

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.14 + phase: 0
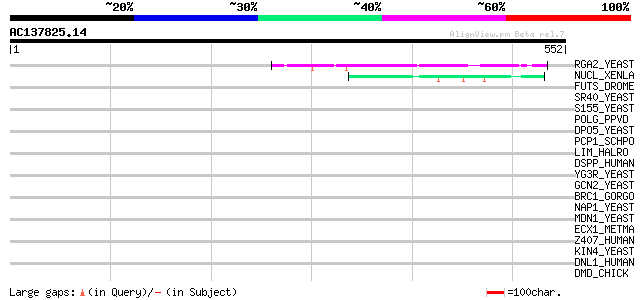
(552 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RGA2_YEAST (Q06407) Rho-type GTPase-activating protein 2 49 4e-05
NUCL_XENLA (P20397) Nucleolin (Protein C23) 48 8e-05
FUTS_DROME (Q9W596) Microtubule-associated protein futsch 43 0.002
SR40_YEAST (P32583) Suppressor protein SRP40 42 0.003
S155_YEAST (P43612) SIT4-associating protein SAP155 41 0.009
POLG_PPVD (P13529) Genome polyprotein [Contains: P1 proteinase (... 40 0.012
DPO5_YEAST (P39985) DNA polymerase V (EC 2.7.7.7) (POL V) 39 0.027
PCP1_SCHPO (Q92351) Spindle pole body protein pcp1 39 0.035
LIM_HALRO (Q25132) LIM/homeobox protein LIM (HRLIM) 38 0.060
DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contai... 38 0.060
YG3R_YEAST (P53288) Hypothetical 22.2 kDa protein in NSR1-TIF463... 37 0.10
GCN2_YEAST (P15442) Serine/threonine-protein kinase GCN2 (EC 2.7... 37 0.10
BRC1_GORGO (Q6J6I8) Breast cancer type 1 susceptibility protein ... 37 0.13
NAP1_YEAST (P25293) Nucleosome assembly protein 37 0.17
MDN1_YEAST (Q12019) Midasin (MIDAS-containing protein) 37 0.17
ECX1_METMA (Q8PTT8) Probable exosome complex exonuclease 1 (EC 3... 37 0.17
Z407_HUMAN (Q9C0G0) Zinc finger protein 407 (Fragment) 36 0.23
KIN4_YEAST (Q01919) Serine/threonine-protein kinase KIN4 (EC 2.7... 36 0.23
DNL1_HUMAN (P18858) DNA ligase I (EC 6.5.1.1) (Polydeoxyribonucl... 36 0.23
DMD_CHICK (P11533) Dystrophin 36 0.23
>RGA2_YEAST (Q06407) Rho-type GTPase-activating protein 2
Length = 1009
Score = 48.5 bits (114), Expect = 4e-05
Identities = 69/284 (24%), Positives = 123/284 (43%), Gaps = 29/284 (10%)
Query: 261 ECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEE-----EVDSILSVSAQTNQII 315
+C + K LL L+ K S+K + L + ++ ++ VS I
Sbjct: 121 DCHEKLLRKKQLL--LENQTKNSSKEDFPIKLPERSVKRPLSPTRINGKSDVSTNNTAIS 178
Query: 316 QDLLPDYEFDQGFTDAYM--EEQSEDSDNESDKDEDDSQCSENSLHC-SVTPSECLNSDS 372
++L+ E DQ T + +E+ E S N+++ +++ E S H +V+ + LNS
Sbjct: 179 KNLVSSNE-DQQLTPQVLVSQERDESSLNDNNDNDNSKDREETSSHARTVSIDDILNSTL 237
Query: 373 DKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEF 432
+ + ++I +S + N Y N+ E++T + P R N DS V + P+ N F
Sbjct: 238 EHDSNSIEEQSLVDNEDYINKMGEDVTYRLLKP-QRANRDSIVVKDPRIPNSNSNANRFF 296
Query: 433 HESYTEAAPRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDK-TSMLAYNLIGRMLEEFA 491
E +D ++ E + + T +++ DK TS L + +M EE
