

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138131.4 + phase: 0 /pseudo
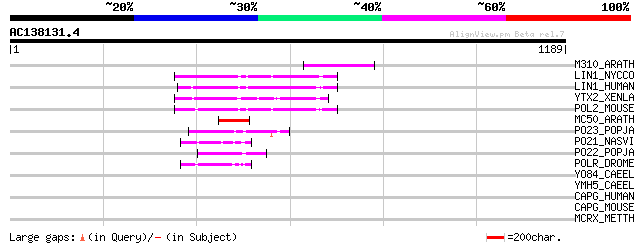
(1189 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310... 130 3e-29
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 108 7e-23
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 103 3e-21
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 101 1e-20
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 100 2e-20
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 89 7e-17
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 63 5e-09
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 52 1e-05
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 49 1e-04
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 48 2e-04
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 38 0.19
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 37 0.42
CAPG_HUMAN (P40121) Macrophage capping protein (Actin-regulatory... 36 0.71
CAPG_MOUSE (P24452) Macrophage capping protein (Myc basic motif ... 35 0.93
MCRX_METTH (P21110) Methyl-coenzyme M reductase II alpha subunit... 33 4.6
>M310_ARATH (P93295) Hypothetical mitochondrial protein AtMg00310
(ORF154)
Length = 154
Score = 130 bits (326), Expect = 3e-29
Identities = 62/154 (40%), Positives = 92/154 (59%), Gaps = 2/154 (1%)
Query: 629 SIPTYFMSLFTLPLSLCDEIEKMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKD-GGMSFK 687
++P Y MS F L LC ++ M FWW S NK+ I W++W KL K+D GG+ F+
Sbjct: 2 ALPVYAMSCFRLSKLLCKKLTSAMTEFWWS-SCENKRKISWVAWQKLCKSKEDDGGLGFR 60
Query: 688 NLGTFNLAMLGKQGWRILTQSDTLIARIYKARYFPRCNFFESELGHNPSFVWRSICKSKF 747
+LG FN A+L KQ +RI+ Q TL++R+ ++RYFP + E +G PS+ WRSI +
Sbjct: 61 DLGWFNQALLAKQSFRIIHQPHTLLSRLLRSRYFPHSSMMECSVGTRPSYAWRSIIHGRE 120
Query: 748 ILKAGSRWKIGDGTSISVWNNYWMRNDVTIFPLD 781
+L G IGDG VW + W+ ++ + PL+
Sbjct: 121 LLSRGLLRTIGDGIHTKVWLDRWIMDETPLPPLN 154
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 108 bits (271), Expect = 7e-23
Identities = 90/353 (25%), Positives = 160/353 (44%), Gaps = 18/353 (5%)
Query: 354 RLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDR 413
R++ + K I Q F+PG + N+ +I +I H+ K+K N + L +D KA+D
Sbjct: 549 RIQQHIKKIIHHDQVGFIPGSQGWFNIRKSINVIQHIN-KLK-NKDHMILSIDAEKAFDN 606
Query: 414 IEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYL 473
I+ ++ + + K+G ++K I + +++NG G RQG PLSP L
Sbjct: 607 IQHPFMIRTLKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLL 666
Query: 474 FIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNIL 533
F I E L+ IR+ +A I G+ + + +S LFADD ++ + K+ ++
Sbjct: 667 FNIVMEVLAIAIREEKA---IKGIHIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVI 721
Query: 534 SVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLG--LPSMIGRSK 591
Y SG +N KS F N + A+ K + + + V KYLG L +
Sbjct: 722 KEYSNVSGYKINTHKSVAFIYTNNNQAE-KTVKDSIPFTVVPKKMKYLGVYLTKDVKDLY 780
Query: 592 KATFNFIKDRVWKKINSWSSKCLSKAGREVLIKFVLQSIPTYFMSLFTL--PLSLCDEIE 649
K + ++ + + +N W + S GR ++K + Y + + PLS ++E
Sbjct: 781 KENYETLRKEIAEDVNKWKNIPCSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLE 840
Query: 650 KMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGW 702
K++ F W ++ LS K GG++ +L + +++ K W
Sbjct: 841 KIILHFIWNQKKPQ------IAKTLLSNKNKAGGITLPDLRLYYKSIVIKTAW 887
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 103 bits (257), Expect = 3e-21
Identities = 90/348 (25%), Positives = 159/348 (44%), Gaps = 18/348 (5%)
