

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137986.10 - phase: 0
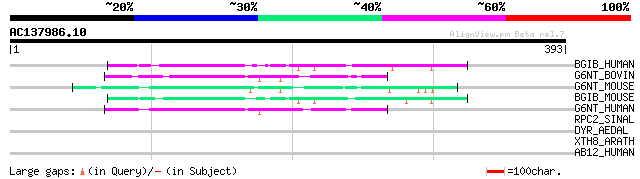
(393 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
BGIB_HUMAN (Q06430) N-acetyllactosaminide beta-1,6-N-acetylgluco... 52 2e-06
G6NT_BOVIN (Q92180) Beta-1,3-galactosyl-O-glycosyl-glycoprotein ... 50 8e-06
G6NT_MOUSE (Q09324) Beta-1,3-galactosyl-O-glycosyl-glycoprotein ... 50 1e-05
BGIB_MOUSE (P97402) N-acetyllactosaminide beta-1,6-N-acetylgluco... 50 1e-05
G6NT_HUMAN (Q02742) Beta-1,3-galactosyl-O-glycosyl-glycoprotein ... 49 3e-05
RPC2_SINAL (Q9THV5) DNA-directed RNA polymerase beta'' chain (EC... 35 0.33
DYR_AEDAL (P28019) Dihydrofolate reductase (EC 1.5.1.3) 33 1.6
XTH8_ARATH (Q8L9A9) Probable xyloglucan endotransglucosylase/hyd... 31 6.2
AB12_HUMAN (Q86UK0) ATP-binding cassette, sub-family A, member 1... 31 6.2
>BGIB_HUMAN (Q06430) N-acetyllactosaminide
beta-1,6-N-acetylglucosaminyl-transferase (EC 2.4.1.150)
(N-acetylglucosaminyltransferase) (I-branching enzyme)
(IGNT)
Length = 400
Score = 52.4 bits (124), Expect = 2e-06
Identities = 67/285 (23%), Positives = 117/285 (40%), Gaps = 51/285 (17%)
Query: 70 RLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLVTYRGPT 129
RL + +Y P N Y +H+D A E KD E + S N ++ K V Y G +
Sbjct: 110 RLFRAIYMPQNIYCVHVDEKA-TTEFKDAVE-----QLLSCFPNAFLASKMEPVVYGGIS 163
Query: 130 MLATTLHAMAMLLK-TCHWDWFINLSASDYPLVTQDGMVPRDINFIQHTSRLGWKFNKRG 188
L L+ + L W + IN D+PL T +V +++ G+K K
Sbjct: 164 RLQADLNCIRDLSAFEVSWKYVINTCGQDFPLKTNKEIV----QYLK-----GFK-GKNI 213
Query: 189 KPMIIDPGLYSLNKS----------DIWWIIKQRNL----PTSFKLYTGSAWTIVSRSFS 234
P ++ P +++ ++ ++ ++I+ L P + +Y GSA+ +SR F+
Sbjct: 214 TPGVLPPA-HAIGRTKYVHQEHLGKELSYVIRTTALKPPPPHNLTIYFGSAYVALSREFA 272
Query: 235 EYCIMGWENLPRTL-LLYYTNFVSSPEGYFQTVICN----SQEYKNTTANHDLHYITWDN 289
+ + + PR + LL ++ SP+ +F + N + +L I W +
Sbjct: 273 NFVL----HDPRAVDLLQWSKDTFSPDEHFWVTLNRIPGVPGSMPNASWTGNLRAIKWSD 328
Query: 290 PPKQHPRS----------LGLKDYRKMVLSSRPFARKFKRNNIVL 324
+H G D + +V S FA KF+ N L
Sbjct: 329 MEDRHGGCHGHYVHGICIYGNGDLKWLVNSPSLFANKFELNTYPL 373
>G6NT_BOVIN (Q92180) Beta-1,3-galactosyl-O-glycosyl-glycoprotein
beta-1,6-N-acetylglucosaminyltransferase (EC 2.4.1.102)
(Core 2 branching enzyme) (Core2-GlcNAc-transferase)
(C2GNT) (Core 2 GNT)
Length = 427
Score = 50.4 bits (119), Expect = 8e-06
Identities = 52/208 (25%), Positives = 91/208 (43%), Gaps = 25/208 (12%)
