

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137831.14 + phase: 0
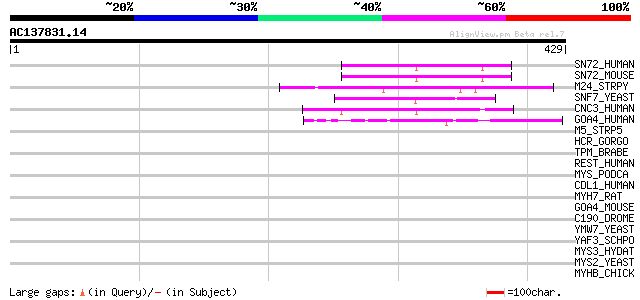
(429 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SN72_HUMAN (Q9NZZ3) SNF7 domain containing protein 2 (CGI-34) (H... 53 2e-06
SN72_MOUSE (Q9D7S9) SNF7 domain containing protein 2 52 3e-06
M24_STRPY (P12379) M protein, serotype 24 precursor 49 2e-05
SNF7_YEAST (P39929) Nuclear protein SNF7 47 7e-05
CNC3_HUMAN (Q9BY43) Protein C14orf123 (HSPC134) (CDA04) 47 9e-05
GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member... 46 2e-04
M5_STRP5 (P02977) M protein, serotype 5 precursor 44 0.001
HCR_GORGO (Q8HZ59) Alpha helical coiled-coil rod protein 42 0.002
TPM_BRABE (Q9NDS0) Tropomyosin 42 0.004
REST_HUMAN (P30622) Restin (Cytoplasmic linker protein-170 alpha... 41 0.005
MYS_PODCA (Q05000) Myosin heavy chain (Fragment) 41 0.005
CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2... 41 0.005
MYH7_RAT (P02564) Myosin heavy chain, cardiac muscle beta isofor... 41 0.007
GOA4_MOUSE (Q91VW5) Golgi autoantigen, golgin subfamily A member... 41 0.007
C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 1... 41 0.007
YMW7_YEAST (Q04272) Hypothetical 25.6 kDa protein in ABF2-CHL12 ... 40 0.009
YAF3_SCHPO (Q09857) Hypothetical protein C29E6.03c in chromosome I 40 0.009
MYS3_HYDAT (P39922) Myosin heavy chain, clone 203 (Fragment) 40 0.009
MYS2_YEAST (P19524) Myosin-2 (Class V unconventional myosin MYO2... 40 0.009
MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle 40 0.009
>SN72_HUMAN (Q9NZZ3) SNF7 domain containing protein 2 (CGI-34)
(HSPC177)
Length = 219
Score = 52.8 bits (125), Expect = 2e-06
Identities = 29/137 (21%), Positives = 72/137 (52%), Gaps = 5/137 (3%)
Query: 257 RKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIKE--N 314
++ ALR ++ ++ Q R+ + +E+ + + + TKT +AM++G + +K+
Sbjct: 62 KQKALRVLKQKRMYEQQRDNLAQQSFNMEQANYTIQSLKDTKTTVDAMKLGVKEMKKAYK 121
Query: 315 KISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELE---LALEKEA 371
++ ++ ++ L++ ++ E+++AL ++ +++++D+E EL+ L LA E +
Sbjct: 122 QVKIDQIEDLQDQLEDMMEDANEIQEALSRSYGTPELDEDDLEAELDALGDELLADEDSS 181
Query: 372 QVDTLEKTTTSEEGNAT 388
+D EG T
Sbjct: 182 YLDEAASAPAIPEGVPT 198
>SN72_MOUSE (Q9D7S9) SNF7 domain containing protein 2
Length = 219
Score = 52.0 bits (123), Expect = 3e-06
Identities = 29/137 (21%), Positives = 72/137 (52%), Gaps = 5/137 (3%)
Query: 257 RKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIKE--N 314
++ ALR ++ ++ Q R+ + +E+ + + + TKT +AM++G + +K+
Sbjct: 62 KQKALRVLKQKRMYEQQRDNLAQQSFNMEQANYTIQSLKDTKTTVDAMKLGVKEMKKAYK 121
Query: 315 KISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELE---LALEKEA 371
++ ++ ++ L++ ++ E+++AL ++ +++++D+E EL+ L LA E +
Sbjct: 122 EVKIDQIEDLQDQLEDMMEDANEIQEALGRSYGTPELDEDDLEAELDALGDELLADEDSS 181
Query: 372 QVDTLEKTTTSEEGNAT 388
+D EG T
Sbjct: 182 YLDEAASAPAIPEGVPT 198
