

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138017.6 - phase: 0
(300 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
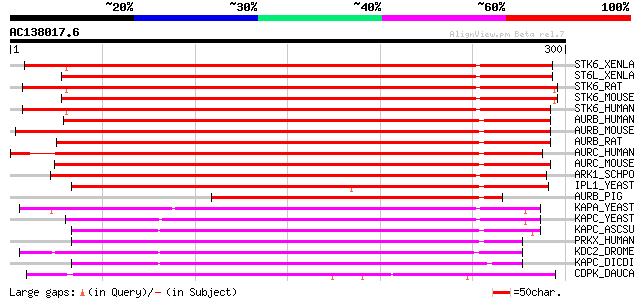
Score E
Sequences producing significant alignments: (bits) Value
STK6_XENLA (Q91820) Serine/threonine-protein kinase Eg2 (EC 2.7.... 376 e-104
ST6L_XENLA (Q91819) Serine/threonine-protein kinase Eg2-like (EC... 376 e-104
STK6_RAT (P59241) Serine/threonine-protein kinase 6 (EC 2.7.1.37... 372 e-103
STK6_MOUSE (P97477) Serine/threonine-protein kinase 6 (EC 2.7.1.... 371 e-102
STK6_HUMAN (O14965) Serine/threonine-protein kinase 6 (EC 2.7.1.... 368 e-102
AURB_HUMAN (Q96GD4) Serine/threonine-protein kinase 12 (EC 2.7.1... 364 e-100
AURB_MOUSE (O70126) Serine/threonine-protein kinase 12 (EC 2.7.1... 361 1e-99
AURB_RAT (O55099) Serine/threonine-protein kinase 12 (EC 2.7.1.3... 358 1e-98
AURC_HUMAN (Q9UQB9) Serine/threonine-protein kinase 13 (EC 2.7.1... 353 4e-97
AURC_MOUSE (O88445) Serine/threonine-protein kinase 13 (EC 2.7.1... 336 5e-92
ARK1_SCHPO (O59790) Serine/threonine-protein kinase ark1 (EC 2.7... 323 4e-88
IPL1_YEAST (P38991) Serine/threonine-protein kinase IPL1 (EC 2.7... 264 2e-70
AURB_PIG (Q9N0X0) Serine/threonine-protein kinase 12 (EC 2.7.1.3... 228 1e-59
KAPA_YEAST (P06244) cAMP-dependent protein kinase type 1 (EC 2.7... 192 6e-49
KAPC_YEAST (P05986) cAMP-dependent protein kinase type 3 (EC 2.7... 186 5e-47
KAPC_ASCSU (P49673) cAMP-dependent protein kinase catalytic subu... 184 3e-46
PRKX_HUMAN (P51817) Serine/threonine-protein kinase PRKX (EC 2.7... 180 3e-45
KDC2_DROME (P16912) Protein kinase DC2 (EC 2.7.1.-) 179 6e-45
KAPC_DICDI (P34099) cAMP-dependent protein kinase catalytic subu... 178 2e-44
CDPK_DAUCA (P28582) Calcium-dependent protein kinase (EC 2.7.1.-... 177 3e-44
>STK6_XENLA (Q91820) Serine/threonine-protein kinase Eg2 (EC
2.7.1.37) (pEg2) (p46Eg265)
Length = 407
Score = 376 bits (966), Expect = e-104
Identities = 179/289 (61%), Positives = 228/289 (77%), Gaps = 6/289 (2%)
Query: 9 PQPHQQHHTASSEVSGSAKDQ---RRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVAL 65
P ++ T + S K++ ++W L DF+IG+PLG+GKFG+VYLARE+ S I+AL
Sbjct: 109 PNVEKKGSTDQGKTSAVPKEEGKKKQWCLEDFEIGRPLGKGKFGNVYLARERESKFILAL 168
Query: 66 KVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKE 125
KVLFKSQL+++ VEHQLRREVEIQSHLRHP+ILRLYGYF+D RVYLIL+YAP GEL++E
Sbjct: 169 KVLFKSQLEKAGVEHQLRREVEIQSHLRHPNILRLYGYFHDASRVYLILDYAPGGELFRE 228
Query: 126 LQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHT 185
LQKC F ++R+A Y+ LA AL+YCH K VIHRDIKPENLL+G+ GELKIADFGWSVH
Sbjct: 229 LQKCTRFDDQRSAMYIKQLAEALLYCHSKKVIHRDIKPENLLLGSNGELKIADFGWSVHA 288
Query: 186 -FNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRI 244
+RR T+CGTLDYLPPEM+E HD +VD+WSLGVLCYEFL G PPFE H +TYRRI
Sbjct: 289 PSSRRTTLCGTLDYLPPEMIEGRMHDETVDLWSLGVLCYEFLVGKPPFETDTHQETYRRI 348
Query: 245 IQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAE 293
+V+ ++P P VS A+DL+S++L + + RLPL +LEHPWI++N++
Sbjct: 349 SKVEFQYP--PYVSEEARDLVSKLLKHNPNHRLPLKGVLEHPWIIKNSQ 395
>ST6L_XENLA (Q91819) Serine/threonine-protein kinase Eg2-like (EC
2.7.1.37) (p46XlEg22)
Length = 408
Score = 376 bits (966), Expect = e-104
Identities = 177/266 (66%), Positives = 219/266 (81%), Gaps = 3/266 (1%)