Sbjct: 297 SIYDKEETDKDDTDNKENEIIVNT-----------PRNSTDKITSPLNSPMAVQMNEEVE 345
Query: 492 IAEDLNLDLSKRSYLNCDKQSEDIKETEEQSSSRKRKGGPPIVR 535
L L LS+ + N +K S+ I + S+S+ PI R
Sbjct: 346 PPHGLALTLSEATKEN-NKSSQGI----QTSTSKSMNHVSPITR 384
>NUCL_XENLA (P20397) Nucleolin (Protein C23)
Length = 650
Score = 47.8 bits (112), Expect = 8e-05
Identities = 54/204 (26%), Positives = 80/204 (38%), Gaps = 23/204 (11%)
Query: 338 EDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEE 397
EDS+ E D +EDDS E + TP++ + + K TP N Q +
Sbjct: 25 EDSEEEEDMEEDDSSDEEVEVPVKKTPAKKTATPAKATPGKAATPGKKGATPAKNGKQAK 84
Query: 398 ITEQFSTPMSRKNCDSSAVSLDKEPDEN--IVKRHEFHESYTEAAPRDKSNFCEE----K 451
E + DS + D++P +N + K+ + +E D+ EE K
Sbjct: 85 KQES-----EEEEDDSDEEAEDQKPIKNKPVAKKAVAKKEESEEDDDDEDESEEEKAVAK 139
Query: 452 KPIPTKYSGHKNQYLAAQD---ACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNC 508
KP P K K Q +D + D+ +A L G+ + A +D D
Sbjct: 140 KPTPAKKPAGKKQESEEEDDEESEDEPMEVAPALKGKKTAQAAEEDDEEED--------- 190
Query: 509 DKQSEDIKETEEQSSSRKRKGGPP 532
D ED + EEQ S KRK P
Sbjct: 191 DDDEEDDDDEEEQQGSAKRKKEMP 214
>FUTS_DROME (Q9W596) Microtubule-associated protein futsch
Length = 5412
Score = 43.1 bits (100), Expect = 0.002
Identities = 59/318 (18%), Positives = 130/318 (40%), Gaps = 51/318 (16%)
Query: 236 IEPKATVVNKSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKE 295
+E K V+ S+++ + T S ++ + D + + + KS+ +T K
Sbjct: 4201 VESKVEVLESSVKQVEEKVQT----------SVKQAETTVTDSLEQLTKKSSEQLTEIKS 4250
Query: 296 HIEEEVDSILSVSAQTNQIIQ------DLLPDYEFDQGFTDAYMEEQ---SEDSDNESDK 346
++ + + + A ++++ D++PD F + +EE+ + D++ ESDK
Sbjct: 4251 VLDTNFEEVAKIVADVAKVLKSDKDITDIIPD------FDERQLEEKLKSTADTEEESDK 4304
Query: 347 DEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESK----------MRNTPYNNQCQE 396
D + E S+ + E S D+ I++E K +R + E
Sbjct: 4305 STRDEKSLEISVKVEI---ESEKSSPDQKSGPISIEEKDKIEQSEKAQLRQGILTSSRPE 4361
Query: 397 EITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPIPT 456
+ Q P S + SA S + + V+ E H++ + P ++ EK +
Sbjct: 4362 SVASQ---PESVPSPSQSAASHEHKE----VELSESHKAEKSSRPESVASQVSEKDMKTS 4414
Query: 457 KYSGHKNQYLAAQDACDKTSMLAYNLI------GRMLEEFAIAEDLNLDLSKRSYLNCDK 510
+ + +Q+ + + T L ++L + +EE + E ++ ++K + L+
Sbjct: 4415 RPASSTSQFSTKEGDEETTESLLHSLTTTETVETKQMEEKSSFESVSTSVTKSTVLSSQS 4474
Query: 511 QSEDIKETEEQSSSRKRK 528
+ +E+ +S S K
Sbjct: 4475 TVQLREESTSESLSSSLK 4492
>SR40_YEAST (P32583) Suppressor protein SRP40
Length = 406
Score = 42.4 bits (98), Expect = 0.003
