

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC138011.4 + phase: 0
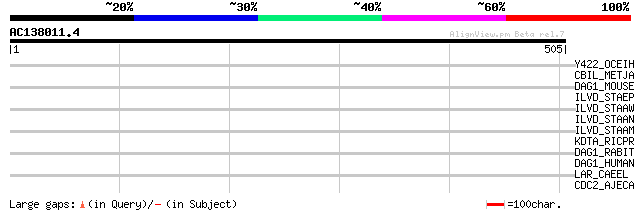
(505 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
Y422_OCEIH (Q8ET41) Putative nucleotidase OB0422 (EC 3.1.3.-) 34 0.77
CBIL_METJA (Q58181) Probable cobalt-precorrin-2 C(20)-methyltran... 34 1.0
DAG1_MOUSE (Q62165) Dystroglycan precursor (Dystrophin-associate... 33 1.7
ILVD_STAEP (Q8CNL6) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD) 33 2.3
ILVD_STAAW (P65158) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD) 32 3.8
ILVD_STAAN (P65157) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD) 32 3.8
ILVD_STAAM (P65156) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD) 32 3.8
KDTA_RICPR (Q9ZE58) 3-deoxy-D-manno-octulosonic-acid transferase... 32 5.0
DAG1_RABIT (Q28685) Dystroglycan precursor (Dystrophin-associate... 32 5.0
DAG1_HUMAN (Q14118) Dystroglycan precursor (Dystrophin-associate... 32 5.0
LAR_CAEEL (Q9BMN8) Protein-tyrosine phosphatase Lar-like precurs... 31 8.6
CDC2_AJECA (P54119) Cell division control protein 2 (EC 2.7.1.37... 31 8.6
>Y422_OCEIH (Q8ET41) Putative nucleotidase OB0422 (EC 3.1.3.-)
Length = 188
Score = 34.3 bits (77), Expect = 0.77
Identities = 14/39 (35%), Positives = 25/39 (63%)
Query: 413 EDIPTILNDLDKFGYDLYIVGQGHHRNVRAFSSLLEWCD 451
+D PT+L+ L K L I Q ++RN ++F+ +++W D
Sbjct: 144 DDKPTVLDTLTKTNTKLLIKDQSYNRNKKSFARIIDWHD 182
>CBIL_METJA (Q58181) Probable cobalt-precorrin-2
C(20)-methyltransferase (EC 2.1.1.151)
(S-adenosyl-L-methionine--cobalt-precorrin-2
methyltransferase)
Length = 230
Score = 33.9 bits (76), Expect = 1.0
Identities = 31/133 (23%), Positives = 56/133 (41%), Gaps = 12/133 (9%)
Query: 307 LGSITKMNFRICMIFIGGPDDHEALAVAWRMVGHPGTRLSLVRML--LFDEAAEVNI--I 362
L + K + + +I IG P + + W+++ G + +V + +F AA +NI +
Sbjct: 83 LEKVLKEDGEVAIITIGDPTLYSTFSYVWKLLKERGVEVEIVNGISSIFASAAALNIPLV 142
Query: 363 SHYEVQGILSVVMDNEKQKD-------LDEEYVNSFRLKAVNNNDSISYSEVDVHSSEDI 415
E IL D EK D + + +N +L + N D V + ED
Sbjct: 143 EGDEKLCILPQGKDLEKYIDEFDTIIIMKTKNLNE-KLSVIKNRDDYIIGLVKRATFEDE 201
Query: 416 PTILNDLDKFGYD 428
++ LD+ +D
Sbjct: 202 KVVIGKLDEINFD 214
>DAG1_MOUSE (Q62165) Dystroglycan precursor (Dystrophin-associated
glycoprotein 1) [Contains: Alpha-dystroglycan
(Alpha-DG); Beta-dystroglycan (Beta-DG)]
Length = 893
Score = 33.1 bits (74), Expect = 1.7
Identities = 20/80 (25%), Positives = 41/80 (51%), Gaps = 9/80 (11%)
