

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138015.8 + phase: 0
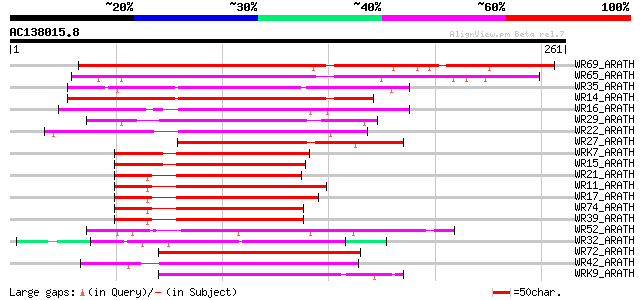
(261 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
WR69_ARATH (Q93WV5) Probable WRKY transcription factor 69 (WRKY ... 234 2e-61
WR65_ARATH (Q9LP56) Probable WRKY transcription factor 65 (WRKY ... 211 1e-54
WR35_ARATH (O64747) Probable WRKY transcription factor 35 (WRKY ... 149 9e-36
WR14_ARATH (Q9SA80) Probable WRKY transcription factor 14 (WRKY ... 138 2e-32
WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY ... 116 6e-26
WR29_ARATH (Q9SUS1) Probable WRKY transcription factor 29 (WRKY ... 114 2e-25
WR22_ARATH (O04609) WRKY transcription factor 22 (WRKY DNA-bindi... 114 3e-25
WR27_ARATH (Q9FLX8) Probable WRKY transcription factor 27 (WRKY ... 112 9e-25
WRK7_ARATH (Q9STX0) Probable WRKY transcription factor 7 (WRKY D... 105 8e-23
WR15_ARATH (O22176) Probable WRKY transcription factor 15 (WRKY ... 105 8e-23
WR21_ARATH (O04336) Probable WRKY transcription factor 21 (WRKY ... 105 1e-22
WR11_ARATH (Q9SV15) Probable WRKY transcription factor 11 (WRKY ... 102 9e-22
WR17_ARATH (Q9SJA8) Probable WRKY transcription factor 17 (WRKY ... 100 3e-21
WR74_ARATH (Q93WU6) Probable WRKY transcription factor 74 (WRKY ... 100 4e-21
WR39_ARATH (Q9SR07) Probable WRKY transcription factor 39 (WRKY ... 100 5e-21
WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY ... 99 1e-20
WR32_ARATH (P59583) Probable WRKY transcription factor 32 (WRKY ... 92 1e-18
WR72_ARATH (Q9LXG8) Probable WRKY transcription factor 72 (WRKY ... 89 8e-18
WR42_ARATH (Q9XEC3) Probable WRKY transcription factor 42 (WRKY ... 89 8e-18
WRK9_ARATH (Q9C9F0) Probable WRKY transcription factor 9 (WRKY D... 87 3e-17
>WR69_ARATH (Q93WV5) Probable WRKY transcription factor 69 (WRKY
DNA-binding protein 69)
Length = 271
Score = 234 bits (597), Expect = 2e-61
Identities = 129/240 (53%), Positives = 158/240 (65%), Gaps = 22/240 (9%)
Query: 33 PASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKP 92
P SPSSCED+KI + +PKKRR ++KRVV++PIADVEGSKSRGE YPPSDSWAWRKYGQKP
Sbjct: 23 PESPSSCEDSKISKPTPKKRRNVEKRVVSVPIADVEGSKSRGEVYPPSDSWAWRKYGQKP 82
Query: 93 IKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPL----PKSHHSS 148
IKGSPYPRGYYRCSSSKGCPARKQVERSRVDP+ L++TYA +HNH P KSHH S
Sbjct: 83 IKGSPYPRGYYRCSSSKGCPARKQVERSRVDPSKLMITYACDHNHPFPSSSANTKSHHRS 142
Query: 149 TNAVTATVTAAVDTPSPVESPAATPQPEDRP---IFVTHPDFDLT--GDHHAV--FGWFD 201
+ TA + E T + P + ++H D L G + + FGWF
Sbjct: 143 S---VVLKTAKKEEEYEEEEEELTVTAAEEPPAGLDLSHVDSPLLLGGCYSEIGEFGWFY 199
Query: 202 DIVSTGVLVSPICGGVEDVTLTM-----REEDESLFADLGELPECSTVFRQRNIPSVFQY 256
D + S DVTL +EEDESLF DLG+LP+C++VFR+ + + Q+
Sbjct: 200 D---ASISSSSGSSNFLDVTLERGFSVGQEEDESLFGDLGDLPDCASVFRRGTVATEEQH 256
>WR65_ARATH (Q9LP56) Probable WRKY transcription factor 65 (WRKY
DNA-binding protein 65)