Query: 68 LKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLVTYRG 127
L RLL+ +Y P N+Y IH+D +E +A V FS NV++ + V Y
Sbjct: 136 LDRLLRAIYMPQNFYCIHVD---AKSEKSFLAAAVGIASCFS---NVFVASQLESVVYAS 189
Query: 128 PTMLATTLHAMAMLLK-TCHWDWFINLSASDYPLVTQDGMVPRDINFIQ-----HTSRLG 181
+ + L+ M L + W + INL D+P+ T +V R + + T ++
Sbjct: 190 WSRVQADLNCMQDLYQMNAGWKYLINLCGMDFPIKTNLEIV-RKLKLLMGENNLETEKMP 248
Query: 182 WKFNKRGKP--MIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIM 239
+R K +++ L ++ I + P L++GSA +VSR + EY +
Sbjct: 249 SHKKERWKKHYEVVNGKLTNMGTDKI-------HPPLETPLFSGSAHFVVSREYVEYVLQ 301
Query: 240 GWENLPRTLLLYYTNFVSSPEGYFQTVI 267
+N+ + + + SP+ Y I
Sbjct: 302 N-QNIQK--FMEWAKDTYSPDEYLWATI 326
>G6NT_MOUSE (Q09324) Beta-1,3-galactosyl-O-glycosyl-glycoprotein
beta-1,6-N-acetylglucosaminyltransferase (EC 2.4.1.102)
(Core 2 branching enzyme) (Core2-GlcNAc-transferase)
(C2GNT)
Length = 428
Score = 50.1 bits (118), Expect = 1e-05
Identities = 69/308 (22%), Positives = 124/308 (39%), Gaps = 56/308 (18%)
Query: 45 RTNITYPITFAYLISASKGDTLKLKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVAN 104
+ + +PI ++ ++ L RLL+ +Y P N+Y IH+D A + + A
Sbjct: 116 KEEVGFPIAYSIVVHHK---IEMLDRLLRAIYMPQNFYCIHVDRKAEE------SFLAAV 166
Query: 105 DPVFSQVGNVWIVGKPNLVTYRGPTMLATTLHAMAMLLK-TCHWDWFINLSASDYPLVTQ 163
+ S NV++ + V Y T + L+ M L + +W + INL D+P+ T
Sbjct: 167 QGIASCFDNVFVASQLESVVYASWTRVKADLNCMKDLYRMNANWKYLINLCGMDFPIKTN 226
Query: 164 DGMVPR------DINFIQHTSRLGWKFNKRGKP--MIIDPGLYSLNKSDIWWIIKQRNLP 215
+V + + N T ++ +R K ++D L + I + P
Sbjct: 227 LEIVRKLKCSTGENNL--ETEKMPPNKEERWKKRYAVVDGKLTNTG-------IVKAPPP 277
Query: 216 TSFKLYTGSAWTIVSRSFSEYCIMGWENLPRTLLLYYTNFVSSPEGYFQTVI-------- 267
L++GSA+ +V+R + Y ++ EN+ + L+ + SP+ + I
Sbjct: 278 LKTPLFSGSAYFVVTREYVGY-VLENENIQK--LMEWAQDTYSPDEFLWATIQRIPEVPG 334
Query: 268 --CNSQEYKNTTANHDLHYITWD-----------NPPKQ--HPRSL---GLKDYRKMVLS 309
+S +Y + N ++ W PP H RS+ G D M+
Sbjct: 335 SFPSSNKYDLSDMNAIARFVKWQYFEGDVSNGAPYPPCSGVHVRSVCVFGAGDLSWMLRQ 394
Query: 310 SRPFARKF 317
FA KF
Sbjct: 395 HHLFANKF 402
>BGIB_MOUSE (P97402) N-acetyllactosaminide
beta-1,6-N-acetylglucosaminyl-transferase (EC 2.4.1.150)
(N-acetylglucosaminyltransferase) (I-branching enzyme)
(IGNT) (Large I antigen-forming
beta-1,6-N-acetylglucosaminyltransferase)
Length = 400
Score = 50.1 bits (118), Expect = 1e-05
Identities = 65/285 (22%), Positives = 116/285 (39%), Gaps = 51/285 (17%)