Query: 29 QRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEI 88
+++W L DF+IG+PLG+GKFG+VYLARE+ S I+ALKVLFKSQL+++ VEHQLRREVEI
Sbjct: 132 KKQWCLEDFEIGRPLGKGKFGNVYLARERESKFILALKVLFKSQLEKAGVEHQLRREVEI 191
Query: 89 QSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARAL 148
QSHLRHP+ILRLYGYF+D RVYLIL+YAP GEL++ELQKC F ++R+A Y+ LA AL
Sbjct: 192 QSHLRHPNILRLYGYFHDASRVYLILDYAPGGELFRELQKCTRFDDQRSALYIKQLAEAL 251
Query: 149 IYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHT-FNRRRTMCGTLDYLPPEMVESV 207
+YCH K VIHRDIKPENLL+G+ GELKIADFGWSVH +RR T+CGTLDYLPPEM+E
Sbjct: 252 LYCHSKKVIHRDIKPENLLLGSNGELKIADFGWSVHAPSSRRTTLCGTLDYLPPEMIEGR 311
Query: 208 EHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQ 267
HD VD+WSLGVLCYEFL G PPFE H +TYRRI +V+ ++P P VS AKDL+S+
Sbjct: 312 MHDEKVDLWSLGVLCYEFLVGKPPFETDTHQETYRRISKVEFQYP--PYVSEEAKDLVSK 369
Query: 268 MLVKDSSERLPLHKLLEHPWIVQNAE 293
+L + + RLPL +LEHPWIV+N++
Sbjct: 370 LLKHNPNHRLPLKGVLEHPWIVKNSQ 395
>STK6_RAT (P59241) Serine/threonine-protein kinase 6 (EC 2.7.1.37)
(Aurora-A) (ratAurA)
Length = 397
Score = 372 bits (956), Expect = e-103
Identities = 186/296 (62%), Positives = 225/296 (75%), Gaps = 9/296 (3%)
Query: 8 QPQPHQQHHTASSEVSGSAKDQ----RRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIV 63
QPQP + + E + K + R+W L DFDIG+PLG+GKFG+VYLAREK S I+
Sbjct: 93 QPQPAASGNNSEKEQTSIQKTEDSKKRQWTLEDFDIGRPLGKGKFGNVYLAREKQSKFIL 152
Query: 64 ALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELY 123
ALKVLFK QL+++ VEHQLRREVEIQSHLRHP+ILRLYGYF+D RVYLILEYAP G +Y
Sbjct: 153 ALKVLFKVQLEKAGVEHQLRREVEIQSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVY 212
Query: 124 KELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSV 183
+ELQK F E+R ATY+ LA AL YCH K VIHRDIKPENLL+G+ GELKIADFGWSV
Sbjct: 213 RELQKLSKFDEQRTATYITELANALSYCHSKRVIHRDIKPENLLLGSNGELKIADFGWSV 272
Query: 184 HT-FNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYR 242
H +RR T+CGTLDY PPEM+E HD VD+WSLGVLCYEFL G+PPFEA + +TYR
Sbjct: 273 HAPSSRRTTLCGTLDYQPPEMIEGRMHDEKVDLWSLGVLCYEFLVGMPPFEAHTYQETYR 332
Query: 243 RIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAE--PSG 296
RI +V+ FP V+ A+DLIS++L +SS+RL L ++LEHPWI N+ P+G
Sbjct: 333 RISRVEFTFP--DFVTEGARDLISRLLKHNSSQRLTLAEVLEHPWIKANSSKPPTG 386
>STK6_MOUSE (P97477) Serine/threonine-protein kinase 6 (EC 2.7.1.37)
(Aurora-family kinase 1) (Aurora/IPL1-related kinase 1)
(Ipl1- and aurora-related kinase 1) (Aurora-A)
(Serine/threonine kinase Ayk1)
Length = 395
Score = 371 bits (952), Expect = e-102
Identities = 180/271 (66%), Positives = 218/271 (80%), Gaps = 5/271 (1%)
Query: 29 QRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEI 88
+R+W L DFDIG+PLG+GKFG+VYLARE+ S I+ALKVLFK+QL+++ VEHQLRREVEI
Sbjct: 116 KRQWTLEDFDIGRPLGKGKFGNVYLARERQSKFILALKVLFKTQLEKANVEHQLRREVEI 175
Query: 89 QSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARAL 148
QSHLRHP+ILRLYGYF+D RVYLILEYAP G +Y+ELQK F E+R ATY+ LA AL
Sbjct: 176 QSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVYRELQKLSKFDEQRTATYITELANAL 235
Query: 149 IYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHT-FNRRRTMCGTLDYLPPEMVESV 207
YCH K VIHRDIKPENLL+G+ GELKIADFGWSVH +RR TMCGTLDYLPPEM+E
Sbjct: 236 SYCHSKRVIHRDIKPENLLLGSNGELKIADFGWSVHAPSSRRTTMCGTLDYLPPEMIEGR 295
Query: 208 EHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQ 267
