrolidone-carboxylate peptidase (EC 3.4.19.... 29 6.7
GLMU_BUCAI (P57139) Bifunctional glmU protein [Includes: UDP-N-a... 29 6.7
UNG_STRMU (Q8DTV8) Uracil-DNA glycosylase (EC 3.2.2.-) (UDG) 29 8.8
RS1_MYCTU (O06147) 30S ribosomal protein S1 29 8.8
NLG3_MACMU (Q8WMH2) Neuroligin 3 (Gliotactin homolog) (Fragment) 29 8.8
MUTL_YERPE (Q8ZIW4) DNA mismatch repair protein mutL 29 8.8
FA9B_HUMAN (Q8IZU0) Protein FAM9B 29 8.8
A2S3_HUMAN (O60296) Amyotrophic lateral sclerosis 2 chromosomal ... 29 8.8
>RT23_SCHPO (Q9UR07) Retrotransposable element Tf2 155 kDa protein
type 3
Length = 1333
Score = 59.3 bits (142), Expect = 6e-09
Identities = 34/103 (33%), Positives = 54/103 (52%), Gaps = 5/103 (4%)
Query: 33 QETTEKVKMIREKMKASQSRQKSYHDKRRKALE-FQEGDHVFLRVTPMTGVGRALKSRKL 91
QET + + ++E + + + K Y D + + +E FQ GD V ++ T G KS KL
Sbjct: 1158 QETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVK---RTKTGFLHKSNKL 1214
Query: 92 TPKFIGPYQISERIGTVAYRVGLPPHLSNL-HDVFHVSQLRKY 133
P F GP+ + ++ G Y + LP + ++ FHVS L KY
Sbjct: 1215 APSFAGPFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>RT22_SCHPO (Q9C0R2) Retrotransposable element Tf2 155 kDa protein
type 2
Length = 1333
Score = 59.3 bits (142), Expect = 6e-09
Identities = 34/103 (33%), Positives = 54/103 (52%), Gaps = 5/103 (4%)
Query: 33 QETTEKVKMIREKMKASQSRQKSYHDKRRKALE-FQEGDHVFLRVTPMTGVGRALKSRKL 91
QET + + ++E + + + K Y D + + +E FQ GD V ++ T G KS KL
Sbjct: 1158 QETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVK---RTKTGFLHKSNKL 1214
Query: 92 TPKFIGPYQISERIGTVAYRVGLPPHLSNL-HDVFHVSQLRKY 133
P F GP+ + ++ G Y + LP + ++ FHVS L KY
Sbjct: 1215 APSFAGPFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 59.3 bits (142), Expect = 6e-09
Identities = 34/103 (33%), Positives = 54/103 (52%), Gaps = 5/103 (4%)
Query: 33 QETTEKVKMIREKMKASQSRQKSYHDKRRKALE-FQEGDHVFLRVTPMTGVGRALKSRKL 91
QET + + ++E + + + K Y D + + +E FQ GD V ++ T G KS KL
Sbjct: 1158 QETIQVFQTVKEHLNTNNIKMKKYFDMKIQEIEEFQPGDLVMVK---RTKTGFLHKSNKL 1214
Query: 92 TPKFIGPYQISERIGTVAYRVGLPPHLSNL-HDVFHVSQLRKY 133
P F GP+ + ++ G Y + LP + ++ FHVS L KY
Sbjct: 1215 APSFAGPFYVLQKSGPNNYELDLPDSIKHMFSSTFHVSHLEKY 1257
>CWFD_SCHPO (Q09882) Cell cycle control protein cwf13
(Transcriptional coregulator Snw1)
Length = 557
Score = 31.6 bits (70), Expect = 1.4
Identities = 17/59 (28%), Positives = 31/59 (51%), Gaps = 4/59 (6%)
Query: 33 QETTEKVKMIREKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRKL 91
QE K K+ +K+ + Q Q + + A+ Q D V++R TP +G+AL +++
Sbjct: 147 QEVANKTKLALQKILSKQIAQS----QPKSAVVQQRDDPVYIRYTPSNQMGQALSKQRI 201