Query: 70 RLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLVTYRGPT 129
RL + ++ P N Y +H+D A AE K E + S NV++ K V Y G +
Sbjct: 110 RLFRAIFMPQNIYCVHVDEKAT-AEFKGAVEQLV-----SCFPNVFLASKMEPVVYGGIS 163
Query: 130 MLATTLHAMAMLLKT-CHWDWFINLSASDYPLVTQDGMVPRDINFIQHTSRLGWKFNKRG 188
L L+ + L + W + IN D+PL T +V Q+ L K
Sbjct: 164 RLQADLNCIKDLSTSEVPWKYAINTCGQDFPLKTNKEIV-------QYLKGLK---GKNL 213
Query: 189 KPMIIDPGLYSLNKS----------DIWWIIKQRNL----PTSFKLYTGSAWTIVSRSFS 234
P ++ P +++ ++ ++ ++I+ L P + +Y GSA+ +SR F+
Sbjct: 214 TPGVLPPA-HAIGRTRYVHREHLSKELSYVIRTTALKPPPPHNLTIYFGSAYVALSREFA 272
Query: 235 EYCIMGWENLPRTL-LLYYTNFVSSPEGYFQTVICNSQEYKNTTAN----HDLHYITWDN 289
+ + PR + LL+++ SP+ +F + + N +L + W +
Sbjct: 273 NFVLRD----PRAVDLLHWSKDTFSPDEHFWVTLNRIPGVPGSPPNASWTGNLRAVKWMD 328
Query: 290 PPKQHPRS----------LGLKDYRKMVLSSRPFARKFKRNNIVL 324
+H G D + ++ S FA KF+ N L
Sbjct: 329 MEAKHGGCQGHYVHGICIYGNGDLQWLINSQSLFANKFELNTYPL 373
>G6NT_HUMAN (Q02742) Beta-1,3-galactosyl-O-glycosyl-glycoprotein
beta-1,6-N-acetylglucosaminyltransferase (EC 2.4.1.102)
(Core 2 branching enzyme) (Core2-GlcNAc-transferase)
(C2GNT) (Core 2 GNT)
Length = 428
Score = 48.5 bits (114), Expect = 3e-05
Identities = 48/206 (23%), Positives = 85/206 (40%), Gaps = 21/206 (10%)
Query: 68 LKRLLKVLYHPNNYYLIHMDYGAPDAEHKDVAEYVANDPVFSQVGNVWIVGKPNLVTYRG 127
L RLL+ +Y P N+Y +H+D + D + A + S NV++ + V Y
Sbjct: 136 LDRLLRAIYMPQNFYCVHVDTKSED------SYLAAVMGIASCFSNVFVASRLESVVYAS 189
Query: 128 PTMLATTLHAMA-MLLKTCHWDWFINLSASDYPLVTQDGMVPRDINFIQ-----HTSRLG 181
+ + L+ M + + +W + INL D+P+ T +V R + + T R+
Sbjct: 190 WSRVQADLNCMKDLYAMSANWKYLINLCGMDFPIKTNLEIV-RKLKLLMGENNLETERMP 248
Query: 182 WKFNKRGKPMIIDPGLYSLNKSDIWWIIKQRNLPTSFKLYTGSAWTIVSRSFSEYCIMGW 241
+R K N + + P L++GSA+ +VSR + Y +
Sbjct: 249 SHKEERWKKRYEVVNGKLTNTGTVKML-----PPLETPLFSGSAYFVVSREYVGYVL--- 300
Query: 242 ENLPRTLLLYYTNFVSSPEGYFQTVI 267
+N L+ + SP+ Y I
Sbjct: 301 QNEKIQKLMEWAQDTYSPDEYLWATI 326
>RPC2_SINAL (Q9THV5) DNA-directed RNA polymerase beta'' chain (EC
2.7.7.6) (PEP) (Plastid-encoded RNA polymerase beta''
subunit) (RNA polymerase beta'' subunit)
Length = 1384
Score = 35.0 bits (79), Expect = 0.33
Identities = 33/131 (25%), Positives = 54/131 (41%), Gaps = 18/131 (13%)
Query: 15 ILFFFLFIPTRLTLQISTLKPSAMDYFNVLRTNITYPITFAYLISASKGDTLKLKRLLKV 74
IL F F T+L QIS +K Y ++ +N+ +P+ +YLI G L +
Sbjct: 989 ILIFIHFAFTKLINQISCIK-----YLHLQTSNVFFPVIHSYLID-ENGRIFNLDPYSNL 1042