Query: 370 ILSVVMDNEKQKDLDEEYVNSFRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDL 429
+L+V++D + K ++ ++ N S+SEV++H+ + +P + N L F
Sbjct: 188 VLTVILDADLTKMTPKQRIDLL-------NRMQSFSEVELHNMKLVPVVNNRL--FDMSA 238
Query: 430 YIVGQGHHRNVRAFSSLLEW 449
++ G G+ + V +LL W
Sbjct: 239 FMAGPGNAKKVVENGALLSW 258
>ILVD_STAEP (Q8CNL6) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD)
Length = 562
Score = 32.7 bits (73), Expect = 2.3
Identities = 39/160 (24%), Positives = 67/160 (41%), Gaps = 24/160 (15%)
Query: 283 AYKDINQNVMQTSPCSVGIFVDRNLGSITKMNFRICMIFIGGPDDHEALAVAWR---MVG 339
A+ D+ QN T G+F + MN + ++ + P + ALAV+ + M+
Sbjct: 179 AFLDMEQNACPTCGSCAGMF------TANSMNCLMEVLGLALPYNGTALAVSDQRREMIR 232
Query: 340 HPGTRL-----------SLVRMLLFDEAAEVNIISHYEVQGIL-SVVMDNEKQKDLDEEY 387
RL ++ D+A +++ +L ++ + NE D D E
Sbjct: 233 QAAFRLVENIKNDIKPRDIITQDAIDDAFALDMAMGGSTNTVLHTLAIANEAGIDYDLER 292
Query: 388 VNSFRLKAVNNND---SISYSEVDVHSSEDIPTILNDLDK 424
+N K + S SYS DVH + +P I+N+L K
Sbjct: 293 INEIAKKTPYLSKIAPSSSYSMHDVHEAGGVPAIINELMK 332
>ILVD_STAAW (P65158) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD)
Length = 562
Score = 32.0 bits (71), Expect = 3.8
Identities = 41/173 (23%), Positives = 76/173 (43%), Gaps = 27/173 (15%)
Query: 273 SLGALETTSVA---YKDINQNVMQTSPCSVGIFVDRNLGSITKMNFRICMIFIGGPDDHE 329
++GA + S++ + D+ QN T G+F + MN + ++ + P +
Sbjct: 166 AVGAFKEGSISKEEFLDMEQNACPTCGSCAGMF------TANSMNCLMEVLGLALPYNGT 219
Query: 330 ALAV-----------AWRMVGHPGTRLS---LVRMLLFDEAAEVNIISHYEVQGIL-SVV 374
ALAV A+++V + L +V D+A +++ +L ++
Sbjct: 220 ALAVSDQRREMIRQAAFKLVENIKNDLKPRDIVTREAIDDAFALDMAMGGSTNTVLHTLA 279
Query: 375 MDNEKQKDLDEEYVNSFRLKAVNNND---SISYSEVDVHSSEDIPTILNDLDK 424
+ NE D D E +N+ + + S SYS DVH + +P I+N+L K
Sbjct: 280 IANEAGIDYDLERINAIAKRTPYLSKIAPSSSYSMHDVHEAGGVPAIINELMK 332
>ILVD_STAAN (P65157) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD)
Length = 562
Score = 32.0 bits (71), Expect = 3.8
Identities = 41/173 (23%), Positives = 76/173 (43%), Gaps = 27/173 (15%)
Query: 273 SLGALETTSVA---YKDINQNVMQTSPCSVGIFVDRNLGSITKMNFRICMIFIGGPDDHE 329
++GA + S++ + D+ QN T G+F + MN + ++ + P +
Sbjct: 166 AVGAFKEGSISKEEFLDMEQNACPTCGSCAGMF------TANSMNCLMEVLGLALPYNGT 219
Query: 330 ALAV-----------AWRMVGHPGTRLS---LVRMLLFDEAAEVNIISHYEVQGIL-SVV 374
ALAV A+++V + L +V D+A +++ +L ++
Sbjct: 220 ALAVSDQRREMIRQAAFKLVENIKNDLKPRDIVTREAIDDAFALDMAMGGSTNTVLHTLA 279