HD VD+WSLGVLCYEFL G+PPFEA + +TYRRI +V+ FP V+ A+DLIS+
Sbjct: 296 MHDEKVDLWSLGVLCYEFLVGMPPFEAHTYQETYRRISRVEFTFP--DFVTEGARDLISR 353
Query: 268 MLVKDSSERLPLHKLLEHPWIVQNAE--PSG 296
+L ++S+RL L ++LEHPWI N+ P+G
Sbjct: 354 LLKHNASQRLTLAEVLEHPWIKANSSKPPTG 384
>STK6_HUMAN (O14965) Serine/threonine-protein kinase 6 (EC 2.7.1.37)
(Serine/threonine kinase 15) (Aurora/IPL1-related kinase
1) (Aurora-related kinase 1) (hARK1) (Aurora-A)
(Breast-tumor-amplified kinase)
Length = 403
Score = 368 bits (945), Expect = e-102
Identities = 180/290 (62%), Positives = 221/290 (76%), Gaps = 7/290 (2%)
Query: 8 QPQPHQQHHTASSEVSGSAKDQ----RRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIV 63
QP P + E++ K++ R+W L DF+IG+PLG+GKFG+VYLAREK S I+
Sbjct: 100 QPLPSAPENNPEEELASKQKNEESKKRQWALEDFEIGRPLGKGKFGNVYLAREKQSKFIL 159
Query: 64 ALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELY 123
ALKVLFK+QL+++ VEHQLRREVEIQSHLRHP+ILRLYGYF+D RVYLILEYAP G +Y
Sbjct: 160 ALKVLFKAQLEKAGVEHQLRREVEIQSHLRHPNILRLYGYFHDATRVYLILEYAPLGTVY 219
Query: 124 KELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSV 183
+ELQK F E+R ATY+ LA AL YCH K VIHRDIKPENLL+G+ GELKIADFGWSV
Sbjct: 220 RELQKLSKFDEQRTATYITELANALSYCHSKRVIHRDIKPENLLLGSAGELKIADFGWSV 279
Query: 184 HT-FNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYR 242
H +RR T+CGTLDYLPPEM+E HD VD+WSLGVLCYEFL G PPFEA + +TY+
Sbjct: 280 HAPSSRRTTLCGTLDYLPPEMIEGRMHDEKVDLWSLGVLCYEFLVGKPPFEANTYQETYK 339
Query: 243 RIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNA 292
RI +V+ FP V+ A+DLIS++L + S+R L ++LEHPWI N+
Sbjct: 340 RISRVEFTFP--DFVTEGARDLISRLLKHNPSQRPMLREVLEHPWITANS 387
>AURB_HUMAN (Q96GD4) Serine/threonine-protein kinase 12 (EC
2.7.1.37) (Aurora- and Ipl1-like midbody-associated
protein 1) (AIM-1) (Aurora/IPL1-related kinase 2)
(Aurora-related kinase 2) (STK-1) (Aurora-B)
Length = 344
Score = 364 bits (935), Expect = e-100
Identities = 174/264 (65%), Positives = 217/264 (81%), Gaps = 3/264 (1%)
Query: 30 RRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQ 89
R + ++DF+IG+PLG+GKFG+VYLAREK S+ IVALKVLFKSQ+++ VEHQLRRE+EIQ
Sbjct: 70 RHFTIDDFEIGRPLGKGKFGNVYLAREKKSHFIVALKVLFKSQIEKEGVEHQLRREIEIQ 129
Query: 90 SHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALI 149
+HL HP+ILRLY YFYD++R+YLILEYAP+GELYKELQK F E+R AT + LA AL+
Sbjct: 130 AHLHHPNILRLYNYFYDRRRIYLILEYAPRGELYKELQKSCTFDEQRTATIMEELADALM 189
Query: 150 YCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFN-RRRTMCGTLDYLPPEMVESVE 208
YCHGK VIHRDIKPENLL+G +GELKIADFGWSVH + RR+TMCGTLDYLPPEM+E
Sbjct: 190 YCHGKKVIHRDIKPENLLLGLKGELKIADFGWSVHAPSLRRKTMCGTLDYLPPEMIEGRM 249
Query: 209 HDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQM 268
H+ VD+W +GVLCYE L G PPFE+ H++TYRRI++VDLKFP V + A+DLIS++
Sbjct: 250 HNEKVDLWCIGVLCYELLVGNPPFESASHNETYRRIVKVDLKFPAS--VPTGAQDLISKL 307
Query: 269 LVKDSSERLPLHKLLEHPWIVQNA 292
L + SERLPL ++ HPW+ N+
Sbjct: 308 LRHNPSERLPLAQVSAHPWVRANS 331
>AURB_MOUSE (O70126) Serine/threonine-protein kinase 12 (EC
2.7.1.37) (Aurora-related kinase 2)
(Serine/threonine-protein kinase 5) (STK-1) (Aurora-B)
Length = 345
Score = 361 bits (927), Expect = 1e-99
Identities = 173/290 (59%), Positives = 227/290 (77%), Gaps = 3/290 (1%)
Query: 4 ATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIV 63
+T Q ++ + S S +++++ + +++F+IG+PLG+GKFG+VYLAREK S IV