Query: 359 LDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDRIEWNY 418
+ K I Q F+P + N+ +I II H I + + + +D KA+D+I+ +
Sbjct: 554 IKKLIHHDQVGFIPAMQGWFNIRKSINIIQH--INRTKDTNHMIISIDAEKAFDKIQQPF 611
Query: 419 LRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICA 478
+ + ++K+G ++K I + + +++NG G RQG PLSP L I
Sbjct: 612 MLKPLNKLGIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPLSPLLPNIVL 671
Query: 479 EGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNILSVYEK 538
E L+ IR+ + +I G+++ +S LFADD ++ + AQ + ++S + K
Sbjct: 672 EVLARAIRQEK---EIKGIQLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSK 726
Query: 539 ASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLG--LPSMIGRSKKATFN 596
SG +N+QKS+ F N + I + L KYLG L + K +
Sbjct: 727 VSGYKINVQKSQAFLYTN-NRQTESQIMSELPFTIASKRIKYLGIQLTRDVKDLFKENYK 785
Query: 597 FIKDRVWKKINSWSSKCLSKAGREVLIKF-VLQSIPTYFMSL-FTLPLSLCDEIEKMMNS 654
+ + + + N W + S GR ++K +L + F ++ LP++ E+EK
Sbjct: 786 PLLNEIKEDTNKWKNIPCSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLK 845
Query: 655 FWWGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGW 702
F W N+K H ++ LS K GG++ + + A + K W
Sbjct: 846 FIW-----NQKRAH-IAKSTLSQKNKAGGITLPDFKLYYKATVTKTAW 887
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 101 bits (252), Expect = 1e-20
Identities = 93/340 (27%), Positives = 158/340 (46%), Gaps = 35/340 (10%)
Query: 354 RLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDR 413
RLK VL + I QS VPGR+IFDNV +++H + + + L LD KA+DR
Sbjct: 545 RLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFAR---RTGLSLAFLSLDQEKAFDR 601
Query: 414 IEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYL 473
++ YL + F Q++ ++ + + + V +N ++T P+ GRG+RQG PLS L
Sbjct: 602 VDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPLSGQL 661
Query: 474 FIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFR--ANLNKAQKMRN 531
+ + E L+RK + G+ + + +ADD L + +L +AQ+ +
Sbjct: 662 YSLAIEPFLCLLRKR-----LTGLVLKEPDMRVVLSAYADDVILVAQDLVDLERAQECQ- 715
Query: 532 ILSVYEKASGQAVNLQKSEFFCSRNVS----PADHKNIANILGVKTVLGTGKYLGLPSMI 587
VY AS +N KS ++ P ++I+ K + KYLG+ +
Sbjct: 716 --EVYAAASSARINWSKSSGLLEGSLKVDFLPPAFRDIS--WESKII----KYLGV-YLS 766
Query: 588 GRSKKATFNFI--KDRVWKKINSWS--SKCLSKAGREVLIKFVLQSIPTYFMSLFTLPLS 643
+ NFI ++ V ++ W +K LS GR ++I ++ S Y + +
Sbjct: 767 AEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGRALVINQLVASQIWYRLICLSPTQE 826
Query: 644 LCDEIEKMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKDGG 683
+I++ + F W G HW+S S+ K+GG
Sbjct: 827 FIAKIQRRLLDFLW-------IGKHWVSAGVSSLPLKEGG 859
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 100 bits (249), Expect = 2e-20
Identities = 89/354 (25%), Positives = 160/354 (45%), Gaps = 20/354 (5%)
Query: 354 RLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHM-KIKVKGNVGEVALKLDISKAYD 412
R++ + I Q F+PG + N+ +I +IH++ K+K K ++ + LD KA+D
Sbjct: 576 RIQEHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKDKNHM---IISLDAEKAFD 632
Query: 413 RIEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPY 472
+I+ ++ +V+++ G ++ I + + VNG I G RQG PLSPY
Sbjct: 633 KIQHPFMIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPY 692
Query: 473 LFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRANLNKAQKMRNI 532
LF I E L+ IR+ + +I G+++ IS L ADD ++ N +++ N+
Sbjct: 693 LFNIVLEVLARAIRQQK---EIKGIQIGKEEVKIS--LLADDMIVYISDPKNSTRELLNL 747
Query: 533 LSVYEKASGQAVNLQKSEFFCSRNVSPADHKNIANILGVKTVLGTGKYLG--LPSMIGRS 590
