

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138087.10 + phase: 0
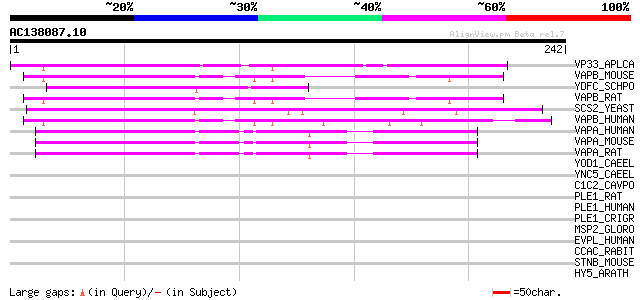
(242 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
VP33_APLCA (Q16943) Vesicle-associated membrane protein/synaptob... 82 1e-15
VAPB_MOUSE (Q9QY76) Vesicle-associated membrane protein-associat... 78 2e-14
YDFC_SCHPO (Q10484) Hypothetical protein C17C9.12 in chromosome I 76 8e-14
VAPB_RAT (Q9Z269) Vesicle-associated membrane protein-associated... 76 8e-14
SCS2_YEAST (P40075) SCS2 protein 75 1e-13
VAPB_HUMAN (O95292) Vesicle-associated membrane protein-associat... 75 1e-13
VAPA_HUMAN (Q9P0L0) Vesicle-associated membrane protein-associat... 74 2e-13
VAPA_MOUSE (Q9WV55) Vesicle-associated membrane protein-associat... 73 7e-13
VAPA_RAT (Q9Z270) Vesicle-associated membrane protein-associated... 70 6e-12
YOD1_CAEEL (P34593) Hypothetical protein ZC262.1 in chromosome III 40 0.005
YNC5_CAEEL (P34538) Hypothetical protein R05D3.5 in chromosome III 38 0.025
C1C2_CAVPO (Q9QZY7) T-cell surface glycoprotein CD1c2 precursor ... 37 0.056
PLE1_RAT (P30427) Plectin 1 (PLTN) (PCN) 36 0.095
PLE1_HUMAN (Q15149) Plectin 1 (PLTN) (PCN) (Hemidesmosomal prote... 36 0.095
PLE1_CRIGR (Q9JI55) Plectin 1 (PLTN) (PCN) (300-kDa intermediate... 36 0.095
MSP2_GLORO (P53022) Major sperm protein 2 34 0.36
EVPL_HUMAN (Q92817) Envoplakin (210 kDa paraneoplastic pemphigus... 33 0.62
CCAC_RABIT (P15381) Voltage-dependent L-type calcium channel alp... 33 0.62
STNB_MOUSE (Q8BZ60) Stonin 2 (Stoned B) 33 0.80
HY5_ARATH (O24646) Transcription factor HY5 (LONG HYPOCOL5 prote... 33 0.80
>VP33_APLCA (Q16943) Vesicle-associated membrane
protein/synaptobrevin binding protein (VAP-33)
Length = 260
Score = 82.0 bits (201), Expect = 1e-15
Identities = 67/219 (30%), Positives = 102/219 (45%), Gaps = 8/219 (3%)
Query: 1 MSPDGDLLSIEPL-ELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGI 59
M+ L +EP EL+F ++ L+LSN TD + FKVKTT P++YCVRPN+GI
Sbjct: 1 MASHEQALILEPAGELRFKGPFTDVVTADLKLSNPTDRRICFKVKTTAPKRYCVRPNSGI 60
Query: 60 VLPRSTCDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEA-GHVVE 118
+ P+++ V V +Q P + K KF++QS+ D V S E+ K+A +
Sbjct: 61 LEPKTSIAVAVMLQPFNYDPNEKN-KHKFMVQSMYAPDHVVE---SQELLWKDAPPESLM 116
Query: 119 ECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKS 178
+ KLR V+ P + + FSE+ P A + + P+
Sbjct: 117 DTKLRCVFEMPDGSHQAPASDASRATDAGAHFSESA-LEDPTVAS-RKTETQSPKRVGAV 174
Query: 179 AEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNR 217
A + +L E A + + L+ E LK EG R R
Sbjct: 175 GSAGEDVKKLQHELKKAQSEITSLKGENSQLKDEGIRLR 213
>VAPB_MOUSE (Q9QY76) Vesicle-associated membrane protein-associated