Length = 259
Score = 211 bits (537), Expect = 1e-54
Identities = 120/239 (50%), Positives = 143/239 (59%), Gaps = 27/239 (11%)
Query: 30 EDGPASPSSCE----DTKIEESSPKK-RREMKKRVVTIPIADVEGSKSRGETYPPSDSWA 84
E GP SP+S I SPK+ RR ++KRVV +P+ ++EGS+ +G+T PPSDSWA
Sbjct: 19 ETGPESPNSSTFNGMKALISSHSPKRSRRSVEKRVVNVPMKEMEGSRHKGDTTPPSDSWA 78
Query: 85 WRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKS 144
WRKYGQKPIKGSPYPRGYYRCSS+KGCPARKQVERSR DPT +L+TY EHNH PL S
Sbjct: 79 WRKYGQKPIKGSPYPRGYYRCSSTKGCPARKQVERSRDDPTMILITYTSEHNHPWPLTSS 138
Query: 145 HHSSTNAVTATVTAAVDTPSPVESPAATP--QPEDRPIFVTHPDFDLTGDHHAVFGWFDD 202
T P P P P + ED V + T F WF +
Sbjct: 139 --------TRNGPKPKPEPKPEPEPEVEPEAEEEDNKFMVLGRGIETTPSCVDEFAWFTE 190
Query: 203 IVSTG--VLVSPI--------CGGVEDVTL--TMREEDESLFADLGELPECSTVFRQRN 249
+ +T +L SPI G +DV + M EEDESLFADLGELPECS VFR R+
Sbjct: 191 METTSSTILESPIFSSEKKTAVSGADDVAVFFPMGEEDESLFADLGELPECSVVFRHRS 249
>WR35_ARATH (O64747) Probable WRKY transcription factor 35 (WRKY
DNA-binding protein 35)
Length = 427
Score = 149 bits (375), Expect = 9e-36
Identities = 84/182 (46%), Positives = 104/182 (56%), Gaps = 25/182 (13%)
Query: 28 ILEDGPASPSSCEDTKIEESSP-----KKRREMKKRVVTIPIADVEGSKSRGETYPPSDS 82
+L D + SSC +I SSP K+R+ K+VV IP S+S GE P SD
Sbjct: 160 LLVDNNNNTSSCSQVQIS-SSPRNLGIKRRKSQAKKVVCIPAPAAMNSRSSGEVVP-SDL 217
Query: 83 WAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLP 142
WAWRKYGQKPIKGSPYPRGYYRCSSSKGC ARKQVERSR DP L++TY EHNH P P
Sbjct: 218 WAWRKYGQKPIKGSPYPRGYYRCSSSKGCSARKQVERSRTDPNMLVITYTSEHNH--PWP 275
Query: 143 KSHHSSTNAVTATVTAAVDTPSPVESPAATPQPEDR----------------PIFVTHPD 186
++ + ++ +++++ S + AAT P R P THP
Sbjct: 276 TQRNALAGSTRSSSSSSLNPSSKSSTAAATTSPSSRVFQNNSSKDEPNNSNLPSSSTHPP 335
Query: 187 FD 188
FD
Sbjct: 336 FD 337
>WR14_ARATH (Q9SA80) Probable WRKY transcription factor 14 (WRKY
DNA-binding protein 14) (AR411)
Length = 430
Score = 138 bits (347), Expect = 2e-32
Identities = 70/144 (48%), Positives = 90/144 (61%), Gaps = 4/144 (2%)
Query: 28 ILEDGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRK 87
+L DG S + + K+R+ K+VV IP S+S GE P SD WAWRK
Sbjct: 166 LLVDGTTFSSQIQISSPRNLGLKRRKSQAKKVVCIPAPAAMNSRSSGEVVP-SDLWAWRK 224
Query: 88 YGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHS 147
YGQKPIKGSP+PRGYYRCSSSKGC ARKQVERSR DP L++TY EHNH P+ ++ +
Sbjct: 225 YGQKPIKGSPFPRGYYRCSSSKGCSARKQVERSRTDPNMLVITYTSEHNHPWPIQRNALA 284
Query: 148 STNAVTATVTAAVDTPSPVESPAA 171
+ T + T++ P+P + A
Sbjct: 285 GS---TRSSTSSSSNPNPSKPSTA 305
>WR16_ARATH (Q9FL92) Probable WRKY transcription factor 16 (WRKY
DNA-binding protein 16)
Length = 1372
Score = 116 bits (290), Expect = 6e-26
Identities = 68/177 (38%), Positives = 96/177 (53%), Gaps = 22/177 (12%)
Query: 24 KLEPILEDGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGETYPPSDSW 83
K + ++++ +S + ++ +I S +++ +KRVV + GS+S SD W