++ + + G +N KS F A+ K I V KYLG L +
Sbjct: 748 INSFGEVVGYKINSNKSMAFLYTKNKQAE-KEIRETTPFSIVTNNIKYLGVTLTKEVKDL 806
Query: 591 KKATFNFIKDRVWKKINSWSSKCLSKAGREVLIKFVL--QSIPTYFMSLFTLPLSLCDEI 648
F +K + + + W S GR ++K + ++I + +P +E+
Sbjct: 807 YDKNFKSLKKEIKEDLRRWKDLPCSWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNEL 866
Query: 649 EKMMNSFWWGHSGSNKKGIHWMSWDKLSMHKKDGGMSFKNLGTFNLAMLGKQGW 702
E + F W +NKK ++ L + GG++ +L + A++ K W
Sbjct: 867 EGAICKFVW----NNKK--PRIAKSLLKDKRTSGGITMPDLKLYYRAIVIKTAW 914
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 89.0 bits (219), Expect = 7e-17
Identities = 40/67 (59%), Positives = 52/67 (76%)
Query: 448 LVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIIS 507
++NG G + P RGLRQGDPLSPYLFI+C E LS L R+A+ +G + G++V NN+P I+
Sbjct: 13 IINGAPQGLVTPSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRIN 72
Query: 508 HLLFADD 514
HLLFADD
Sbjct: 73 HLLFADD 79
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 62.8 bits (151), Expect = 5e-09
Identities = 61/229 (26%), Positives = 100/229 (43%), Gaps = 25/229 (10%)
Query: 384 IEIIHHMKIK-VKGNVGEVALKLDISKAYDRIEWNYLRQVMDKMGFSEQWIKWIMSCVET 442
I + H++K++ +KG V + LDI KA+D + + + M G + +IMS +
Sbjct: 7 IMLEHYIKLRRLKGKTYNV-VSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITD 65
Query: 443 VDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNN 502
++V G T I G++QGDPLSP LF I + L + R + + + C
Sbjct: 66 AYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDEL--VTRLNDEQPGASMTPACK- 122
Query: 503 APIISHLLFADDCFLFFRANLNKAQKMRNILSVYEKASGQAVNLQKSEFFCSRNVS---- 558
I+ L FADD L +++ + + Y + G +N +K + VS
Sbjct: 123 ---IASLAFADDLLLLEDRDIDVPNSLATTCA-YFRTRGMTLNPEKCASISAATVSGRSV 178
Query: 559 -------PADHKNIANILGVKTVLGTGKYLGLP-SMIGRSKKATFNFIK 599
D + I + G+ T KYLGL S G +K +N +
Sbjct: 179 PRSKPSFTIDGRYIKPLGGINTF----KYLGLTFSSTGVAKPTVYNLTR 223
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 52.0 bits (123), Expect = 1e-05
Identities = 46/154 (29%), Positives = 68/154 (43%), Gaps = 17/154 (11%)
Query: 367 QSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDRIEWNYLRQVMDKM 426
Q AF+ + +N +I ++K+KG + LD+ KA+D +E + + +
Sbjct: 420 QRAFIVADGVAENTSLLSAMIKEARMKIKGLYIAI---LDVKKAFDSVEHRSILDALRRK 476
Query: 427 GFS---EQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSA 483
+I W+ +T V G I P RG+RQGDPLSP LF + +
Sbjct: 477 KLPLEMRNYIMWVYRNSKTRLEVVKTKGRW---IRPARGVRQGDPLSPLLFNCVMD--AV 531
Query: 484 LIRKAEARGDINGVKVCNNAPIISHLLFADDCFL 517
L R E G + G A I L+FADD L
Sbjct: 532 LRRLPENTGFLMG------AEKIGALVFADDLVL 559
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 48.5 bits (114), Expect = 1e-04
Identities = 37/148 (25%), Positives = 68/148 (45%), Gaps = 8/148 (5%)
Query: 403 LKLDISKAYDRIEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVN-GNVTGPIIPGR 461
+ LD+ KA+D + + + + + ++G E +I + + V G+ T I R
Sbjct: 140 VSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYITGSLSDSTTTIRVGPGSQTRKICIRR 199
Query: 462 GLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFLFFRA 521
G++QGDPLSP+LF + L ++ G G + I L FADD L
Sbjct: 200 GVKQGDPLSPFLFNAVLDELLCSLQSTPGIGGTIGEEK------IPVLAFADDLLLLEDN 253
Query: 522 NLNKAQKMRNILSVYEKASGQAVNLQKS 549
++ + + + + + G ++N +KS
Sbjct: 254 DVLLPTTLATVANFF-RLRGMSLNAKKS 280
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 47.8 bits (112), Expect = 2e-04
Identities = 40/152 (26%), Positives = 66/152 (43%), Gaps = 11/152 (7%)