>M24_STRPY (P12379) M protein, serotype 24 precursor
Length = 539
Score = 48.9 bits (115), Expect = 2e-05
Identities = 56/225 (24%), Positives = 103/225 (44%), Gaps = 15/225 (6%)
Query: 209 ALSGASNLDCDVLYLIWTIEKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELK 268
AL GA N I T+E + L+ + +L +S V + + + + L +RE K
Sbjct: 268 ALEGAMNFSTADSAKIKTLEAEKAALEA--EKADLEHQSQVLNANRQSLRRDLDASREAK 325
Query: 269 LVTQSREKCSSLLNRVEEV--HGVVVDAESTKTVSEAMQIGARAIKE-NKISVEDVDICL 325
++ + N++ E + D ++++ + ++ + ++E NKIS
Sbjct: 326 KQLEAEHQKLEEQNKISEASRQSLRRDLDASREAKKQLEAEHQKLEEQNKISEASRQSLR 385
Query: 326 RDLQESIDSQKEVEKALEQTPS---YTDINDEDIEE-----ELEELELALEKEAQVDTL- 376
RDL S +++K+VEKALE+ S + ++++EE E E+ EL + EA+ L
Sbjct: 386 RDLDASREAKKQVEKALEEANSKLAALEKLNKELEESKKLTEKEKAELQAKLEAEAKALK 445
Query: 377 EKTTTSEEGNATLEASELLSDTLSNLKLSDRPV-GKSRTTHAASE 420
EK E A L A + + K ++ V GK + A ++
Sbjct: 446 EKLAKQAEELAKLRAGKASDSQTPDAKPGNKAVPGKGQAPQAGTK 490
>SNF7_YEAST (P39929) Nuclear protein SNF7
Length = 240
Score = 47.4 bits (111), Expect = 7e-05
Identities = 31/126 (24%), Positives = 62/126 (48%), Gaps = 3/126 (2%)
Query: 252 LHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARAI 311
L GN+ +A ++ K + Q K + +E+ + A AMQ GA+A+
Sbjct: 55 LTKGNKVMAKNALKKKKTIEQLLSKVEGTMESMEQQLFSIESANLNLETMRAMQEGAKAM 114
Query: 312 K--ENKISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEK 369
K + + ++ VD + +++E ++ E+ A+ + P T N+ D +E EEL++ ++
Sbjct: 115 KTIHSGLDIDKVDETMDEIREQVELGDEISDAISR-PLITGANEVDEDELDEELDMLAQE 173
Query: 370 EAQVDT 375
A +T
Sbjct: 174 NANQET 179
>CNC3_HUMAN (Q9BY43) Protein C14orf123 (HSPC134) (CDA04)
Length = 222
Score = 47.0 bits (110), Expect = 9e-05
Identities = 39/172 (22%), Positives = 80/172 (45%), Gaps = 12/172 (6%)
Query: 227 IEKLQQQLDVIDRRCELSRKSAVASLHSG------NRKLALRYARELKLVTQSREKCSSL 280
I+KL++ ++ ++ E + L + N++ AL+ R K Q +
Sbjct: 23 IQKLKETEKILIKKQEFLEQKIQQELQTAKKYGTKNKRAALQALRRKKRFEQQLAQTDGT 82
Query: 281 LNRVEEVHGVVVDAESTKTVSEAMQIGARAIKE--NKISVEDVDICLRDLQESIDSQKEV 338
L+ +E + +A + V M++ A+++K+ + ++ VD + D+ E + +++
Sbjct: 83 LSTLEFQREAIENATTNAEVLRTMELAAQSMKKAYQDMDIDKVDELMTDITEQQEVAQQI 142
Query: 339 EKALEQTPSYTDINDED-IEEELEELELALEKEAQVDTLEKTTTSEEGNATL 389
A+ + + D DED + EELEELE ++E + L EE + L
Sbjct: 143 SDAISRPMGFRDDVDEDELLEELEELE---QEELAQELLNVGDKEEEPSVKL 191
>GOA4_HUMAN (Q13439) Golgi autoantigen, golgin subfamily A member 4
(Trans-Golgi p230) (256 kDa golgin) (Golgin-245) (72.1
protein)
Length = 2230
Score = 45.8 bits (107), Expect = 2e-04
Identities = 50/206 (24%), Positives = 96/206 (46%), Gaps = 32/206 (15%)
Query: 228 EKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEV 287
E LQ+QLD +R EL + + LH A + KL+TQ R+ +L+ ++E+
Sbjct: 317 EALQEQLD--ERLQELEK---IKDLHM---------AEKTKLITQLRD-AKNLIEQLEQD 361
Query: 288 HGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQK-----EVEKAL 342
G+V+ AE+ + + E +++ I + + ++ + +L+E + + E+EKAL