Sbjct: 1134 KTDKLVDNIQSSMIATKEIEITRSKSRRKNNKEKRVVCVVD---RGSRS-------SDLW 1183
Query: 84 AWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLP--- 140
WRKYGQKPIK SPYPR YYRC+SSKGC ARKQVERSR DP ++TY EHNH P
Sbjct: 1184 VWRKYGQKPIKSSPYPRSYYRCASSKGCFARKQVERSRTDPNVSVITYISEHNHPFPTLR 1243
Query: 141 --LPKSHHSS-------TNAVTATVTAAVDTPSPVESPAATPQPEDRPIFVTHPDFD 188
L S SS T + ++TV+ + P P++ P + V D +
Sbjct: 1244 NTLAGSTRSSSSKCSDVTTSASSTVSQDKEGPDKSHLPSSPASPPYAAMVVKEEDME 1300
>WR29_ARATH (Q9SUS1) Probable WRKY transcription factor 29 (WRKY
DNA-binding protein 29)
Length = 304
Score = 114 bits (286), Expect = 2e-25
Identities = 65/148 (43%), Positives = 81/148 (53%), Gaps = 27/148 (18%)
Query: 37 SSCEDTKIEESSPKK-RREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKG 95
S + +I ES KK ++ +KRVV + E SD+WAWRKYGQKPIKG
Sbjct: 100 SRADHIRISESKSKKSKKNQQKRVV----------EQVKEENLLSDAWAWRKYGQKPIKG 149
Query: 96 SPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTAT 155
SPYPR YYRCSSSKGC ARKQVER+ +P +TY +EHNH LP + N++ +
Sbjct: 150 SPYPRSYYRCSSSKGCLARKQVERNPQNPEKFTITYTNEHNHELP------TRRNSLAGS 203
Query: 156 VTAAVDTPSP----------VESPAATP 173
A P P V SP + P
Sbjct: 204 TRAKTSQPKPTLTKKSEKEVVSSPTSNP 231
>WR22_ARATH (O04609) WRKY transcription factor 22 (WRKY DNA-binding
protein 22)
Length = 298
Score = 114 bits (284), Expect = 3e-25
Identities = 65/162 (40%), Positives = 88/162 (54%), Gaps = 21/162 (12%)
Query: 17 EEP-EPELKLEPILEDGPASPSSCEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSRGE 75
EEP +P+ + P+ + + + + S K+R+ K+V + +
Sbjct: 75 EEPRKPQNQKRPLSLSASSGSVTSKPSGSNTSRSKRRKIQHKKVCHVAAEALN------- 127
Query: 76 TYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEH 135
SD WAWRKYGQKPIKGSPYPRGYYRCS+SKGC ARKQVER+R DP +VTY EH
Sbjct: 128 ----SDVWAWRKYGQKPIKGSPYPRGYYRCSTSKGCLARKQVERNRSDPKMFIVTYTAEH 183
Query: 136 NHSLPLPKSHHSST---------NAVTATVTAAVDTPSPVES 168
NH P ++ + + + + T T A + SPV S
Sbjct: 184 NHPAPTHRNSLAGSTRQKPSDQQTSKSPTTTIATYSSSPVTS 225
>WR27_ARATH (Q9FLX8) Probable WRKY transcription factor 27 (WRKY
DNA-binding protein 27)
Length = 348
Score = 112 bits (280), Expect = 9e-25
Identities = 58/107 (54%), Positives = 67/107 (62%), Gaps = 4/107 (3%)
Query: 80 SDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
SD WAWRKYGQKPIKGSPYPR YYRCSSSKGC ARKQVERS +DP +VTY EH H
Sbjct: 165 SDLWAWRKYGQKPIKGSPYPRNYYRCSSSKGCLARKQVERSNLDPNIFIVTYTGEHTHPR 224
Query: 140 PLPKSHHSSTNAVTATVTAAVD-TPSPVESPAATPQPEDRPIFVTHP 185
P +H +S T + V+ P P SP + E+ + T P
Sbjct: 225 P---THRNSLAGSTRNKSQPVNPVPKPDTSPLSDTVKEEIHLSPTTP 268
>WRK7_ARATH (Q9STX0) Probable WRKY transcription factor 7 (WRKY
DNA-binding protein 7)
Length = 353
Score = 105 bits (263), Expect = 8e-23
Identities = 50/92 (54%), Positives = 64/92 (69%), Gaps = 6/92 (6%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
KKR+ KRV+ +P + + PSD ++WRKYGQKPIKGSP+PRGYY+CSS +