Query: 75 LYHP--NNYYLIHMDYGAPDAEHKDVA----------EYVANDPVFSQVGNVWIVGKPNL 122
+ +P N+Y +H +Y E +A + + G V IV + ++
Sbjct: 1043 VLNPFKLNWYFLHQNYNNNYCEETSTIISLGQFFCENVCIAKKEPYLKSGQVLIVQRDSV 1102
Query: 123 VTYRGPTMLAT 133
V LAT
Sbjct: 1103 VIRSAKPYLAT 1113
>DYR_AEDAL (P28019) Dihydrofolate reductase (EC 1.5.1.3)
Length = 186
Score = 32.7 bits (73), Expect = 1.6
Identities = 28/86 (32%), Positives = 41/86 (47%), Gaps = 8/86 (9%)
Query: 106 PVFSQVGNVWIVGKPNLVTYRGPTMLATTLHAMAM--LLKTCHWDWFINLSASDYPLVTQ 163
P+ +++ NVWIVG Y+ M + H + + + +T D F SD+ LV
Sbjct: 104 PLVNEIENVWIVG--GNAVYK-EAMQSDRCHRIYLTEIKETFECDAFFPEITSDFQLVKN 160
Query: 164 DGMVPRDI---NFIQHTSRLGWKFNK 186
D VP DI N IQ+ R+ K K
Sbjct: 161 DDDVPEDIQEENGIQYQYRIYEKVPK 186
>XTH8_ARATH (Q8L9A9) Probable xyloglucan
endotransglucosylase/hydrolase protein 8 precursor (EC
2.4.1.207) (At-XTH8) (XTH-8)
Length = 292
Score = 30.8 bits (68), Expect = 6.2
Identities = 24/97 (24%), Positives = 41/97 (41%), Gaps = 12/97 (12%)
Query: 264 QTVICNSQEYKNTTANHDLHYITWDNPPKQ-HPRSLGLKDYRKMVLSSRPFARKFKRNNI 322
Q I + YKN T N ++ + W +P K H S+ +++ + R R +K +
Sbjct: 114 QPYIIQTNVYKNGTGNREMRHSLWFDPTKDYHTYSILWNNHQLVFFVDRVPIRVYKNS-- 171
Query: 323 VLDKI-DRDLLKRYKGGFSFGG------WCSQGGRNK 352
DK+ + D K + F W ++GG K
Sbjct: 172 --DKVPNNDFFPNQKPMYLFSSIWNADDWATRGGLEK 206
>AB12_HUMAN (Q86UK0) ATP-binding cassette, sub-family A, member 12
(ATP-binding cassette transporter 12) (ATP-binding
cassette 12)
Length = 2595
Score = 30.8 bits (68), Expect = 6.2
Identities = 20/75 (26%), Positives = 34/75 (44%), Gaps = 5/75 (6%)
Query: 243 NLPRTLLLYYTNFVSSPEGYFQTVICNSQEYKNTTANHDLHYITWDNPPKQHPRSLGLKD 302
++P T +Y +N +++P+G F T+ TT Y+T P Q P+ KD
Sbjct: 702 SVPLTQAMYRSNRMNTPQGSFSTISQALCSQGITT-----EYLTAMLPSSQRPKGNHTKD 756
Query: 303 YRKMVLSSRPFARKF 317
+ L+ A K+
Sbjct: 757 FLTYKLTKEQIASKY 771
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.325 0.140 0.447
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,948,244
Number of Sequences: 164201
Number of extensions: 2168095
Number of successful extensions: 5474
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 5461
Number of HSP's gapped (non-prelim): 12
length of query: 393
length of database: 59,974,054
effective HSP length: 112
effective length of query: 281
effective length of database: 41,583,542
effective search space: 11684975302
effective search space used: 11684975302
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137986.10