Sbjct: 362 KGMVI-AETKRQMHETLEMKEEEIAQLRSRIKQMTTQGEELREQKEKSERAAFEELEKAL 420
Query: 343 EQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLEASELLSDTLSNL 402
T T+ ++ E++E Q+ T+EKT+ E + E S + + + +
Sbjct: 421 S-TAQKTEEARRKLKAEMDE---------QIKTIEKTSEEERISLQQELSRVKQEVVDVM 470
Query: 403 -KLSDRPVGKSRTTHAASEGHKTANL 427
K S+ + K + H K L
Sbjct: 471 KKSSEEQIAKLQKLHEKELARKEQEL 496
>M5_STRP5 (P02977) M protein, serotype 5 precursor
Length = 492
Score = 43.5 bits (101), Expect = 0.001
Identities = 47/194 (24%), Positives = 90/194 (46%), Gaps = 14/194 (7%)
Query: 228 EKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALR---YARELKLVTQSREKCSSLLNRV 284
+K +Q + + + EL++K + +RK R +RE K ++ + N++
Sbjct: 237 QKSKQNIGALKQ--ELAKKDEANKISDASRKGLRRDLDASREAKKQLEAEHQKLEEQNKI 294
Query: 285 EEVH--GVVVDAESTKTVSEAMQIGARAIKE-NKISVEDVDICLRDLQESIDSQKEVEKA 341
E G+ D ++++ + ++ + ++E NKIS RDL S +++K+VEKA
Sbjct: 295 SEASRKGLRRDLDASREAKKQLEAEQQKLEEQNKISEASRKGLRRDLDASREAKKQVEKA 354
Query: 342 LEQTPSYTDINDEDIEEELEELELALEKE-----AQVDTLEKTTTSEEGNATLEASELLS 396
LE+ S E + +ELEE + EKE A+++ K + E ++L +
Sbjct: 355 LEEANSKLAAL-EKLNKELEESKKLTEKEKAELQAKLEAEAKALKEQLAKQAEELAKLRA 413
Query: 397 DTLSNLKLSDRPVG 410
S+ + D G
Sbjct: 414 GKASDSQTPDTKPG 427
>HCR_GORGO (Q8HZ59) Alpha helical coiled-coil rod protein
Length = 782
Score = 42.4 bits (98), Expect = 0.002
Identities = 53/222 (23%), Positives = 98/222 (43%), Gaps = 34/222 (15%)
Query: 223 LIWTIEKLQQQLDVIDRRCELSR---KSAVASLHSGNRKLALRYARELKLVTQSREKCSS 279
L+ T++ LQ+ D + EL + +S L +L + L + KC S
Sbjct: 273 LLETMQHLQEDRDSLQATAELLQVRVQSLTHILALQEEELTRKVQPSDSLEPEFTRKCQS 332
Query: 280 LLNRVEEVHGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVE 339
LLNR E K + +Q+ A+ + E+ SV+ + + LQE + SQ + +
Sbjct: 333 LLNRWRE-----------KVFALMVQLKAQEL-EHSDSVKQLKGQVASLQEKVTSQSQEQ 380
Query: 340 KALEQTPSYTDINDEDIEEELE-------ELELALEKEAQVDTLEKTTTSEEG-----NA 387
L+Q+ + D+ E E+E +LEL+ +EA+ ++T ++EE NA
Sbjct: 381 AILQQS-----LQDKAAEVEVERMGAKGLQLELSRAQEARCRWQQQTASAEEQLRLVVNA 435
Query: 388 TLEASELLSDTLSNLK--LSDRPVGKSRTTHAASEGHKTANL 427
+ L T++ ++ + P +R ++A + H L
Sbjct: 436 VSSSQIWLETTMAKVEGAAAQLPSLNNRLSYAVRKVHTIRGL 477
>TPM_BRABE (Q9NDS0) Tropomyosin
Length = 284
Score = 41.6 bits (96), Expect = 0.004
Identities = 43/174 (24%), Positives = 79/174 (44%), Gaps = 16/174 (9%)
Query: 228 EKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALRYA------RELKLVTQSREKCSSLL 281
+K++ D +D+ E S K A L + +K A A R ++LV + ++ L
Sbjct: 48 KKIRMVEDELDKAQE-SLKEATEQLEAATKKAADAEAEVASLNRRIQLVEEELDRAQERL 106
Query: 282 NRVEEVHGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQ--ESIDSQKEVE 339
N E + D+E SE AR + EN+ ++ + L D+Q E+ +E +
Sbjct: 107 NSTVEK---LTDSEKAADESER----ARKVLENRQGADEDKMELLDMQLREAKMIAEEAD 159
Query: 340 KALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLEASE 393
+ E+ I + D+E E +LA K +++ KTTT + + +A++
Sbjct: 160 RKYEEVARKLVITEGDLERAEERADLAETKARELEDELKTTTGQLKSMEAQATK 213