>YCO1_ARATH (P61430) Hypothetical protein At1g22275
Length = 856
Score = 30.8 bits (68), Expect = 2.3
Identities = 16/51 (31%), Positives = 25/51 (48%), Gaps = 5/51 (9%)
Query: 87 KSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADP 137
+S +LTPK + P I++ + PH +N+ D+F L Y DP
Sbjct: 807 RSTRLTPKLMTPTIIAKETAMADH-----PHSANIGDLFSEGSLNPYADDP 852
>PCP_BACC1 (Q735N6) Pyrrolidone-carboxylate peptidase (EC 3.4.19.3)
(5-oxoprolyl-peptidase) (Pyroglutamyl-peptidase I)
(PGP-I) (Pyrase)
Length = 215
Score = 30.8 bits (68), Expect = 2.3
Identities = 26/101 (25%), Positives = 48/101 (46%), Gaps = 17/101 (16%)
Query: 88 SRKLTPKFIGPYQI-SERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD--PSHVIPRD 144
++ L K IG Y+I S+++ TV ++ +S L++Y+ + P +I
Sbjct: 24 AKNLHEKTIGEYKIISKQVPTVFHK--------------SISVLKEYIEELAPEFIICIG 69
Query: 145 DVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGGAT 185
R ++T+E + + IDD ++ G + V VV G T
Sbjct: 70 QAGGRPDITIERVAINIDDARIADNEGNQPVDVPVVEEGPT 110
>NLG3_RAT (Q62889) Neuroligin 3 precursor (Gliotactin homolog)
Length = 848
Score = 30.8 bits (68), Expect = 2.3
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Query: 104 RIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADP-----SHVIPR 143
R VA+ L PHL NLHD+FH + V P SH+ R
Sbjct: 617 RATKVAFWKHLVPHLYNLHDMFHYTSTTTKVPPPDTTHSSHITRR 661
>NLG3_MOUSE (Q8BYM5) Neuroligin 3 precursor (Gliotactin homolog)
Length = 825
Score = 30.8 bits (68), Expect = 2.3
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Query: 104 RIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADP-----SHVIPR 143
R VA+ L PHL NLHD+FH + V P SH+ R
Sbjct: 594 RATKVAFWKHLVPHLYNLHDMFHYTSTTTKVPPPDTTHSSHITRR 638
>NLG3_HUMAN (Q9NZ94) Neuroligin 3 precursor (Gliotactin homolog)
Length = 848
Score = 30.8 bits (68), Expect = 2.3
Identities = 18/45 (40%), Positives = 22/45 (48%), Gaps = 5/45 (11%)
Query: 104 RIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADP-----SHVIPR 143
R VA+ L PHL NLHD+FH + V P SH+ R
Sbjct: 617 RATKVAFWKHLVPHLYNLHDMFHYTSTTTKVPPPDTTHSSHITRR 661
>AT7B_SHEEP (Q9XT50) Copper-transporting ATPase 2 (EC 3.6.3.4)
(Copper pump 2) (Wilson disease-associated protein
homolog)
Length = 1505
Score = 30.0 bits (66), Expect = 3.9
Identities = 12/44 (27%), Positives = 23/44 (52%)
Query: 90 KLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKY 133
K P+ IGP I + I + +R L + N H + H +++++
Sbjct: 648 KFDPEIIGPRDIVKLIEEIGFRASLAQRIPNAHHLDHKVEIKQW 691
>SPD5_CAEEL (P91349) Spindle-defective protein 5
Length = 1198
Score = 29.3 bits (64), Expect = 6.7