Sbjct: 49 STAAPGQKLAENKSQGSTASQGSQNKQPFTIDNFEIGRPLGKGKFGNVYLAREKKSRFIV 108
Query: 64 ALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELY 123
ALK+LFKSQ+++ VEHQLRRE+EIQ+HL+HP+IL+LY YFYDQ+R+YLILEYAP+GELY
Sbjct: 109 ALKILFKSQIEKEGVEHQLRREIEIQAHLKHPNILQLYNYFYDQQRIYLILEYAPRGELY 168
Query: 124 KELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSV 183
KELQK + F E+R AT + L+ AL YCH K VIHRDIKPENLL+G QGELKIADFGWSV
Sbjct: 169 KELQKSRTFDEQRTATIMEELSDALTYCHKKKVIHRDIKPENLLLGLQGELKIADFGWSV 228
Query: 184 HTFN-RRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYR 242
H + RR+TMCGTLDYLPPEM+E H+ VD+W +GVLCYE + G PPFE+ HS+TYR
Sbjct: 229 HAPSLRRKTMCGTLDYLPPEMIEGRMHNEMVDLWCIGVLCYELMVGNPPFESPSHSETYR 288
Query: 243 RIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNA 292
RI++VDLKFP V S A+DLIS++L + +RLPL ++ HPW+ N+
Sbjct: 289 RIVKVDLKFPSS--VPSGAQDLISKLLKHNPWQRLPLAEVAAHPWVRANS 336
>AURB_RAT (O55099) Serine/threonine-protein kinase 12 (EC 2.7.1.37)
(Aurora- and Ipl1-like midbody-associated protein 1)
(AIM-1) (Aurora-B)
Length = 343
Score = 358 bits (918), Expect = 1e-98
Identities = 169/268 (63%), Positives = 218/268 (81%), Gaps = 3/268 (1%)
Query: 26 AKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRRE 85
++ ++ + +++F+IG+PLG+GKFG+VYLAREK S IVALK+LFKSQ+++ VEHQLRRE
Sbjct: 69 SQSRQPFTIDNFEIGRPLGKGKFGNVYLAREKKSRFIVALKILFKSQIEKEGVEHQLRRE 128
Query: 86 VEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLA 145
+EIQ+HL+HP+IL+LY YFYDQ+R+YLILEYAP+GELYKELQK F E+R AT + L+
Sbjct: 129 IEIQAHLKHPNILQLYNYFYDQQRIYLILEYAPRGELYKELQKSGTFDEQRTATIMEELS 188
Query: 146 RALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFN-RRRTMCGTLDYLPPEMV 204
AL+YCH K VIHRDIKPENLL+G QGELKIADFGWSVH + RR+TMCGTLDYLPPEM+
Sbjct: 189 DALMYCHKKKVIHRDIKPENLLLGLQGELKIADFGWSVHAPSLRRKTMCGTLDYLPPEMI 248
Query: 205 ESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDL 264
E H+ VD+W +GVLCYE + G PPFE+ HS+TYRRI++VDLKFP + AKDL
Sbjct: 249 EGRMHNEMVDLWCIGVLCYELMVGNPPFESPSHSETYRRIVKVDLKFPSS--MPLGAKDL 306
Query: 265 ISQMLVKDSSERLPLHKLLEHPWIVQNA 292
IS++L + S+RLPL ++ HPW+ N+
Sbjct: 307 ISKLLKHNPSQRLPLEQVSAHPWVRANS 334
>AURC_HUMAN (Q9UQB9) Serine/threonine-protein kinase 13 (EC
2.7.1.37) (Aurora/Ipl1/Eg2 protein 2)
(Aurora/Ipl1-related kinase 3) (Aurora-C)
Length = 309
Score = 353 bits (905), Expect = 4e-97
Identities = 174/289 (60%), Positives = 221/289 (76%), Gaps = 16/289 (5%)
Query: 1 MAIATETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSN 60
+A A +T QP S+ RR ++DF+IG+PLG+GKFG+VYLAR K S+
Sbjct: 20 LATANQTAQQP-------------SSPAMRRLTVDDFEIGRPLGKGKFGNVYLARLKESH 66
Query: 61 HIVALKVLFKSQLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKG 120
IVALKVLFKSQ+++ +EHQLRRE+EIQ+HL+HP+ILRLY YF+D +RVYLILEYAP+G
Sbjct: 67 FIVALKVLFKSQIEKEGLEHQLRREIEIQAHLQHPNILRLYNYFHDARRVYLILEYAPRG 126
Query: 121 ELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFG 180
ELYKELQK + E+R AT + LA AL YCH K VIHRDIKPENLL+G +GE+KIADFG
Sbjct: 127 ELYKELQKSEKLDEQRTATIIEELADALTYCHDKKVIHRDIKPENLLLGFRGEVKIADFG 186
Query: 181 WSVHTFN-RRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSD 239
WSVHT + RR+TMCGTLDYLPPEM+E +D VD+W +GVLCYE L G PPFE+ HS+
Sbjct: 187 WSVHTPSLRRKTMCGTLDYLPPEMIEGRTYDEKVDLWCIGVLCYELLVGYPPFESASHSE 246
Query: 240 TYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWI 288