>REST_HUMAN (P30622) Restin (Cytoplasmic linker protein-170 alpha-2)
(CLIP-170) (Reed-Sternberg intermediate filament
associated protein)
Length = 1427
Score = 41.2 bits (95), Expect = 0.005
Identities = 37/143 (25%), Positives = 66/143 (45%), Gaps = 11/143 (7%)
Query: 273 SREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIKENK---ISVEDVDICLRDLQ 329
+RE+ S LL +EE+ A++ +T +AMQI + KE S+ED LQ
Sbjct: 1050 AREENSGLLQELEELRKQADKAKAAQTAEDAMQIMEQMTKEKTETLASLEDTKQTNAKLQ 1109
Query: 330 ESIDSQKEVE----KALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEG 385
+D+ KE + L ++ + ++ +EE +E+E K+A ++ + +E
Sbjct: 1110 NELDTLKENNLKNVEELNKSKELLTVENQKMEEFRKEIETL--KQAAAQKSQQLSALQEE 1167
Query: 386 NATL--EASELLSDTLSNLKLSD 406
N L E + S+ KL +
Sbjct: 1168 NVKLAEELGRSRDEVTSHQKLEE 1190
Score = 32.0 bits (71), Expect = 3.1
Identities = 35/156 (22%), Positives = 73/156 (46%), Gaps = 10/156 (6%)
Query: 227 IEKLQQQLDVIDRRCELSRKS-AVASLHSGNRKLALRYAREL--KLVTQSREKCSSLLNR 283
+++ QQ ++ + +L R A A+ H G + L AR+ + V + K L
Sbjct: 357 LKEKQQHIEQLLAERDLERAEVAKATSHVGEIEQELALARDGHDQHVLELEAKMDQLRTM 416
Query: 284 VEEVHGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALE 343
VE D E + +++ ++ R +++ + VE+ I DL+ + K LE
Sbjct: 417 VE-----AADREKVELLNQ-LEEEKRKVEDLQFRVEEESITKGDLETQTKLEHARIKELE 470
Query: 344 QTPSYTDINDEDIEEELEELELA-LEKEAQVDTLEK 378
Q+ + + ++ ELE+ +A + +++++ LEK
Sbjct: 471 QSLLFEKTKADKLQRELEDTRVATVSEKSRIMELEK 506
>MYS_PODCA (Q05000) Myosin heavy chain (Fragment)
Length = 692
Score = 41.2 bits (95), Expect = 0.005
Identities = 40/200 (20%), Positives = 88/200 (44%), Gaps = 32/200 (16%)
Query: 227 IEKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEE 286
+EKL+++L + + +++ + A A+L KL L+ E + QS ++ L + EE
Sbjct: 265 VEKLRRKLGMENEELQIALEEAEAALEQEEGKL-LKVQLEYTQLRQSSDR--KLSEKDEE 321
Query: 287 VHGV-------------VVDAEST----------KTVSEAMQIGARAIKENKISVED--- 320
+ G+ +D+ES K ++ M++ ++ N+++ E
Sbjct: 322 LEGLRKNHQRQMESLQNTIDSESRSKAEQQKLRKKYDADMMELESQLESSNRVAAESQKQ 381
Query: 321 ---VDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLE 377
+ +++LQ ID + + + S ++ D+ +L+E +ALE+ + L
Sbjct: 382 MKKLQAQIKELQSMIDDESRGRDDMRDSASRSERRANDLAVQLDEARVALEQAERARKLA 441
Query: 378 KTTTSEEGNATLEASELLSD 397
+ SE + E L ++
Sbjct: 442 ENEKSENSDRVAELQALYNN 461
>CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1) (CLK-1)
(CDK11) (p58 CLK-1)
Length = 795
Score = 41.2 bits (95), Expect = 0.005
Identities = 45/196 (22%), Positives = 87/196 (43%), Gaps = 21/196 (10%)
Query: 231 QQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEVHGV 290
+ +L+ ++R+ E RK +K R A E + ++R + S+ + E +
Sbjct: 165 RDRLEQLERKRERERKMREQQKEQREQKERERRAEERRKEREARREVSAHHRTMREDYSD 224
Query: 291 VVDAES-----TKTVSEAMQIG----------ARA-----IKENKISVEDVDICLRDLQE 330
V A + E ++G ARA +KE K+ D+ L+D+ +
Sbjct: 225 KVKASHWSRSPPRPPRERFELGDGRKPGEARPARAQKPAQLKEEKMEERDLLSDLQDISD 284
Query: 331 SIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLE 390