protein B (VAMP-associated protein B) (VAMP-associated
protein 33b) (VAMP-B) (VAP-B)
Length = 243
Score = 77.8 bits (190), Expect = 2e-14
Identities = 68/213 (31%), Positives = 107/213 (49%), Gaps = 34/213 (15%)
Query: 7 LLSIEPL-ELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LS+EP ELKF ++ +L+L N TD V FKVKTT PR+YCVRPN+G++ ++
Sbjct: 7 VLSLEPQHELKFRGPFTDVVTTNLKLGNPTDRNVCFKVKTTVPRRYCVRPNSGVIDAGAS 66
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDIS-AEMFNKEA-GHVVEECKLR 123
+V V +Q P + + K KF++QS+ +P D S E KEA + + KLR
Sbjct: 67 LNVSVMLQPFDYDPNE-KSKHKFMVQSM-----FAPPDTSDMEAVWKEAKPEDLMDSKLR 120
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V+ P +EN + E ++ SA + EA A A++
Sbjct: 121 CVFELP---------------------AENAKPHDVEINKIIPTSASKTEA---PAAAKS 156
Query: 184 LISRLTE-EKNNAIQQTSRLRQELELLKREGNR 215
L S L + E +++ RL+ E++ L+ E +
Sbjct: 157 LTSPLDDTEVKKVMEECRRLQGEVQRLREESRQ 189
>YDFC_SCHPO (Q10484) Hypothetical protein C17C9.12 in chromosome I
Length = 319
Score = 75.9 bits (185), Expect = 8e-14
Identities = 43/115 (37%), Positives = 63/115 (54%), Gaps = 2/115 (1%)
Query: 17 FPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDVMVTMQAQK 76
FP L + + C L+L N + FKVKTT P++YCVRPN G + S V V +Q
Sbjct: 11 FPRPLTRLVKCDLELRNTAPYPIGFKVKTTAPKQYCVRPNGGRIEANSAVSVEVILQPLD 70
Query: 77 EAPA-DMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSPP 130
PA +C+DKFL+QS + + DI A+++ + + + E K+R VY P
Sbjct: 71 HEPAPGTKCRDKFLVQSTELKPELQGMDI-ADIWTQVSKANISERKIRCVYSEGP 124
>VAPB_RAT (Q9Z269) Vesicle-associated membrane protein-associated
protein B (VAMP-associated protein B) (VAMP-B) (VAP-B)
Length = 243
Score = 75.9 bits (185), Expect = 8e-14
Identities = 67/213 (31%), Positives = 105/213 (48%), Gaps = 34/213 (15%)
Query: 7 LLSIEPL-ELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LS+EP ELKF ++ +L+L N TD V FKVKTT PR+YCVRPN+G++ ++
Sbjct: 7 VLSLEPQHELKFRGPFTDVVTTNLKLGNPTDRNVCFKVKTTAPRRYCVRPNSGVIDAGAS 66
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDIS-AEMFNKEA-GHVVEECKLR 123
+V V +Q P + + K KF++QS+ +P D S E KEA + + KLR
Sbjct: 67 LNVSVMLQPFDYDPNE-KSKHKFMVQSM-----FAPPDTSDMEAVWKEAKPEDLMDSKLR 120
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
V+ P +EN + E ++ SA + EA A+
Sbjct: 121 CVFELP---------------------AENAKPHDVEINKIMPTSASKTEA---PVAAKP 156
Query: 184 LISRLTE-EKNNAIQQTSRLRQELELLKREGNR 215
L S L + E +++ RL+ E++ L+ E +
Sbjct: 157 LTSPLDDAEVKKVMEECRRLQGEVQRLREESRQ 189
>SCS2_YEAST (P40075) SCS2 protein
Length = 244
Score = 75.5 bits (184), Expect = 1e-13
Identities = 62/237 (26%), Positives = 104/237 (43%), Gaps = 12/237 (5%)
Query: 8 LSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCD 67