TYRRI++VD++FP + A+DLIS++L ERLPL ++L+HPW+
Sbjct: 247 TYRRILKVDVRFPLS--MPLGARDLISRLLRYQPLERLPLAQILKHPWV 293
>AURC_MOUSE (O88445) Serine/threonine-protein kinase 13 (EC
2.7.1.37) (Aurora/Ipl1/Eg2 protein 1) (Aurora-C)
Length = 282
Score = 336 bits (861), Expect = 5e-92
Identities = 163/269 (60%), Positives = 209/269 (77%), Gaps = 3/269 (1%)
Query: 25 SAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRR 84
S ++ + +NDF+IG+PLGRGKFG VYLAR K ++ IVALKVLFKS++++ +EHQLRR
Sbjct: 4 STSTRKHFTINDFEIGRPLGRGKFGRVYLARLKENHFIVALKVLFKSEIEKEGLEHQLRR 63
Query: 85 EVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASL 144
EVEIQ+HL+H +ILRLY YFYD R+YLILEYAP GELYKELQ+ + ++R AT + L
Sbjct: 64 EVEIQAHLQHRNILRLYNYFYDDTRIYLILEYAPGGELYKELQRHQKLDQQRTATIIQEL 123
Query: 145 ARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFN-RRRTMCGTLDYLPPEM 203
+ AL YCH K VIHRDIKPENLL+G GE+KI+DFGWSVHT + RR+TMCGTLDYLPPEM
Sbjct: 124 SDALTYCHEKKVIHRDIKPENLLLGLNGEVKISDFGWSVHTPSLRRKTMCGTLDYLPPEM 183
Query: 204 VESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKD 263
+ ++ VD+W +GVLCYE L G PPFE+ S+TYRRI QVD KFP V + A+D
Sbjct: 184 IAQKPYNEMVDLWCIGVLCYELLVGKPPFESSTSSETYRRIRQVDFKFPSS--VPAGAQD 241
Query: 264 LISQMLVKDSSERLPLHKLLEHPWIVQNA 292
LIS++L SERL L ++L+HPW+ +++
Sbjct: 242 LISKLLRYHPSERLSLAQVLKHPWVREHS 270
>ARK1_SCHPO (O59790) Serine/threonine-protein kinase ark1 (EC
2.7.1.37) (Aurora-related kinase 1)
Length = 355
Score = 323 bits (827), Expect = 4e-88
Identities = 159/270 (58%), Positives = 204/270 (74%), Gaps = 4/270 (1%)
Query: 23 SGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQL 82
S + R + + F+IGKPLG+GKFG VYLA+EK + IVALK L KS+L QS++E Q+
Sbjct: 75 SSAGPQWREFHIGMFEIGKPLGKGKFGRVYLAKEKKTGFIVALKTLHKSELVQSKIEKQV 134
Query: 83 RREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVA 142
RRE+EIQS+LRH +ILRLYG+F+D+KR+YLILE+A +GELY+ L++ K FSE A+ Y+
Sbjct: 135 RREIEIQSNLRHKNILRLYGHFHDEKRIYLILEFAGRGELYQHLRRAKRFSEEVASKYIF 194
Query: 143 SLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHT-FNRRRTMCGTLDYLPP 201
+A AL Y H KHVIHRDIKPEN+L+G GE+K++DFGWSVH NRR T+CGTLDYLPP
Sbjct: 195 QMANALSYLHKKHVIHRDIKPENILLGIDGEIKLSDFGWSVHAPSNRRTTLCGTLDYLPP 254
Query: 202 EMVESVEHDASVDIWSLGVLCYEFLYGVPPFE-AKEHSDTYRRIIQVDLKFPPKPIVSSA 260
EMVE EH VD+WSLGVL YEFL G PPFE HS TY+RI +VDLK P V
Sbjct: 255 EMVEGKEHTEKVDLWSLGVLTYEFLVGAPPFEDMSGHSATYKRIAKVDLKIP--SFVPPD 312
Query: 261 AKDLISQMLVKDSSERLPLHKLLEHPWIVQ 290
A+DLIS++L + +R+ L +++ HPWIV+
Sbjct: 313 ARDLISRLLQHNPEKRMSLEQVMRHPWIVK 342
>IPL1_YEAST (P38991) Serine/threonine-protein kinase IPL1 (EC
2.7.1.37)
Length = 367
Score = 264 bits (675), Expect = 2e-70
Identities = 127/260 (48%), Positives = 183/260 (69%), Gaps = 4/260 (1%)
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR 93
L+DF++GK LG+GKFG VY R +++ +I ALKV+ K ++ + ++ Q RREVEIQ+ L
Sbjct: 101 LDDFELGKKLGKGKFGKVYCVRHRSTGYICALKVMEKEEIIKYNLQKQFRREVEIQTSLN 160
Query: 94 HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHG 153
HP++ + YGYF+D+KRVYL++EY GE+YK L+ F++ A+ Y+ +A AL Y H
Sbjct: 161 HPNLTKSYGYFHDEKRVYLLMEYLVNGEMYKLLRLHGPFNDILASDYIYQIANALDYMHK 220
Query: 154 KHVIHRDIKPENLLIGAQGELKIADFGWSV--HTFNRRRTMCGTLDYLPPEMVESVEHDA 211
K++IHRDIKPEN+LIG +K+ DFGWS+ NRR+T+CGT+DYL PEMVES E+D
Sbjct: 221 KNIIHRDIKPENILIGFNNVIKLTDFGWSIINPPENRRKTVCGTIDYLSPEMVESREYDH 280