S E + ++ S ++ +E+ EEE EE + E E + + E+ E G+ + E
Sbjct: 285 SERKTSSAESSSAESGSGSEEEEEEEEEEEEEGSTSEESEEEEEE-EEEEEEETGSNSEE 343
Query: 391 ASELLSDTLSNLKLSD 406
ASE ++ +S ++S+
Sbjct: 344 ASEQSAEEVSEEEMSE 359
>MYH7_RAT (P02564) Myosin heavy chain, cardiac muscle beta isoform
(MyHC-beta)
Length = 1935
Score = 40.8 bits (94), Expect = 0.007
Identities = 61/250 (24%), Positives = 98/250 (38%), Gaps = 46/250 (18%)
Query: 187 TQYLSVLKNEFVEGVKISLSAAALSGASNLDCDVL------------YLIWTIEKLQQQL 234
TQ L LK + E VK + A ++ DCD+L L + K ++
Sbjct: 1309 TQQLEDLKRQLEEEVKAKNALAHALQSARHDCDLLREQYEEETEAKAELQRVLSKANSEV 1368
Query: 235 DVIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSL---LNRVE-EVHGV 290
+ E L +KLA R + V KCSSL +R++ E+ +
Sbjct: 1369 AQWRTKYETDAIQRTEELEEAKKKLAQRLQDAEEAVEAVNAKCSSLEKTKHRLQNEIEDL 1428
Query: 291 VVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEV------------ 338
+VD E + + A+ R +KI VE E SQKE
Sbjct: 1429 MVDVERSNAAAAALDKKQRNF--DKILVEWKQKYEESQSELESSQKEARSLSTELFKLKN 1486
Query: 339 --EKALEQTPSYTDINDEDIEEELEEL------------EL-ALEKEAQVDTLEKTTTSE 383
E++LE ++ N ++++EE+ +L EL + K+ + + LE + E
Sbjct: 1487 AYEESLEHLETFKREN-KNLQEEISDLTEQLGSTGKSIHELEKIRKQLEAEKLELQSALE 1545
Query: 384 EGNATLEASE 393
E A+LE E
Sbjct: 1546 EAEASLEHEE 1555
Score = 31.6 bits (70), Expect = 4.1
Identities = 36/176 (20%), Positives = 75/176 (42%), Gaps = 27/176 (15%)
Query: 227 IEKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEE 286
+++LQ +++ ++ E R + KL +REL+ +++ R+EE
Sbjct: 1108 LKELQARIEELEEELEAERTARAKV-----EKLRSDLSRELEEISE----------RLEE 1152
Query: 287 VHGVV-VDAESTKTVSEAMQIGARAIKENKISVEDVDICLR--------DLQESIDSQKE 337
G V E K Q R ++E + E LR +L E ID+ +
Sbjct: 1153 AGGATSVQIEMNKKREAEFQKMRRDLEEATLQHEATAAALRKKHADSVAELGEQIDNLQR 1212
Query: 338 VEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLEASE 393
V++ LE+ S + +D+ +E++ ++ +A ++ + +T + +A E
Sbjct: 1213 VKQKLEKEKSEFKLELDDVTSNMEQI---IKAKANLEKMCRTLEDQMNEHRSKAEE 1265
>GOA4_MOUSE (Q91VW5) Golgi autoantigen, golgin subfamily A member 4
(tGolgin-1)
Length = 2238
Score = 40.8 bits (94), Expect = 0.007
Identities = 47/177 (26%), Positives = 79/177 (44%), Gaps = 25/177 (14%)
Query: 231 QQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELKLV-----TQSREKCSSLLNRVE 285
QQ+LD++ R CE + A L L L++ LK + TQ +K L V+
Sbjct: 1972 QQELDILKRECE---QEAEEKLKQEQEDLELKHTSTLKQLMREFNTQLAQKEQELERTVQ 2028
Query: 286 EV--HGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALE 343
E V+AE ++ E Q R I E + +D+ R +E +D+++E E
Sbjct: 2029 ETIDKAQEVEAELLESHQEETQQLHRKIAEKE---DDLRRTARRYEEILDAREE-----E 2080
Query: 344 QTPSYTDINDEDIEEELEELELALEK--EAQVDTLEKTTTSEEGNATLEASELLSDT 398
T T D++ +LEEL+ ++ E + T + T E + + L+SD+
Sbjct: 2081 MTGKVT-----DLQTQLEELQKKYQQRLEQEESTKDSVTILELQTQLAQKTTLISDS 2132
>C190_DROME (Q9VJE5) Restin homolog (Cytoplasmic linker protein 190)
(Microtubule binding protein 190) (d-CLIP-190)
Length = 1690