+ I P L + L +Q + +SN +D +AFKVKTT P+ YCVRPN +V P T
Sbjct: 4 VEISPDVLVYKSPLTEQSTEYASISNNSDQTIAFKVKTTAPKFYCVRPNAAVVAPGETIQ 63
Query: 68 VMVTMQAQKEAP-ADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEEC--KLRV 124
V V E P AD +C+DKFL+ ++ + ++ K ++ + EA + K++V
Sbjct: 64 VQVIFLGLTEEPAADFKCRDKFLVITLPSPYDLNGKAVADVWSDLEAEFKQQAISKKIKV 123
Query: 125 VY-VSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAE---RPEAQDKSAE 180
Y +SP P+ E + ++ P AE +P Q K E
Sbjct: 124 KYLISPDVHPAQNQNIQENKETVEPVVQDSEPKEVPAVVNEKEVPAEPETQPPVQVKKEE 183
Query: 181 ARALISRLTEEKN-----NAIQQTSRLRQELELLKREGNRNRGGVSFIIVILIGLLG 232
++ + +N N+ +E + E + G+ ++ +LI +LG
Sbjct: 184 VPPVVQKTVPHENEKQTSNSTPAPQNQIKEAATVPAENESSSMGIFILVALLILVLG 240
>VAPB_HUMAN (O95292) Vesicle-associated membrane protein-associated
protein B/C (VAMP-associated protein B/C)
(VAMP-B/VAMP-C) (VAP-B/VAP-C) (UNQ484/PRO983)
Length = 243
Score = 75.1 bits (183), Expect = 1e-13
Identities = 74/248 (29%), Positives = 119/248 (47%), Gaps = 33/248 (13%)
Query: 7 LLSIEPL-ELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
+LS+EP ELKF ++ +L+L N TD V FKVKTT PR+YCVRPN+GI+ ++
Sbjct: 7 VLSLEPQHELKFRGPFTDVVTTNLKLGNPTDRNVCFKVKTTAPRRYCVRPNSGIIDAGAS 66
Query: 66 CDVMVTMQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDIS-AEMFNKEA-GHVVEECKLR 123
+V V +Q P + + K KF++QS+ +P D S E KEA + + KLR
Sbjct: 67 INVSVMLQPFDYDPNE-KSKHKFMVQSM-----FAPTDTSDMEAVWKEAKPEDLMDSKLR 120
Query: 124 VVYVSPPQPPSP--------VPEGSEEGSSPRGSFSENGNANGPEFAQV------TRGSA 169
V+ P + P + + + +P S S + + + E +V +G
Sbjct: 121 CVFELPAENDKPHDVEINKIISTTASKTETPIVSKSLSSSLDDTEVKKVMEECKRLQGEV 180
Query: 170 ERPEAQDKS-AEARALISRLTEEKNNAIQQTSRLRQELELLKREGNRNRGGVSFIIVILI 228
+R ++K E L R T + N+ I + +E L R +V+L
Sbjct: 181 QRLREENKQFKEEDGLRMRKTVQSNSPISALAPTGKEEGLSTR---------LLALVVLF 231
Query: 229 GLLGLIMG 236
++G+I+G
Sbjct: 232 FIVGVIIG 239
>VAPA_HUMAN (Q9P0L0) Vesicle-associated membrane protein-associated
protein A (VAMP-associated protein A) (VAMP-A) (VAP-A)
(33 kDa Vamp-associated protein) (VAP-33)
Length = 242
Score = 74.3 bits (181), Expect = 2e-13
Identities = 57/201 (28%), Positives = 95/201 (46%), Gaps = 23/201 (11%)
Query: 12 PLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDVMVT 71
P +LKF ++ +L+L N +D V FKVKTT PR+YCVRPN+GI+ P ST V V
Sbjct: 13 PTDLKFKGPFTDVVTTNLKLRNPSDRKVCFKVKTTAPRRYCVRPNSGIIDPGSTVTVSVM 72
Query: 72 MQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSP-- 129
+Q P + + K KF++Q++ S D+ A ++ + + + KLR V+ P
Sbjct: 73 LQPFDYDPNE-KSKHKFMVQTIFAPPNTS--DMEA-VWKEAKPDELMDSKLRCVFEMPNE 128
Query: 130 ------PQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