Query: 212 SVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVK 271
++D W+LGVL +E L G PPFE + TY+RI +D+K P +S A+DLI ++L
Sbjct: 281 TIDAWALGVLAFELLTGAPPFEEEMKDTTYKRIAALDIKMPSN--ISQDAQDLILKLLKY 338
Query: 272 DSSERLPLHKLLEHPWIVQN 291
D +R+ L + HPWI++N
Sbjct: 339 DPKDRMRLGDVKMHPWILRN 358
>AURB_PIG (Q9N0X0) Serine/threonine-protein kinase 12 (EC 2.7.1.37)
(Aurora-B) (Fragment)
Length = 156
Score = 228 bits (582), Expect = 1e-59
Identities = 110/158 (69%), Positives = 129/158 (81%), Gaps = 3/158 (1%)
Query: 110 VYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIG 169
+YLILEYAP+GELYKELQKC+ F E+R AT + LA ALIYCHGK VIHRDIKPENLL+G
Sbjct: 1 IYLILEYAPRGELYKELQKCRTFDEQRTATIMEELADALIYCHGKKVIHRDIKPENLLLG 60
Query: 170 AQGELKIADFGWSVHTFN-RRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYG 228
QGELKIADFGWSVH + RR+TM GTLDYLPPEM+E H+ VD+W +GVLCYE L G
Sbjct: 61 LQGELKIADFGWSVHAPSLRRKTMRGTLDYLPPEMIEGRTHNEKVDLWCIGVLCYELLVG 120
Query: 229 VPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLIS 266
PPFE+ H++TYRRI++VDLKFPP V + A+DLIS
Sbjct: 121 NPPFESASHNETYRRIVKVDLKFPPS--VPAGAQDLIS 156
>KAPA_YEAST (P06244) cAMP-dependent protein kinase type 1 (EC
2.7.1.37) (PKA 1) (CDC25 suppressing protein kinase)
(PK-25)
Length = 397
Score = 192 bits (489), Expect = 6e-49
Identities = 106/294 (36%), Positives = 167/294 (56%), Gaps = 15/294 (5%)
Query: 6 ETQPQPHQQHHTASSE------VSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTS 59
ETQ +P Q H T +E ++ + ++ L DF I + LG G FG V+L R + +
Sbjct: 50 ETQEKPKQPHVTYYNEEQYKQFIAQARVTSGKYSLQDFQILRTLGTGSFGRVHLIRSRHN 109
Query: 60 NHIVALKVLFKS-QLQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAP 118
A+KVL K ++ QVEH + + S + HP I+R++G F D +++++I++Y
Sbjct: 110 GRYYAMKVLKKEIVVRLKQVEHTNDERLML-SIVTHPFIIRMWGTFQDAQQIFMIMDYIE 168
Query: 119 KGELYKELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIAD 178
GEL+ L+K + F A Y A + AL Y H K +I+RD+KPEN+L+ G +KI D
Sbjct: 169 GGELFSLLRKSQRFPNPVAKFYAAEVCLALEYLHSKDIIYRDLKPENILLDKNGHIKITD 228
Query: 179 FGWSVHTFNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHS 238
FG++ + + T+CGT DY+ PE+V + ++ S+D WS G+L YE L G PF
Sbjct: 229 FGFAKYVPDVTYTLCGTPDYIAPEVVSTKPYNKSIDWWSFGILIYEMLAGYTPFYDSNTM 288
Query: 239 DTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDSSERL-----PLHKLLEHPW 287
TY +I+ +L+FP P + KDL+S+++ +D S+RL + HPW
Sbjct: 289 KTYEKILNAELRFP--PFFNEDVKDLLSRLITRDLSQRLGNLQNGTEDVKNHPW 340
>KAPC_YEAST (P05986) cAMP-dependent protein kinase type 3 (EC
2.7.1.37) (PKA 3)
Length = 398
Score = 186 bits (473), Expect = 5e-47
Identities = 102/263 (38%), Positives = 151/263 (56%), Gaps = 9/263 (3%)
Query: 31 RWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQ-SQVEHQLRREVEIQ 89
++ L+DF I + LG G FG V+L R + ALK L K + + QVEH E +
Sbjct: 82 KYSLSDFQILRTLGTGSFGRVHLIRSNHNGRFYALKTLKKHTIVKLKQVEHT-NDERRML 140
Query: 90 SHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALI 149
S + HP I+R++G F D ++V+++++Y GEL+ L+K + F A Y A + AL
Sbjct: 141 SIVSHPFIIRMWGTFQDSQQVFMVMDYIEGGELFSLLRKSQRFPNPVAKFYAAEVCLALE 200
Query: 150 YCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEMVESVEH 209
Y H K +I+RD+KPEN+L+ G +KI DFG++ + + T+CGT DY+ PE+V + +
Sbjct: 201 YLHSKDIIYRDLKPENILLDKNGHIKITDFGFAKYVPDVTYTLCGTPDYIAPEVVSTKPY 260
Query: 210 DASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQML 269