Score = 40.8 bits (94), Expect = 0.007
Identities = 44/212 (20%), Positives = 93/212 (43%), Gaps = 19/212 (8%)
Query: 226 TIEKLQQQLDVIDRRCELSRKSA-------------VASLHSGNRKLALRYARELKLVTQ 272
TI+ LQ++L++ + + K A V ++ N ++ A EL V +
Sbjct: 1081 TIKDLQERLEITNAELQHKEKMASEDAQKIADLKTLVEAIQVANANISATNA-ELSTVLE 1139
Query: 273 SREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESI 332
+ S N + E+ + D S + + + I +KE + +++ +L+E +
Sbjct: 1140 VLQAEKSETNHIFELFEMEADMNSERLIEKVTGI-KEELKETHLQLDERQKKFEELEEKL 1198
Query: 333 DSQKEVEKALEQTPSYTDINDEDIEEELEELELAL-EKEAQVDTLEKTTTSEEGNATLEA 391
++ E+ L+Q + +I++ L+EL+ ++ +KE V LE+ E ++ +EA
Sbjct: 1199 KQAQQSEQKLQQESQTSKEKLTEIQQSLQELQDSVKQKEELVQNLEEKV--RESSSIIEA 1256
Query: 392 SELLSDTLSNLKLSDRPVGKSRTTHAASEGHK 423
+ SN++L ++ T E K
Sbjct: 1257 QNTKLNE-SNVQLENKTSCLKETQDQLLESQK 1287
>YMW7_YEAST (Q04272) Hypothetical 25.6 kDa protein in ABF2-CHL12
intergenic region
Length = 221
Score = 40.4 bits (93), Expect = 0.009
Identities = 33/137 (24%), Positives = 62/137 (45%), Gaps = 4/137 (2%)
Query: 257 RKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARAIKENKI 316
R L R + L+ Q+ ++ +L N V + +V+ K ++ G +K+
Sbjct: 63 RFLLKRIHYQEHLLQQASDQLINLENMVSTLEFKMVE----KQFINGLKNGNEILKKLNK 118
Query: 317 SVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTL 376
+VD + D+Q+ I Q E+ + L ++ T ++D+++EL+ LE L E +
Sbjct: 119 EFSNVDELMDDVQDQIAYQNEINETLSRSLVGTSNYEDDLDKELDALESELNPEKMNNAK 178
Query: 377 EKTTTSEEGNATLEASE 393
S EG +L E
Sbjct: 179 VANMPSTEGLPSLPQGE 195
>YAF3_SCHPO (Q09857) Hypothetical protein C29E6.03c in chromosome I
Length = 1044
Score = 40.4 bits (93), Expect = 0.009
Identities = 35/146 (23%), Positives = 69/146 (46%), Gaps = 18/146 (12%)
Query: 227 IEKLQQQLDVIDRRCEL--SRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRV 284
+++LQ QL+ + E R SA A S + A ELKL +++KCS+L ++
Sbjct: 815 LQELQSQLNQDKNQIETLNERISAAADELSSMESINKNQANELKL---AKQKCSNLQEKI 871
Query: 285 -----------EEVHGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESID 333
E++ + D E+ + + + +K S++ V ++ ++S++
Sbjct: 872 NFGNKLAKEHTEKISSLEKDLEAATKTASTLSKELKTVKSENDSLKSVSNDDQNKEKSVN 931
Query: 334 SQK--EVEKALEQTPSYTDINDEDIE 357
++K EV +AL + + DE+IE
Sbjct: 932 NEKFKEVSQALAEANEKLNARDEEIE 957
>MYS3_HYDAT (P39922) Myosin heavy chain, clone 203 (Fragment)
Length = 539
Score = 40.4 bits (93), Expect = 0.009
Identities = 54/231 (23%), Positives = 106/231 (45%), Gaps = 24/231 (10%)
Query: 211 SGASNLDCDVLYLIWTIEKLQQQLD---VIDRRCELSRK---SAVASLHSGNRKLALRYA 264
S S L + L+ IE+L+++L+ + ++ EL RK S + L +
Sbjct: 302 SQISQLQRKIQELLAKIEELEEELENERKLRQKSELQRKELESRIEELQDQLETAGGATS 361
Query: 265 RELKLVTQSREKCSSLLNRVEEVHGV------VVDAESTKTVSEAMQIGARAIKENKISV 318
++++ + +C+ L +E ++ + A++ T++E +Q A+K+ K +
Sbjct: 362 AQVEVGKKREAECNRLRKEIEALNIANDAAISAIKAKTNATIAE-IQEENEAMKKAKAKL 420
Query: 319 EDVDICLR-DLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTLE 377
E L +L E+ +S +++K ++T S D N +EE++ EL K AQVD L