+P VP + + P P+ V+ E + ++ +
Sbjct: 129 NDKLNDMEPSKAVPLNASKQDGPM-----------PKPHSVSLNDTETRKLMEECKRLQG 177
Query: 184 LISRLTEEKNNAIQQTSRLRQ 204
+ +L+EE + + RLR+
Sbjct: 178 EMMKLSEENRHLRDEGLRLRK 198
>VAPA_MOUSE (Q9WV55) Vesicle-associated membrane protein-associated
protein A (VAMP-associated protein A) (VAMP-A) (VAP-A)
(33 kDa Vamp-associated protein) (VAP-33)
Length = 242
Score = 72.8 bits (177), Expect = 7e-13
Identities = 56/201 (27%), Positives = 95/201 (46%), Gaps = 23/201 (11%)
Query: 12 PLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDVMVT 71
P +LKF ++ +L+L N +D V FKVKTT PR+YCVRPN+GI+ P S V V
Sbjct: 13 PSDLKFKGPFTDVVTTNLKLQNPSDRKVCFKVKTTAPRRYCVRPNSGIIDPGSIVTVSVM 72
Query: 72 MQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSP-- 129
+Q P + + K KF++Q++ +S D+ A ++ + + + KLR V+ P
Sbjct: 73 LQPFDYDPNE-KSKHKFMVQTIFAPPNIS--DMEA-VWKEAKPDELMDSKLRCVFEMPNE 128
Query: 130 ------PQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
+P VP + + P P+ V+ E + ++ +
Sbjct: 129 NDKLNDMEPSKAVPLNASKQDGPL-----------PKPHSVSLNDTETRKLMEECKRLQG 177
Query: 184 LISRLTEEKNNAIQQTSRLRQ 204
+ +L+EE + + RLR+
Sbjct: 178 EMMKLSEENRHLRDEGLRLRK 198
>VAPA_RAT (Q9Z270) Vesicle-associated membrane protein-associated
protein A (VAMP-associated protein A) (VAMP-A) (VAP-A)
(33 kDa Vamp-associated protein) (VAP-33)
Length = 242
Score = 69.7 bits (169), Expect = 6e-12
Identities = 54/201 (26%), Positives = 94/201 (45%), Gaps = 23/201 (11%)
Query: 12 PLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDVMVT 71
P +LKF ++ +L+L N +D V FKVKTT PR+YCVRPN+G++ P S V V
Sbjct: 13 PSDLKFKGPFTDVVTTNLKLQNPSDRKVCFKVKTTAPRRYCVRPNSGVIDPGSIVTVSVM 72
Query: 72 MQAQKEAPADMQCKDKFLLQSVKTNDGVSPKDISAEMFNKEAGHVVEECKLRVVYVSP-- 129
+Q P + + K KF++Q++ +S D+ A ++ + + + K R V+ P
Sbjct: 73 LQPFDYDPNE-KSKHKFMVQTIFAPPNIS--DMEA-VWKEAKPDELMDSKPRCVFEMPNE 128
Query: 130 ------PQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARA 183
+P VP + + P P+ V+ E + ++ +
Sbjct: 129 NDKLNDMEPSKAVPLNASKQDGPL-----------PKPHSVSLNDTETRKLMEECKRLQG 177
Query: 184 LISRLTEEKNNAIQQTSRLRQ 204
+ +L+EE + + RLR+
Sbjct: 178 EMMKLSEENRHLRDEGLRLRK 198
>YOD1_CAEEL (P34593) Hypothetical protein ZC262.1 in chromosome
III
Length = 131
Score = 40.0 bits (92), Expect = 0.005
Identities = 26/75 (34%), Positives = 41/75 (54%), Gaps = 1/75 (1%)
Query: 7 LLSIEPLELKFPF-ELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRST 65
LL LE++F E +K IS +L+L N T V++KV+ T+ + V+P G V P T
Sbjct: 15 LLIYSSLEVEFKCTEDRKPISVNLKLHNPTAVTVSYKVRCTSADIFRVQPPLGFVKPSET 74
Query: 66 CDVMVTMQAQKEAPA 80
+++ Q Q + A
Sbjct: 75 VSIVIWYQNQDKKDA 89
>YNC5_CAEEL (P34538) Hypothetical protein R05D3.5 in chromosome III