Sbjct: 257 KKRKSRVKRVIRVPAVSSKMADI------PSDEFSWRKYGQKPIKGSPHPRGYYKCSSVR 310
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSLPL 141
GCPARK VER+ D L+VTY +HNH+L L
Sbjct: 311 GCPARKHVERALDDAMMLIVTYEGDHNHALVL 342
>WR15_ARATH (O22176) Probable WRKY transcription factor 15 (WRKY
DNA-binding protein 15)
Length = 317
Score = 105 bits (263), Expect = 8e-23
Identities = 48/90 (53%), Positives = 64/90 (70%), Gaps = 6/90 (6%)
Query: 50 KKRREMKKRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSK 109
KKR+ ++R++ +P + S P D ++WRKYGQKPIKGSP+PRGYY+CSS +
Sbjct: 216 KKRKIKQRRIIRVPAISAKMSDV------PPDDYSWRKYGQKPIKGSPHPRGYYKCSSVR 269
Query: 110 GCPARKQVERSRVDPTNLLVTYAHEHNHSL 139
GCPARK VER+ D + L+VTY +HNHSL
Sbjct: 270 GCPARKHVERAADDSSMLIVTYEGDHNHSL 299
>WR21_ARATH (O04336) Probable WRKY transcription factor 21 (WRKY
DNA-binding protein 21)
Length = 380
Score = 105 bits (262), Expect = 1e-22
Identities = 50/93 (53%), Positives = 61/93 (64%), Gaps = 16/93 (17%)
Query: 50 KKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYR 104
KKR+ +R + +P +AD+ P D ++WRKYGQKPIKGSPYPRGYY+
Sbjct: 289 KKRKHRVRRSIRVPAISNKVADI-----------PPDDYSWRKYGQKPIKGSPYPRGYYK 337
Query: 105 CSSSKGCPARKQVERSRVDPTNLLVTYAHEHNH 137
CSS +GCPARK VER DP L+VTY EHNH
Sbjct: 338 CSSMRGCPARKHVERCLEDPAMLIVTYEAEHNH 370
>WR11_ARATH (Q9SV15) Probable WRKY transcription factor 11 (WRKY
DNA-binding protein 11)
Length = 325
Score = 102 bits (254), Expect = 9e-22
Identities = 51/105 (48%), Positives = 66/105 (62%), Gaps = 16/105 (15%)
Query: 50 KKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYR 104
K R+ KR V +P IAD+ P D ++WRKYGQKPIKGSP+PRGYY+
Sbjct: 222 KSRKNRMKRTVRVPAISAKIADI-----------PPDEYSWRKYGQKPIKGSPHPRGYYK 270
Query: 105 CSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSST 149
CS+ +GCPARK VER+ DP L+VTY EH H+ + + SS+
Sbjct: 271 CSTFRGCPARKHVERALDDPAMLIVTYEGEHRHNQSAMQENISSS 315
>WR17_ARATH (Q9SJA8) Probable WRKY transcription factor 17 (WRKY
DNA-binding protein 17)
Length = 321
Score = 100 bits (250), Expect = 3e-21
Identities = 50/101 (49%), Positives = 62/101 (60%), Gaps = 16/101 (15%)
Query: 50 KKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYR 104
K R+ KR V +P IAD+ P D ++WRKYGQKPIKGSP+PRGYY+
Sbjct: 219 KSRKNRMKRTVRVPAVSAKIADI-----------PPDEYSWRKYGQKPIKGSPHPRGYYK 267
Query: 105 CSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSH 145
CS+ +GCPARK VER+ D T L+VTY EH H + H
Sbjct: 268 CSTFRGCPARKHVERALDDSTMLIVTYEGEHRHHQSTMQEH 308
>WR74_ARATH (Q93WU6) Probable WRKY transcription factor 74 (WRKY
DNA-binding protein 74)
Length = 330
Score = 100 bits (249), Expect = 4e-21
Identities = 50/94 (53%), Positives = 62/94 (65%), Gaps = 16/94 (17%)
Query: 50 KKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYR 104
KKR+ KR + +P IAD+ P D ++WRKYGQKPIKGSP+PRGYY+
Sbjct: 238 KKRKLRVKRSIKVPAISNKIADI-----------PPDEYSWRKYGQKPIKGSPHPRGYYK 286