Sbjct: 421 EKEKSALNNELNETKNSLDQIKK--QKTNS--DKNSRMLEEQINELN---SKLAQVDELH 473
Query: 378 KTTTSEEGNATLEASEL---LSDTLSNLKLSDRPVGKSRTTHAASEGHKTA 425
+ S+ E L LS++ NL ++ + + + A S+ A
Sbjct: 474 SQSESKNSKVNSELLALNSQLSESEHNLGIATKNIKTLESQLAESKNFNEA 524
Score = 32.3 bits (72), Expect = 2.4
Identities = 21/115 (18%), Positives = 60/115 (51%), Gaps = 9/115 (7%)
Query: 273 SREKCSSLLNRVEEVHGVVVDAESTKTVSEAMQIGARA-IKENKISVEDVDICLRDLQES 331
+R+K S R++E +V E + + E + G + I + + ++++ + +L+E
Sbjct: 265 NRDKLSETETRLKETQDLVTKREKSISDLENAKEGLESQISQLQRKIQELLAKIEELEEE 324
Query: 332 IDSQKEVEKALEQTPSYTDINDEDIEEELE--------ELELALEKEAQVDTLEK 378
+++++++ + E + E+++++LE ++E+ ++EA+ + L K
Sbjct: 325 LENERKLRQKSELQRKELESRIEELQDQLETAGGATSAQVEVGKKREAECNRLRK 379
Score = 32.0 bits (71), Expect = 3.1
Identities = 36/165 (21%), Positives = 72/165 (42%), Gaps = 5/165 (3%)
Query: 245 RKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVE----EVHGVVVDAESTKTV 300
++ ASL + +L + L L T + SSL +E E H V+V E +
Sbjct: 87 KEKLYASLQAETDRLITIEDKLLNLQTVKDKLESSLNEALEKLDGEEHSVLVLEEKIQEA 146
Query: 301 SEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEEL 360
E + +E + ++ ++ ++ + ID+ E + ++T S + + ++EEL
Sbjct: 147 EEKIDELTEKTEELQSNISRLETEKQNRDKQIDTLNEDIRKQDETISKMNAEKKHVDEEL 206
Query: 361 EELELALE-KEAQVDTLEKTTTSEEGNATLEASELLSDTLSNLKL 404
++ L+ E + + L KT E + +L + S +KL
Sbjct: 207 KDRTEQLQAAEDKCNNLNKTKNKLESSIREIEQDLKKEKDSKMKL 251
>MYS2_YEAST (P19524) Myosin-2 (Class V unconventional myosin MYO2)
(Type V myosin heavy chain MYO2) (Myosin V MYO2) (Cell
divison control protein 66)
Length = 1574
Score = 40.4 bits (93), Expect = 0.009
Identities = 46/191 (24%), Positives = 92/191 (48%), Gaps = 22/191 (11%)
Query: 249 VASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTK------TVSE 302
+AS N+++ R +EL++ + K L +++ H + +D + +K T+
Sbjct: 954 LASKVKENKEMTERI-KELQVQVEESAKLQETLENMKKEHLIDIDNQKSKDMELQKTIEN 1012
Query: 303 AMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALEQTP----SYTDINDEDIEE 358
+Q + +K+ ++ +ED+ +L+E +S+K++E+ LEQT Y +N D++
Sbjct: 1013 NLQSTEQTLKDAQLELEDMVKQHDELKE--ESKKQLEE-LEQTKKTLVEYQTLNG-DLQN 1068
Query: 359 ELEELELALEKEAQVDTLEKTTTSEEGNATLE------ASELLSDTLSNLKLSDRPVG-K 411
E++ L+ + + +L TTS L+ AS + L N LS + K
Sbjct: 1069 EVKSLKEEIARLQTAMSLGTVTTSVLPQTPLKDVMGGGASNFNNMMLENSDLSPNDLNLK 1128
Query: 412 SRTTHAASEGH 422
SR+T ++ H
Sbjct: 1129 SRSTPSSGNNH 1139
>MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle
Length = 1978
Score = 40.4 bits (93), Expect = 0.009
Identities = 43/163 (26%), Positives = 70/163 (42%), Gaps = 36/163 (22%)
Query: 228 EKLQQQLDVIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNRVEEV 287
EK +Q+L+ I R+ E + LH +L + A + + E+ + L R+E
Sbjct: 1052 EKSRQELEKIKRKLE----GESSDLHEQIAELQAQIAELKAQLAKKEEELQAALARLE-- 1105
Query: 288 HGVVVDAESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKALEQTPS 347
D S K A+K+ + +++ + DLQE ++S+K E+