Length = 179
Score = 37.7 bits (86), Expect = 0.025
Identities = 21/61 (34%), Positives = 34/61 (55%)
Query: 20 ELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLPRSTCDVMVTMQAQKEAP 79
E +K IS +L+L N T V++KV+ T+ + V+P G V P T +++ Q Q +
Sbjct: 77 EDRKPISVNLKLHNPTAVTVSYKVRCTSADIFRVQPPLGFVKPSETVSIVIWYQNQDKKD 136
Query: 80 A 80
A
Sbjct: 137 A 137
>C1C2_CAVPO (Q9QZY7) T-cell surface glycoprotein CD1c2 precursor
(CD1-c2 antigen)
Length = 332
Score = 36.6 bits (83), Expect = 0.056
Identities = 19/69 (27%), Positives = 32/69 (45%), Gaps = 4/69 (5%)
Query: 13 LELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTN----PRKYCVRPNTGIVLPRSTCDV 68
L+ K+PF+++ ++ C L T S++ N K CV G + CD+
Sbjct: 106 LQFKYPFDIQVRLGCELHSRETTKSFLHVAFNGLNFLSFQHKSCVPSPEGETRAQKACDI 165
Query: 69 MVTMQAQKE 77
+ T +A KE
Sbjct: 166 LNTYEATKE 174
>PLE1_RAT (P30427) Plectin 1 (PLTN) (PCN)
Length = 4687
Score = 35.8 bits (81), Expect = 0.095
Identities = 16/42 (38%), Positives = 29/42 (68%)
Query: 171 RPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKRE 212
R A++ A+ RAL ++ +EK A+Q+ +RL+ E ELL+++
Sbjct: 2434 RQLAEEDLAQQRALAEKMLKEKMQAVQEATRLKAEAELLQQQ 2475
>PLE1_HUMAN (Q15149) Plectin 1 (PLTN) (PCN) (Hemidesmosomal protein 1)
(HD1)
Length = 4684
Score = 35.8 bits (81), Expect = 0.095
Identities = 16/42 (38%), Positives = 29/42 (68%)
Query: 171 RPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKRE 212
R A++ A+ RAL ++ +EK A+Q+ +RL+ E ELL+++
Sbjct: 2431 RQLAEEDLAQQRALAEKMLKEKMQAVQEATRLKAEAELLQQQ 2472
>PLE1_CRIGR (Q9JI55) Plectin 1 (PLTN) (PCN) (300-kDa intermediate
filament-associated protein) (IFAP300) (Fragment)
Length = 4473
Score = 35.8 bits (81), Expect = 0.095
Identities = 16/42 (38%), Positives = 29/42 (68%)
Query: 171 RPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKRE 212
R A++ A+ RAL ++ +EK A+Q+ +RL+ E ELL+++
Sbjct: 2220 RQLAEEDLAQQRALAEKMLKEKMQAVQEATRLKAEAELLQQQ 2261
>MSP2_GLORO (P53022) Major sperm protein 2
Length = 125
Score = 33.9 bits (76), Expect = 0.36
Identities = 15/72 (20%), Positives = 37/72 (50%)
Query: 3 PDGDLLSIEPLELKFPFELKKQISCSLQLSNKTDSYVAFKVKTTNPRKYCVRPNTGIVLP 62
P GD+ ++ ++ F + + +++ N + + F KTT P++ + P G++ P
Sbjct: 4 PPGDIATMPNQKVVFNAPFDNKATYYVRVINPGTNRIGFAFKTTKPKRINMNPPNGVLGP 63
Query: 63 RSTCDVMVTMQA 74
+ + +V ++ A
Sbjct: 64 KESVNVAISCDA 75
>EVPL_HUMAN (Q92817) Envoplakin (210 kDa paraneoplastic pemphigus
antigen) (p210) (210 kDa cornified envelope precursor)
Length = 2033
Score = 33.1 bits (74), Expect = 0.62
Identities = 24/82 (29%), Positives = 41/82 (49%), Gaps = 6/82 (7%)
Query: 137 PEGSEEGSSPRGSFSENGNANGPEFAQV---TRGSAERPEAQDKSAEARALISRLTEEKN 193