Query: 375 MDNEKQKDLDEEYVNSFRLKAVNNND---SISYSEVDVHSSEDIPTILNDLDK 424
+ NE D D E +N+ + + S SYS DVH + +P I+N+L K
Sbjct: 280 IANEAGIDYDLERINAIAKRTPYLSKIAPSSSYSMHDVHEAGGVPAIINELMK 332
>ILVD_STAAM (P65156) Dihydroxy-acid dehydratase (EC 4.2.1.9) (DAD)
Length = 562
Score = 32.0 bits (71), Expect = 3.8
Identities = 41/173 (23%), Positives = 76/173 (43%), Gaps = 27/173 (15%)
Query: 273 SLGALETTSVA---YKDINQNVMQTSPCSVGIFVDRNLGSITKMNFRICMIFIGGPDDHE 329
++GA + S++ + D+ QN T G+F + MN + ++ + P +
Sbjct: 166 AVGAFKEGSISKEEFLDMEQNACPTCGSCAGMF------TANSMNCLMEVLGLALPYNGT 219
Query: 330 ALAV-----------AWRMVGHPGTRLS---LVRMLLFDEAAEVNIISHYEVQGIL-SVV 374
ALAV A+++V + L +V D+A +++ +L ++
Sbjct: 220 ALAVSDQRREMIRQAAFKLVENIKNDLKPRDIVTREAIDDAFALDMAMGGSTNTVLHTLA 279
Query: 375 MDNEKQKDLDEEYVNSFRLKAVNNND---SISYSEVDVHSSEDIPTILNDLDK 424
+ NE D D E +N+ + + S SYS DVH + +P I+N+L K
Sbjct: 280 IANEAGIDYDLERINAIAKRTPYLSKIAPSSSYSMHDVHEAGGVPAIINELMK 332
>KDTA_RICPR (Q9ZE58) 3-deoxy-D-manno-octulosonic-acid transferase
(EC 2.-.-.-) (KDO transferase)
Length = 461
Score = 31.6 bits (70), Expect = 5.0
Identities = 22/90 (24%), Positives = 46/90 (50%), Gaps = 7/90 (7%)
Query: 344 RLSLVRMLLFDEAAEVNIISHYEVQGILSVVMDNEKQKDLDEEYV----NSFRLKAVNNN 399
+LS + + L + V +H E + ++ +++N K++ +D + + R+K++ NN
Sbjct: 259 KLSKLILHLDNRRVLVFASTHPEDEEVILPIINNLKEQFIDCYIILIPRHPERIKSIINN 318
Query: 400 DSISYSEVDVHSSEDIPTILNDL---DKFG 426
+ + S D+P + NDL D+FG
Sbjct: 319 CKLHHLSATAKSQNDLPVLNNDLYIVDRFG 348
>DAG1_RABIT (Q28685) Dystroglycan precursor (Dystrophin-associated
glycoprotein 1) [Contains: Alpha-dystroglycan
(Alpha-DG); Beta-dystroglycan (Beta-DG)]
Length = 895
Score = 31.6 bits (70), Expect = 5.0
Identities = 19/80 (23%), Positives = 40/80 (49%), Gaps = 9/80 (11%)
Query: 370 ILSVVMDNEKQKDLDEEYVNSFRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDL 429
+L+V++D + K ++ ++ S+SEV++H+ + +P + N L F
Sbjct: 190 VLTVILDADLTKMTPKQRIDLLHRMQ-------SFSEVELHNMKLVPVVNNRL--FDMSA 240
Query: 430 YIVGQGHHRNVRAFSSLLEW 449
++ G G+ + V +LL W
Sbjct: 241 FMAGPGNAKKVVENGALLSW 260
>DAG1_HUMAN (Q14118) Dystroglycan precursor (Dystrophin-associated
glycoprotein 1) [Contains: Alpha-dystroglycan
(Alpha-DG); Beta-dystroglycan (Beta-DG)]
Length = 895
Score = 31.6 bits (70), Expect = 5.0
Identities = 19/80 (23%), Positives = 40/80 (49%), Gaps = 9/80 (11%)
Query: 370 ILSVVMDNEKQKDLDEEYVNSFRLKAVNNNDSISYSEVDVHSSEDIPTILNDLDKFGYDL 429
+L+V++D + K ++ ++ S+SEV++H+ + +P + N L F