Sbjct: 1106 -----DETSQKN---------NALKK----IRELESHISDLQEDLESEKAARNKAEK--- 1144
Query: 348 YTDINDEDIEEELEELELALEKEAQVDTLEKTTTSEEGNATLE 390
D+ EELE L+ LE DTL+ T T +E A E
Sbjct: 1145 ----QKRDLSEELEALKTELE-----DTLDTTATQQELRAKRE 1178
Score = 37.4 bits (85), Expect = 0.075
Identities = 41/175 (23%), Positives = 81/175 (45%), Gaps = 22/175 (12%)
Query: 266 ELKLVTQSREKCSSLLNRVEEVHGVVVDAES---------TKTVSEAMQIGARAIKENKI 316
EL+ + ++K + L +E+ H + + ++ T+ +EA ++ R + K
Sbjct: 865 ELQRTKERQQKAEAELKELEQKHTQLCEEKNLLQEKLQAETELYAEAEEMRVR-LAAKKQ 923
Query: 317 SVEDVDICLRDLQESIDSQKEVEKALEQTPSYTDINDEDIEEELEELELALEKEAQVDTL 376
+E++ L +++ I+ ++E + L+ D+EE+LEE E A +K L
Sbjct: 924 ELEEI---LHEMEARIEEEEERSQQLQAEKKKMQQQMLDLEEQLEEEEAARQKL----QL 976
Query: 377 EKTTTSEEGNATLEASELLSDTLSNLKLSDRPVGKSR----TTHAASEGHKTANL 427
EK T++ +E L+ + +N +R + + R TT+ A E K NL
Sbjct: 977 EK-VTADGKIKKMEDDILIMEDQNNKLTKERKLLEERVSDLTTNLAEEEEKAKNL 1030
Score = 32.3 bits (72), Expect = 2.4
Identities = 39/162 (24%), Positives = 67/162 (41%), Gaps = 21/162 (12%)
Query: 259 LALRYARELKLVTQSREKCSSLLNRVEEVHGVVVDAESTK--------TVSEAMQIGARA 310
L +Y+ ++ T+ EK L VE V ++ +AES T+ +Q
Sbjct: 1269 LQSKYSDGERVRTELNEKVHKLQIEVENVTSLLNEAESKNIKLTKDVATLGSQLQDTQEL 1328
Query: 311 IKENKISVEDVDICLRDL-------QESIDSQKEVEKALEQTPSYTDINDEDIEEELEEL 363
++E +V LR L QE +D + E ++ LE+ S I D +++L+E
Sbjct: 1329 LQEETRQKLNVTTKLRQLEDDKNSLQEQLDEEVEAKQNLERHISTLTIQLSDSKKKLQEF 1388
Query: 364 ELALE------KEAQVDTLEKTTTSEEGNATLEASELLSDTL 399
+E K+ Q + T EE A+ + E + L
Sbjct: 1389 TATVETMEEGKKKLQREIESLTQQFEEKAASYDKLEKTKNRL 1430
Score = 30.8 bits (68), Expect = 7.0
Identities = 35/153 (22%), Positives = 68/153 (43%), Gaps = 9/153 (5%)
Query: 226 TIEKLQQQLD--VIDRRCELSRKSAVASLHSGNRKLALRYARELKLVTQSREKCSSLLNR 283
T +LQQ+LD V+D +++ V++L +K A E + ++ ++
Sbjct: 1426 TKNRLQQELDDLVVDLD---NQRQLVSNLEKKQKKFDQMLAEEKNISSKYADERDRAEAE 1482
Query: 284 VEEVHGVVVD-AESTKTVSEAMQIGARAIKENKISVEDVDICLRDLQESIDSQKEVEKAL 342
E + A + + EA + R K K +ED+ D+ +++ ++ ++ L
Sbjct: 1483 AREKETKALSLARALEEALEAKEELERTNKMLKAEMEDLVSSKDDVGKNVHELEKSKRTL 1542
Query: 343 EQTPSYTDINDEDIEEEL---EELELALEKEAQ 372
EQ E++E+EL E+ +L LE Q
Sbjct: 1543 EQQVEEMKTQLEELEDELQAAEDAKLRLEVNMQ 1575
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.132 0.368
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 46,808,163
Number of Sequences: 164201
Number of extensions: 1928410
Number of successful extensions: 10389
Number of sequences better than 10.0: 377
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 335
Number of HSP's that attempted gapping in prelim test: 9531
Number of HSP's gapped (non-prelim): 923
length of query: 429
length of database: 59,974,054
effective HSP length: 113
effective length of query: 316
effective length of database: 41,419,341
effective search space: 13088511756
effective search space used: 13088511756
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137831.14