Identities = 17/46 (36%), Positives = 26/46 (55%), Gaps = 1/46 (2%)
Query: 27 LGPDLVQETTEKVKMIREKMKASQSRQKSYHDK-RRKALEFQEGDH 71
LG + VQE E+ K++ E + A QSR ++ D R+ EF+ H
Sbjct: 113 LGQENVQEKMEQYKLMEEDLLAMQSRIETSEDNFARQMKEFEAQKH 158
>SMC4_MICAR (Q9ERA5) Structural maintenance of chromosomes 4-like 1
protein (Chromosome-associated polypeptide C) (XCAP-C
homolog) (Fragment)
Length = 1243
Score = 29.3 bits (64), Expect = 6.7
Identities = 20/62 (32%), Positives = 27/62 (43%)
Query: 33 QETTEKVKMIREKMKASQSRQKSYHDKRRKALEFQEGDHVFLRVTPMTGVGRALKSRKLT 92
+E TEK M+ +MKA S K K KA +F E + R + V K + T
Sbjct: 304 KEITEKSNMLSNEMKAKNSAVKDIEKKLHKATKFIEENKEKFRQLDLEDVQVREKLKHAT 363
Query: 93 PK 94
K
Sbjct: 364 SK 365
>PCP_BACAN (Q81NT5) Pyrrolidone-carboxylate peptidase (EC 3.4.19.3)
(5-oxoprolyl-peptidase) (Pyroglutamyl-peptidase I)
(PGP-I) (Pyrase)
Length = 215
Score = 29.3 bits (64), Expect = 6.7
Identities = 25/99 (25%), Positives = 47/99 (47%), Gaps = 17/99 (17%)
Query: 88 SRKLTPKFIGPYQI-SERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVAD--PSHVIPRD 144
++ L K IG Y+I S+++ TV ++ +S L++Y+ + P +I
Sbjct: 24 AKSLHEKTIGEYKIISKQVPTVFHK--------------SISVLKEYIEELAPEFIICIG 69
Query: 145 DVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGG 183
R ++T+E + + IDD ++ G + V VV G
Sbjct: 70 QAGGRPDITIERVAINIDDARIADNEGNQPVDVPVVEEG 108
>GLMU_BUCAI (P57139) Bifunctional glmU protein [Includes:
UDP-N-acetylglucosamine pyrophosphorylase (EC 2.7.7.23)
(N-acetylglucosamine-1-phosphate uridyltransferase);
Glucosamine-1-phosphate N-acetyltransferase (EC
2.3.1.157)]
Length = 459
Score = 29.3 bits (64), Expect = 6.7
Identities = 16/45 (35%), Positives = 23/45 (50%), Gaps = 6/45 (13%)
Query: 118 LSNLHDVFHVSQLRKY------VADPSHVIPRDDVQVRDNLTVET 156
LSNL +F Q+ K + DPSH I R +Q N+ ++T
Sbjct: 232 LSNLEKIFQKKQINKLLINGVTIKDPSHFIFRGTLQHGQNVEIDT 276
>UNG_STRMU (Q8DTV8) Uracil-DNA glycosylase (EC 3.2.2.-) (UDG)
Length = 217
Score = 28.9 bits (63), Expect = 8.8
Identities = 24/100 (24%), Positives = 38/100 (38%), Gaps = 17/100 (17%)
Query: 97 GPYQISERIGTVAYRVGLPPHLSNL-------------HDVFHVSQLRKYVADPSHVIPR 143
GP Q +V +V PP L N+ HD+ ++ + + S +P
Sbjct: 66 GPNQAQGLSFSVPDQVPAPPSLQNILKELADDIGQKQSHDLTSWAKQGVLLLNASLTVPE 125
Query: 144 DDVQVRDNLTVETLPLRIDDRKVKALRGKEIPLVRVVWGG 183
N E D +K + KE P+V ++WGG
Sbjct: 126 HQANAHANGIWEPFT----DAVIKVVNQKETPVVFILWGG 161
>RS1_MYCTU (O06147) 30S ribosomal protein S1
Length = 481
Score = 28.9 bits (63), Expect = 8.8
Identities = 19/50 (38%), Positives = 28/50 (56%), Gaps = 6/50 (12%)