Identities = 38/150 (25%), Positives = 61/150 (40%), Gaps = 3/150 (2%)
Query: 306 SVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPS 365
S S+ ++ D D E D + + S DS ++SD DS +S S + S
Sbjct: 175 SDSSSSSSSSSDSESDSESD---SQSSSSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSS 231
Query: 366 ECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDEN 425
+SDSD + D+ + S ++ ++ E + S S + SS+ KE +
Sbjct: 232 SDSDSDSDSSSDSDSSGSSDSSSSSDSSSDESTSSDSSDSDSDSDSGSSSELETKEATAD 291
Query: 426 IVKRHEFHESYTEAAPRDKSNFCEEKKPIP 455
K E S E+ P S+ K IP
Sbjct: 292 ESKAEETPASSNESTPSASSSSSANKLNIP 321
Score = 34.7 bits (78), Expect = 0.66
Identities = 45/211 (21%), Positives = 79/211 (37%), Gaps = 12/211 (5%)
Query: 245 KSLRKAFVNLLTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSI 304
K +KA + E S S+ S + + S+ S+ + + E + S
Sbjct: 139 KETKKAKTEPESSSSSESSSSGSSSSSESESGSESDSDSSSSSSSSSDSESDSESDSQSS 198
Query: 305 LSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDED-------DSQCSENS 357
S S+ + D + + S DSD++SD D DS S +S
Sbjct: 199 SSSSSSDSSSDSDSSSSDSSSDSDSSSSSSSSSSDSDSDSDSSSDSDSSGSSDSSSSSDS 258
Query: 358 LHCSVTPSECLNSDSDKNC-DTINLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAV 416
T S+ +SDSD + + LE+K + + + STP + + ++ +
Sbjct: 259 SSDESTSSDSSDSDSDSDSGSSSELETKEATADESKAEETPASSNESTPSASSSSSANKL 318
Query: 417 SLDKEPDENIVKRHEFHESYTEAAPRDKSNF 447
++ DE +K E + R K NF
Sbjct: 319 NIPAGTDE--IK--EGQRKHFSRVDRSKINF 345
>S155_YEAST (P43612) SIT4-associating protein SAP155
Length = 1000
Score = 40.8 bits (94), Expect = 0.009
Identities = 33/139 (23%), Positives = 62/139 (43%), Gaps = 7/139 (5%)
Query: 242 VVNKSLRKAFVNLL---TEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIE 298
V K+L +F+ + TE L E S N T + +K +NK +++ ++
Sbjct: 132 VTTKNLDNSFIERMLVETELLNELSRQNKTLLDFICFGFFFDKKTNKKVNNMEYL---VD 188
Query: 299 EEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSL 358
+ ++ I + T + +L+ DY+ Q D+ E+ +SD E +++++D S N
Sbjct: 189 QLMECISKIKTATTVDLNNLI-DYQEQQQLDDSSQEDVYVESDTEQEEEKEDDNNSNNKK 247
Query: 359 HCSVTPSECLNSDSDKNCD 377
S N D + N D
Sbjct: 248 RRKRGSSSFGNDDINNNDD 266
>POLG_PPVD (P13529) Genome polyprotein [Contains: P1 proteinase
(N-terminal protein); Helper component proteinase (EC
3.4.22.45) (HC-pro); Protein P3; 6 kDa protein 1 (6K1);
Cytoplasmic inclusion protein (CI); 6 kDa protein 2
(6K2); Viral genome-linked pr
Length = 3141
Score = 40.4 bits (93), Expect = 0.012