Query: 105 CSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHS 138
CSS +GCPARK VER + + L+VTY EHNHS
Sbjct: 287 CSSVRGCPARKHVERCVEETSMLIVTYEGEHNHS 320
>WR39_ARATH (Q9SR07) Probable WRKY transcription factor 39 (WRKY
DNA-binding protein 39)
Length = 330
Score = 100 bits (248), Expect = 5e-21
Identities = 50/94 (53%), Positives = 62/94 (65%), Gaps = 16/94 (17%)
Query: 50 KKRREMKKRVVTIP-----IADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYR 104
KKR+ KR + +P IAD+ P D ++WRKYGQKPIKGSP+PRGYY+
Sbjct: 238 KKRKLRVKRSIKVPAISNKIADI-----------PPDEYSWRKYGQKPIKGSPHPRGYYK 286
Query: 105 CSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHS 138
CSS +GCPARK VER + + L+VTY EHNHS
Sbjct: 287 CSSVRGCPARKHVERCIDETSMLIVTYEGEHNHS 320
>WR52_ARATH (Q9FH83) Probable WRKY transcription factor 52 (WRKY
DNA-binding protein 52) (Disease resistance protein RRS1)
(Resistance to Ralstonia solanacearum 1 protein)
Length = 1378
Score = 98.6 bits (244), Expect = 1e-20
Identities = 69/194 (35%), Positives = 95/194 (48%), Gaps = 36/194 (18%)
Query: 37 SSCEDTKIEESSP-----KKRREMK-KRVVTIPIADVEGSKSRGETYPPSDSWAWRKYGQ 90
S CE + E S KK RE K K+VV+IP D EG D W WRKYGQ
Sbjct: 1173 SDCESSMTENLSDVPKKEKKHRESKVKKVVSIPAID-EG-----------DLWTWRKYGQ 1220
Query: 91 KPIKGSPYPRGYYRCS--SSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLP-----LPK 143
K I GS +PRGYYRC+ + GC A KQV+RS D L +TY EHNH P L
Sbjct: 1221 KDILGSRFPRGYYRCAYKFTHGCKATKQVQRSETDSNMLAITYLSEHNHPRPTKRKALAD 1280
Query: 144 SHHSSTNAVTATVTAAV--------DTPSPVESPAATPQPEDRPIFVTHPDFDLTGDHHA 195
S S+++++ + +T + D P+ P+++ P + + D + D+
Sbjct: 1281 STRSTSSSICSAITTSASSRVFQNKDEPNQPHLPSSSTPPRNAAVLFKMTDMEEFQDNME 1340
Query: 196 VFGWFDDIVSTGVL 209
V +D+V T L
Sbjct: 1341 V---DNDVVDTRTL 1351
>WR32_ARATH (P59583) Probable WRKY transcription factor 32 (WRKY
DNA-binding protein 32)
Length = 466
Score = 92.4 bits (228), Expect = 1e-18
Identities = 45/126 (35%), Positives = 70/126 (54%), Gaps = 8/126 (6%)
Query: 39 CEDTKIEESSPKKRREMKKRVVTIPIADVEGSKSR------GETYPPSDSWAWRKYGQKP 92
CE+ +EE PK+R + K + G K++ G+ D + WRKYGQK
Sbjct: 285 CENEAVEEPEPKRRLK-KDNSQSSDSVSKPGKKNKFVVHAAGDVGICGDGYRWRKYGQKM 343
Query: 93 IKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHSSTNAV 152
+KG+P+PR YYRC+S+ GCP RK +E + + +++TY HNH +P+PK H +++
Sbjct: 344 VKGNPHPRNYYRCTSA-GCPVRKHIETAVENTKAVIITYKGVHNHDMPVPKKRHGPPSSM 402
Query: 153 TATVTA 158
A
Sbjct: 403 LVAAAA 408
Score = 55.8 bits (133), Expect = 1e-07
Identities = 44/175 (25%), Positives = 70/175 (39%), Gaps = 14/175 (8%)
Query: 4 RRFNTKPLYVADREEPEPELKLEPILEDGPASPSSCEDTKIEESSPKKRREMKKRVVT-I 62
R+ T P A + L + P L PA+ S+ +D + K+ + VV +
Sbjct: 102 RQVETSPSLAASSDS----LTVTPCLSLDPATASTAQDLPLVSVPTKQEQRSDSPVVNRL 157
Query: 63 PIADVEGSKSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRV 122
+ V + +R D + WRKYGQK +K R YYRC+ ++ C K++E S