P+GS SP+GS S + A E A + + +A++ E + R RL E++
Sbjct: 15 PKGSPAKGSPKGSPSRHSRAATQELALLISRMQANADQVERDILETQKRLQQDRLNSEQS 74
Query: 194 NAI---QQTSRLRQELELLKRE 212
A+ Q+T R +E E+L ++
Sbjct: 75 QALQHQQETGRSLKEAEVLLKD 96
>CCAC_RABIT (P15381) Voltage-dependent L-type calcium channel
alpha-1C subunit (Voltage-gated calcium channel alpha
subunit Cav1.2) (Calcium channel, L type, alpha-1
polypeptide, isoform 1, cardiac muscle) (Smooth muscle
calcium channel blocker receptor)
Length = 2171
Score = 33.1 bits (74), Expect = 0.62
Identities = 34/119 (28%), Positives = 50/119 (41%), Gaps = 16/119 (13%)
Query: 101 PKDISAEMFNKEAGHVVEECKLRVVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNAN--- 157
P +SAE + G VV E +L Y+SP G SPR + + N NAN
Sbjct: 17 PSHLSAETESTCKGTVVHEAQLNHFYISP---------GGSNYGSPRPAHA-NMNANAAA 66
Query: 158 --GPEFAQVTRGSAERPEAQDKSAEARALISRLTEEKNNAIQQTSRLRQELELLKREGN 214
PE T G+A +A +A L+ + + T R RQ+ K++G+
Sbjct: 67 GLAPEHIP-TPGAALSWQAAIDAARQAKLMGSAGNATISTVSSTQRKRQQYGKPKKQGS 124
>STNB_MOUSE (Q8BZ60) Stonin 2 (Stoned B)
Length = 895
Score = 32.7 bits (73), Expect = 0.80
Identities = 24/80 (30%), Positives = 35/80 (43%), Gaps = 1/80 (1%)
Query: 124 VVYVSPPQPPSPVPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEA-QDKSAEAR 182
V + P P+P+ G+EE S SE ++ E V GS + + QD S+E
Sbjct: 16 VSFSEEPLFPTPLEGGTEEHFPGLSSSSERSESSSGENHVVDEGSQDLSHSEQDDSSEKM 75
Query: 183 ALISRLTEEKNNAIQQTSRL 202
LIS + +Q T L
Sbjct: 76 GLISEAASPPGSPVQPTPDL 95
>HY5_ARATH (O24646) Transcription factor HY5 (LONG HYPOCOL5 protein)
(AtbZIP56)
Length = 168
Score = 32.7 bits (73), Expect = 0.80
Identities = 21/75 (28%), Positives = 33/75 (44%)
Query: 136 VPEGSEEGSSPRGSFSENGNANGPEFAQVTRGSAERPEAQDKSAEARALISRLTEEKNNA 195
VPE E S E+G+A G E Q T G ++R + + + + RL + +A
Sbjct: 43 VPEFGGEAVGKETSGRESGSATGQERTQATVGESQRKRGRTPAEKENKRLKRLLRNRVSA 102
Query: 196 IQQTSRLRQELELLK 210
Q R + L L+
Sbjct: 103 QQARERKKAYLSELE 117
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.132 0.367
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,774,031
Number of Sequences: 164201
Number of extensions: 1235503
Number of successful extensions: 4777
Number of sequences better than 10.0: 71
Number of HSP's better than 10.0 without gapping: 29
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 4699
Number of HSP's gapped (non-prelim): 107
length of query: 242
length of database: 59,974,054
effective HSP length: 107
effective length of query: 135
effective length of database: 42,404,547
effective search space: 5724613845
effective search space used: 5724613845
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 64 (29.3 bits)
Medicago: description of AC138087.10