Sbjct: 190 VLTVILDADLTKMTPKQRIDLLHRMR-------SFSEVELHNMKLVPVVNNRL--FDMSA 240
Query: 430 YIVGQGHHRNVRAFSSLLEW 449
++ G G+ + V +LL W
Sbjct: 241 FMAGPGNPKKVVENGALLSW 260
>LAR_CAEEL (Q9BMN8) Protein-tyrosine phosphatase Lar-like precursor
(EC 3.1.3.48) (Protein-tyrosine-phosphate
phosphohydrolase ptp-3)
Length = 2200
Score = 30.8 bits (68), Expect = 8.6
Identities = 32/175 (18%), Positives = 66/175 (37%), Gaps = 12/175 (6%)
Query: 173 SPIHVFALYLIELKGRTGALVAAHMDKPSNQPGAQNLTRSQVEQESINNTFEALG--EAY 230
+P +F + L + +H + GA + +E+ +NT + G A
Sbjct: 1819 TPFLIFLKRVKTLNPNDAGPIISHCSAGIGRTGAFIVIDCMLERLRYDNTVDIYGCVTAL 1878
Query: 231 DAVRVETLNVVSSYSTIHEDIYNSANEKRTSMIILPFHKHL-----SSLGALETTSVAYK 285
A R + Y IH+ + ++ N T + H+HL S L + ++
Sbjct: 1879 RAQRSYMVQTEEQYIFIHDAVLDAVNSGSTEVPASRLHQHLHILSQPSADQLSGIDMEFR 1938
Query: 286 DINQNVMQTSPCSVGIF-----VDRNLGSITKMNFRICMIFIGGPDDHEALAVAW 335
+ ++ C+V +R L ++ + R+ M + G D + + +W
Sbjct: 1939 HLTTLKWTSNRCTVANLPVNRPKNRMLSAVPYDSNRVIMRLLPGADGSDYINASW 1993
>CDC2_AJECA (P54119) Cell division control protein 2 (EC 2.7.1.37)
(Cyclin-dependent protein kinase)
Length = 324
Score = 30.8 bits (68), Expect = 8.6
Identities = 31/146 (21%), Positives = 60/146 (40%), Gaps = 21/146 (14%)
Query: 117 TINVIYKPRKRFEQNKLKTIQKLRLDA-----------ELRILTCVHNARQATGVISLVE 165
T V+YK R N++ ++K+RL+A E+ +L +H+ ++ L+
Sbjct: 14 TYGVVYKARDLTHPNRIVALKKIRLEAEDEGVPSTAIREISLLKEMHD----PNIVRLLN 69
Query: 166 SFNATRISPIHVFALYLIELKGRTGALVAAHMDKPSNQPGAQNLTRSQVEQESINNTFEA 225
+A VF ++LK AL + + P L +++ EA
Sbjct: 70 IVHADGHKLYLVFEFLDLDLKKYMEALPVSEGGRGKALPDGSTLDMNRL------GLGEA 123
Query: 226 LGEAYDAVRVETLNVVSSYSTIHEDI 251
+ + + A VE + S+ +H D+
Sbjct: 124 MVKKFMAQLVEGIRYCHSHRVLHRDL 149
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.135 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,469,769
Number of Sequences: 164201
Number of extensions: 2066687
Number of successful extensions: 5765
Number of sequences better than 10.0: 12
Number of HSP's better than 10.0 without gapping: 1
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 5756
Number of HSP's gapped (non-prelim): 20
length of query: 505
length of database: 59,974,054
effective HSP length: 115
effective length of query: 390
effective length of database: 41,090,939
effective search space: 16025466210
effective search space used: 16025466210
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC138011.4