Sbjct: 158 SVTPVPRTPAR-------DGYNWRKYGQKQVKSPKGSRSYYRCTYTECC--AKKIECSND 208
Query: 123 DPTNLLVTYAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSPVESPAATPQPED 177
+ + H H P S VT + + + VE + P D
Sbjct: 209 SGNVVEIVNKGLHTHEPPRKTSFSPREIRVTTAIRPVSEDDTVVEELSIVPSGSD 263
>WR72_ARATH (Q9LXG8) Probable WRKY transcription factor 72 (WRKY
DNA-binding protein 72)
Length = 548
Score = 89.4 bits (220), Expect = 8e-18
Identities = 42/95 (44%), Positives = 60/95 (62%)
Query: 71 KSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVT 130
++R +T +D WRKYGQK KG+P PR YYRC+ + GCP RKQV+R D + L+ T
Sbjct: 218 RARCDTPTMNDGCQWRKYGQKIAKGNPCPRAYYRCTVAPGCPVRKQVQRCADDMSILITT 277
Query: 131 YAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSP 165
Y H+HSLPL + +ST + A++ + + SP
Sbjct: 278 YEGTHSHSLPLSATTMASTTSAAASMLLSGSSSSP 312
>WR42_ARATH (Q9XEC3) Probable WRKY transcription factor 42 (WRKY
DNA-binding protein 42)
Length = 528
Score = 89.4 bits (220), Expect = 8e-18
Identities = 53/137 (38%), Positives = 70/137 (50%), Gaps = 14/137 (10%)
Query: 34 ASPSSCEDTKIEESSPKKRRE------MKKRVVTIPIADVEGSKSRGETYPPSDSWAWRK 87
AS S C E +S K + M+K V++ ++R E SD WRK
Sbjct: 248 ASSSICGGNGSENASSKVIEQAAAEATMRKARVSV--------RARSEAPMLSDGCQWRK 299
Query: 88 YGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVTYAHEHNHSLPLPKSHHS 147
YGQK KG+P PR YYRC+ + GCP RKQV+R D T L+ TY HNH LP + +
Sbjct: 300 YGQKMAKGNPCPRAYYRCTMAVGCPVRKQVQRCAEDRTILITTYEGNHNHPLPPAAMNMA 359
Query: 148 STNAVTATVTAAVDTPS 164
ST A++ + T S
Sbjct: 360 STTTAAASMLLSGSTMS 376
>WRK9_ARATH (Q9C9F0) Probable WRKY transcription factor 9 (WRKY
DNA-binding protein 9)
Length = 374
Score = 87.4 bits (215), Expect = 3e-17
Identities = 47/117 (40%), Positives = 69/117 (58%), Gaps = 5/117 (4%)
Query: 71 KSRGETYPPSDSWAWRKYGQKPIKGSPYPRGYYRCSSSKGCPARKQVERSRVDPTNLLVT 130
++R ET +D WRKYGQK KG+P PR YYRC+ + GCP RKQV+R D + L+ T
Sbjct: 226 RARCETATMNDGCQWRKYGQKTAKGNPCPRAYYRCTVAPGCPVRKQVQRCLEDMSILITT 285
Query: 131 YAHEHNHSLPLPKSHHSSTNAVTATVTAAVDTPSPVESPA--ATPQPEDRPIFVTHP 185
Y HNH LP+ + +ST + + + +D+ + P+ TPQ D + +T+P
Sbjct: 286 YEGTHNHPLPVGATAMASTASTSPFL--LLDSSDNLSHPSYYQTPQAIDSSL-ITYP 339
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.313 0.132 0.402
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 35,367,251
Number of Sequences: 164201
Number of extensions: 1661028
Number of successful extensions: 5523
Number of sequences better than 10.0: 114
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 5255
Number of HSP's gapped (non-prelim): 169
length of query: 261
length of database: 59,974,054
effective HSP length: 108
effective length of query: 153
effective length of database: 42,240,346
effective search space: 6462772938
effective search space used: 6462772938
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 65 (29.6 bits)
Medicago: description of AC138015.8