+ SVD WS GVL YE L G PF TY I+ +LKFP P A+DL+ +++
Sbjct: 261 NKSVDWWSFGVLIYEMLAGYTPFYNSNTMKTYENILNAELKFP--PFFHPDAQDLLKKLI 318
Query: 270 VKDSSERL-----PLHKLLEHPW 287
+D SERL + HPW
Sbjct: 319 TRDLSERLGNLQNGSEDVKNHPW 341
>KAPC_ASCSU (P49673) cAMP-dependent protein kinase catalytic subunit
(EC 2.7.1.37) (PKA C)
Length = 337
Score = 184 bits (466), Expect = 3e-46
Identities = 98/260 (37%), Positives = 148/260 (56%), Gaps = 9/260 (3%)
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVL-FKSQLQQSQVEHQLRREVEIQSHL 92
++DFD +G G FG VYL + + S ALK + + + Q EH + E + S L
Sbjct: 26 VDDFDRICTIGTGSFGRVYLVQHRASEQYFALKKMAIREVVSMRQTEH-VHSEKRLLSRL 84
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
HP I+++Y +D+ +Y++ EY GEL+ L+ + FS A Y A + AL Y H
Sbjct: 85 SHPFIVKMYCASWDKYNLYMLFEYLAGGELFSYLRASRTFSNSMARFYAAEIVCALQYLH 144
Query: 153 GKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEMVESVEHDAS 212
K++ +RD+KPENL++ +G LK+ DFG++ +R TMCGT +YL PE++ + HD +
Sbjct: 145 SKNIAYRDLKPENLMLNKEGHLKMTDFGFAKEVIDRTWTMCGTPEYLAPEVIGNKGHDTA 204
Query: 213 VDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKD 272
VD WSLGVL YE + G+PPF K + Y +II L+F AKDL+ ++L D
Sbjct: 205 VDWWSLGVLIYEMMIGIPPFRGKTLDEIYEKIILGKLRFTRS--FDLFAKDLVKKLLQVD 262
Query: 273 SSERLPLHK-----LLEHPW 287
++RL K ++ H W
Sbjct: 263 RTQRLGNQKDGAADVMNHKW 282
>PRKX_HUMAN (P51817) Serine/threonine-protein kinase PRKX (EC
2.7.1.37) (Protein kinase PKX1)
Length = 358
Score = 180 bits (457), Expect = 3e-45
Identities = 89/244 (36%), Positives = 143/244 (58%), Gaps = 2/244 (0%)
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQSQVEHQLRREVEIQSHLR 93
L DFD +G G FG V+L +EKT+ H ALKV+ + + + E + E + +
Sbjct: 46 LQDFDTLATVGTGTFGRVHLVKEKTAKHFFALKVMSIPDVIRLKQEQHVHNEKSVLKEVS 105
Query: 94 HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCHG 153
HP ++RL+ ++D++ +Y+++EY P GEL+ L+ FS Y A + A+ Y H
Sbjct: 106 HPFLIRLFWTWHDERFLYMLMEYVPGGELFSYLRNRGRFSSTTGLFYSAEIICAIEYLHS 165
Query: 154 KHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEMVESVEHDASV 213
K +++RD+KPEN+L+ G +K+ DFG++ +R T+CGT +YL PE+++S H +V
Sbjct: 166 KEIVYRDLKPENILLDRDGHIKLTDFGFAKKLVDRTWTLCGTPEYLAPEVIQSKGHGRAV 225
Query: 214 DIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKDS 273
D W+LG+L +E L G PPF Y++I+ + FP + KDLI ++LV D
Sbjct: 226 DWWALGILIFEMLSGFPPFFDDNPFGIYQKILAGKIDFPRH--LDFHVKDLIKKLLVVDR 283
Query: 274 SERL 277
+ RL
Sbjct: 284 TRRL 287
>KDC2_DROME (P16912) Protein kinase DC2 (EC 2.7.1.-)
Length = 502
Score = 179 bits (455), Expect = 6e-45
Identities = 99/273 (36%), Positives = 158/273 (57%), Gaps = 5/273 (1%)
Query: 6 ETQPQPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVAL 65
ET + SS V +AK R++ L+D+ I K +G G FG V L R++ S A+
Sbjct: 163 ETDDEEDDDESEESSSVQ-TAKGVRKYHLDDYQIIKTVGTGTFGRVCLCRDRISEKYCAM 221
Query: 66 KVLFKSQ-LQQSQVEHQLRREVEIQSHLRHPHILRLYGYFYDQKRVYLILEYAPKGELYK 124
K+L ++ ++ Q+EH ++ E I +RHP ++ L D +Y+I +Y GEL+
Sbjct: 222 KILAMTEVIRLKQIEH-VKNERNILREIRHPFVISLEWSTKDDSNLYMIFDYVCGGELFT 280
Query: 125 ELQKCKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGELKIADFGWSVH 184
L+ F+ + + Y A + AL Y H +++RD+KPENLLI G LKI DFG++
Sbjct: 281 YLRNAGKFTSQTSNFYAAEIVSALEYLHSLQIVYRDLKPENLLINRDGHLKITDFGFAKK 340
Query: 185 TFNRRRTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRI 244