Query: 118 LSNLHDVFHVSQLR-KYVADPSHVIPRDDVQVRDNLTVETLPLRIDDRKV 166
L + + HVS+L K++ PS V VQV D +TVE L + +D +V
Sbjct: 228 LGGVDGLVHVSELSWKHIDHPSEV-----VQVGDEVTVEVLDVDMDRERV 272
>NLG3_MACMU (Q8WMH2) Neuroligin 3 (Gliotactin homolog) (Fragment)
Length = 202
Score = 28.9 bits (63), Expect = 8.8
Identities = 13/23 (56%), Positives = 15/23 (64%)
Query: 104 RIGTVAYRVGLPPHLSNLHDVFH 126
R VA+ L PHL NLHD+FH
Sbjct: 122 RATKVAFWKHLVPHLYNLHDMFH 144
>MUTL_YERPE (Q8ZIW4) DNA mismatch repair protein mutL
Length = 635
Score = 28.9 bits (63), Expect = 8.8
Identities = 15/55 (27%), Positives = 27/55 (48%)
Query: 89 RKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHVSQLRKYVADPSHVIPR 143
+KL P+ +G E I L HL + H+V++VSQ + + + + P+
Sbjct: 556 QKLIPELLGYLSQHEEISPDTLATWLARHLGSEHEVWNVSQAIQLLTEVERLCPQ 610
>FA9B_HUMAN (Q8IZU0) Protein FAM9B
Length = 186
Score = 28.9 bits (63), Expect = 8.8
Identities = 13/43 (30%), Positives = 25/43 (57%)
Query: 30 DLVQETTEKVKMIREKMKASQSRQKSYHDKRRKALEFQEGDHV 72
D ++ T K K ++ S+++ KS H R+K L+ Q+ D++
Sbjct: 58 DTAEDLTAKRKRMKMDKTCSKTKNKSKHALRKKQLKRQKRDYI 100
>A2S3_HUMAN (O60296) Amyotrophic lateral sclerosis 2 chromosomal
region candidate gene protein 3
Length = 914
Score = 28.9 bits (63), Expect = 8.8
Identities = 45/208 (21%), Positives = 86/208 (40%), Gaps = 36/208 (17%)
Query: 12 CRTPLCWFESGESVVLGPDLVQETTEKVKMIREKMKASQSRQKSYHDK----RRKALEFQ 67
C TPL + ES L L+Q ++M++EK+K + + K + + + ++
Sbjct: 191 CSTPLRF---NESFSLSQGLLQ-----LEMLQEKLKELEEENMALRSKACHIKTETVTYE 242
Query: 68 EGDHVFLRVTPMTGVGRALKSRKLTPKFIGPYQISERIGTVAYRVGLPPHLSNLHDVFHV 127
E + + + ++T + G S+ + + Y+ L LS + D+ H
Sbjct: 243 EKEQQLVSDCVKELRETNAQMSRMTEELSGK---SDEL--IRYQEELSSLLSQIVDLQH- 296
Query: 128 SQLRKYVADPS----HVIPRDDVQVRDNLTVETLP---------LRIDDRKVKALRGKEI 174
+L+++V + H+ D Q + + + L L ++K LR +
Sbjct: 297 -KLKEHVIEKEELKLHLQASKDAQRQLTMELHELQDRNMECLGMLHESQEEIKELRSRSG 355
Query: 175 PLVRVV----WGGATGESLTWELESKMR 198
P + +G TGESL E+E MR
Sbjct: 356 PTAHLYFSQSYGAFTGESLAAEIEGTMR 383
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,828,064
Number of Sequences: 164201
Number of extensions: 919400
Number of successful extensions: 2811
Number of sequences better than 10.0: 20
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 2796
Number of HSP's gapped (non-prelim): 21
length of query: 206
length of database: 59,974,054
effective HSP length: 105
effective length of query: 101
effective length of database: 42,732,949
effective search space: 4316027849
effective search space used: 4316027849
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC138133.11