Query: 367 QSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYDRIEWNYLRQVMDKM 426
Q F+P DN ++ H + LD+SKA+D + + +
Sbjct: 446 QRGFLPTDGCADNATIVDLVLRHSHKHFRSCY---IANLDVSKAFDSLSHASIYDTLRAY 502
Query: 427 GFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLRQGDPLSPYLFIICAEGLSALIR 486
G + ++ ++ + E + +G + +P RG++QGDPLSP LF + + L+R
Sbjct: 503 GAPKGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPLSPILFNLV---MDRLLR 559
Query: 487 KAEARGDINGVKVCNNAPIISHLLFADDCFLF 518
+ G KV N I + FADD LF
Sbjct: 560 TLPSE---IGAKVGN--AITNAAAFADDLVLF 586
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 37.7 bits (86), Expect = 0.19
Identities = 22/79 (27%), Positives = 39/79 (48%)
Query: 405 LDISKAYDRIEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIPGRGLR 464
LD SKA+D++ + L + + ++ I+W+ + + V V ++ P G+
Sbjct: 23 LDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTNRSFKVKVGNTLSEPKKTVCGVP 82
Query: 465 QGDPLSPYLFIICAEGLSA 483
QG +SP LF I +SA
Sbjct: 83 QGSVISPVLFGIFVNEISA 101
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 36.6 bits (83), Expect = 0.42
Identities = 28/125 (22%), Positives = 53/125 (42%), Gaps = 4/125 (3%)
Query: 353 TRLKVVLDKCISEFQSAFVPGRSIFDNVMAAIEIIHHMKIKVKGNVGEVALKLDISKAYD 412
+R++ +S Q F+ RS +++ +I + H + +K L D +KA+D
Sbjct: 704 SRIRSEYSHLLSPHQHGFLNFRSCPSSLVRSISLYHSI---LKNEKSLDILFFDFAKAFD 760
Query: 413 RIEWNYLRQVMDKMGFSEQWIKWIMSCVETVDYFVLVNGNVTGPIIP-GRGLRQGDPLSP 471
++ L + + G + W + + V +N V+ P G+ QG P
Sbjct: 761 KVSHPILLKKLALFGLDKLTCSWFKEFLHLRTFSVKINKFVSSNAYPISSGVPQGSVSGP 820
Query: 472 YLFII 476
LFI+
Sbjct: 821 LLFIL 825
>CAPG_HUMAN (P40121) Macrophage capping protein (Actin-regulatory
protein CAP-G)
Length = 348
Score = 35.8 bits (81), Expect = 0.71
Identities = 18/59 (30%), Positives = 31/59 (52%)
Query: 459 PGRGLRQGDPLSPYLFIICAEGLSALIRKAEARGDINGVKVCNNAPIISHLLFADDCFL 517
P L++G+P +AL + ++A G +N KV +++P LL +DDCF+
Sbjct: 226 PKPALKEGNPEEDLTADKANAQAAALYKVSDATGQMNLTKVADSSPFALELLISDDCFV 284
>CAPG_MOUSE (P24452) Macrophage capping protein (Myc basic motif
homolog-1) (Actin-capping protein GCAP39)
Length = 352
Score = 35.4 bits (80), Expect = 0.93
Identities = 15/36 (41%), Positives = 24/36 (66%)
Query: 482 SALIRKAEARGDINGVKVCNNAPIISHLLFADDCFL 517
+AL + ++A G +N KV +++P S LL DDCF+
Sbjct: 253 AALYKVSDATGQMNLTKVADSSPFASELLIPDDCFV 288
>MCRX_METTH (P21110) Methyl-coenzyme M reductase II alpha subunit
(EC 2.8.4.1) (Coenzyme-B sulfoethylthiotransferase
alpha) (MCR II alpha)
Length = 553
Score = 33.1 bits (74), Expect = 4.6
Identities = 13/43 (30%), Positives = 24/43 (55%)
Query: 922 YISYSNVQVAEIYGVCGARPILLITYSISTKMSRISYSEYFRY 964
++ Y V + YG+CG +P + + IST+++ S +Y Y
Sbjct: 348 FVYYGMEYVDDKYGICGTKPTMDVVRDISTEVTLYSLEQYEEY 390
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.342 0.150 0.492
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 123,513,743
Number of Sequences: 164201
Number of extensions: 4786758
Number of successful extensions: 14212
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 1
Number of HSP's that attempted gapping in prelim test: 14180
Number of HSP's gapped (non-prelim): 19
length of query: 1189
length of database: 59,974,054
effective HSP length: 121
effective length of query: 1068
effective length of database: 40,105,733
effective search space: 42832922844
effective search space used: 42832922844
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (22.0 bits)
S2: 72 (32.3 bits)
Medicago: description of AC138131.4