+R T+CGT +Y+ PE+++S H+ +VD W+LGVL YE L G PPF ++ Y +I
Sbjct: 341 LRDRTWTLCGTPEYIAPEIIQSKGHNKAVDWWALGVLIYEMLVGYPPFYDEQPFGIYEKI 400
Query: 245 IQVDLKFPPKPIVSSAAKDLISQMLVKDSSERL 277
+ +++ + + AKDLI ++LV D ++RL
Sbjct: 401 LSGKIEW--ERHMDPIAKDLIKKLLVNDRTKRL 431
>KAPC_DICDI (P34099) cAMP-dependent protein kinase catalytic subunit
(EC 2.7.1.37)
Length = 648
Score = 178 bits (451), Expect = 2e-44
Identities = 100/245 (40%), Positives = 143/245 (57%), Gaps = 4/245 (1%)
Query: 34 LNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLFKSQLQQ-SQVEHQLRREVEIQSHL 92
L +F + LG G FG VYL + A+K L K+ + Q QVEH L E I S +
Sbjct: 333 LKEFKQIRVLGTGTFGKVYLIQNTKDGCYYAMKCLNKAYVVQLKQVEH-LNSEKSILSSI 391
Query: 93 RHPHILRLYGYFYDQKRVYLILEYAPKGELYKELQKCKYFSERRAATYVASLARALIYCH 152
HP I+ LY F D+K++YL+ EY GE++ L+K FS A Y A + AL + H
Sbjct: 392 HHPFIVNLYQAFQDEKKLYLLFEYVAGGEVFTHLRKSMKFSNSTAKFYAAEIVLALEFLH 451
Query: 153 GKHVIHRDIKPENLLIGAQGELKIADFGWSVHTFNRRRTMCGTLDYLPPEMVESVEHDAS 212
+++++RD+KPENLLI QG +KI DFG++ +R T+CGT +YL PE+++S H +
Sbjct: 452 KQNIVYRDLKPENLLIDNQGHIKITDFGFAKRVEDRTFTLCGTPEYLAPEIIQSKGHGKA 511
Query: 213 VDIWSLGVLCYEFLYGVPPFEAKEHSDTYRRIIQVDLKFPPKPIVSSAAKDLISQMLVKD 272
VD W+LG+L +E L G PPF + Y +I+ + FP V AKDLI ++L D
Sbjct: 512 VDWWALGILIFEMLAGYPPFYDDDTFAIYNKILAGRITFPLGFDVD--AKDLIKRLLTAD 569
Query: 273 SSERL 277
+ RL
Sbjct: 570 RTRRL 574
>CDPK_DAUCA (P28582) Calcium-dependent protein kinase (EC 2.7.1.-)
(CDPK)
Length = 532
Score = 177 bits (449), Expect = 3e-44
Identities = 101/294 (34%), Positives = 162/294 (54%), Gaps = 12/294 (4%)
Query: 10 QPHQQHHTASSEVSGSAKDQRRWILNDFDIGKPLGRGKFGHVYLAREKTSNHIVALKVLF 69
+P Q H S+ + G + R + +GK LGRG+FG VY E +S + A K +
Sbjct: 57 KPRQVHRPESNTILGKPFEDIR---GKYTLGKELGRGQFGCVYQCTENSSGQLYACKSIL 113
Query: 70 KSQLQQSQVEHQLRREVEIQSHLR-HPHILRLYGYFYDQKRVYLILEYAPKGELYKELQK 128
K +L + ++RE++I HL P+I+ G F D++ V+L++E GEL+ +
Sbjct: 114 KRKLVSKNDKEDIKREIQILQHLSGQPNIVEFKGVFEDRQSVHLVMELCAGGELFDRIIA 173
Query: 129 CKYFSERRAATYVASLARALIYCHGKHVIHRDIKPENLLIGAQGE---LKIADFGWSVHT 185
++SER AAT + + CH V+HRD+KPEN L+ ++ + LK DFG SV
Sbjct: 174 QGHYSERAAATICRQIVNVVHVCHFMGVMHRDLKPENFLLSSKDKDAMLKATDFGLSVFI 233
Query: 186 FNRR--RTMCGTLDYLPPEMVESVEHDASVDIWSLGVLCYEFLYGVPPFEAKEHSDTYRR 243
+ R + G+ Y+ PE++ + +DIWS GV+ Y L GVPPF A+ +
Sbjct: 234 EEGKVYRNIVGSAYYVAPEVLRR-SYGKEIDIWSAGVILYILLSGVPPFWAENEKGIFDA 292
Query: 244 IIQ--VDLKFPPKPIVSSAAKDLISQMLVKDSSERLPLHKLLEHPWIVQNAEPS 295
I++ +D + P P VS++AKDL+ +ML +D R+ ++L+HPW+ + E S
Sbjct: 293 ILEGVIDFESEPWPSVSNSAKDLVRKMLTQDPRRRITSAQVLDHPWMREGGEAS 346
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.137 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 36,218,426
Number of Sequences: 164201
Number of extensions: 1483043
Number of successful extensions: 8625
Number of sequences better than 10.0: 1695
Number of HSP's better than 10.0 without gapping: 1536
Number of HSP's successfully gapped in prelim test: 159
Number of HSP's that attempted gapping in prelim test: 3437
Number of HSP's gapped (non-prelim): 2066
length of query: 300
length of database: 59,974,054
effective HSP length: 110
effective length of query: 190
effective length of database: 41,911,944
effective search space: 7963269360
effective search space used: 7963269360
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC138017.6