Identities = 38/137 (27%), Positives = 67/137 (48%), Gaps = 21/137 (15%)
Query: 275 ILDVINKCSNKSTHDVTL---QKEHIEEEVDSILSVSA-QTNQIIQDLLPDYEFDQGFTD 330
+++ IN +D L + +H VDS VS Q+N+II P +F GFT+
Sbjct: 262 VVNPINLSDPMQVYDTDLFIVRGKHNSILVDSRCKVSKKQSNEIIHYSDPGKQFSDGFTN 321
Query: 331 AYMEEQSEDSDNESDKDEDDSQCSE-NSLHC-SVTPSECLNSDSDKNCDTIN-LESKMRN 387
++M+ + ++D++S D D +C + +L C ++ P C I L+ +
Sbjct: 322 SFMQCKLRETDHQSTSDLDVKECGDVAALVCQAIIP-----------CGKITCLQCAQK- 369
Query: 388 TPYNNQCQEEITEQFST 404
Y+ Q+EI ++FST
Sbjct: 370 --YSYMSQQEIRDRFST 384
>DPO5_YEAST (P39985) DNA polymerase V (EC 2.7.7.7) (POL V)
Length = 1022
Score = 39.3 bits (90), Expect = 0.027
Identities = 47/208 (22%), Positives = 90/208 (42%), Gaps = 25/208 (12%)
Query: 202 PEVKSLQSDIMQAILSVEKKEVVPL--PVLRELQLLIEPKATVVNKSLRK------AFVN 253
PE+ S Q ++ IL +E V L P+ L+ + + ++K R
Sbjct: 551 PEINSWQYLTLKLILDIENSHVGDLINPLDENLENIKNEAISCLSKVCRSRTAQSWGLST 610
Query: 254 LLTEFLFECSDMNSTPKSLLQILDVINKCSNKSTHDVT-------LQKEHIEEEVDSIL- 305
LL+ L + ++ S+++ L +K N S +T QK+ + ++ I+
Sbjct: 611 LLSMCLVQLYAGDTDSISVIEELCEFSKHENNSMVGITEILLSLLAQKKALLRKLSLIIW 670
Query: 306 -----SVSAQTNQIIQDLLPDYEFDQGFTDAYM-EEQSEDSDNESDKDEDDSQC---SEN 356
V + QI+ D+L E QGF + EE+ E+ E+D ED+S+ SE+
Sbjct: 671 QQFIEEVGLEELQILLDILKARENKQGFAQLFEGEEEFEEIKEENDASEDESKTGSESES 730
Query: 357 SLHCSVTPSECLNSDSDKNCDTINLESK 384
++ + + + N D +N++ +
Sbjct: 731 ESESDSDDADEKDEEDEANEDILNIDKE 758
>PCP1_SCHPO (Q92351) Spindle pole body protein pcp1
Length = 1208
Score = 38.9 bits (89), Expect = 0.035
Identities = 55/259 (21%), Positives = 104/259 (39%), Gaps = 27/259 (10%)
Query: 273 LQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFD----QGF 328
L+ L + N HD+ + E + ++DSI + ++ + YE + G
Sbjct: 508 LETLRMKNSNDLNEIHDLREENEGLTLKIDSITKEKDRLINELEQRIKSYEVNVSELNGT 567
Query: 329 TDAYMEEQSEDSDNESDKDEDDSQCSENSL---HCSVTPSECLNSDSDKNCDTINLE-SK 384
D Y + +D + ++ + Q +N L H S+ + + N + NL S
Sbjct: 568 IDEY-RNKLKDKEETYNEVMNAFQYKDNDLRRFHESINKLQDREKELTSNLEKKNLVISS 626
Query: 385 MRNTPYNNQCQEEITEQFSTPMSRKNCDSSAV-------------SLDKEPDENIVKRHE 431
+R T + + E +++ + + K+ D++ + L DE V E
Sbjct: 627 LRETVAMLEKERESIKKYLSG-NAKDLDNTNLMEILNDKISVLQRQLTDVKDELDVSEEE 685
Query: 432 FHESYTEAAPRDKSNF---CEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLE 488
E+ A + ++F EK+ + KYS KN+ + AQ+ D+ L ++ E
Sbjct: 686 REEAIV-AGQKLSASFELMSNEKQALELKYSSLKNELINAQNLLDRREEELSELSKKLFE 744
Query: 489 EFAIAEDLNLDLSKRSYLN 507
E I N D+ K +N
Sbjct: 745 ERKIRSGSNDDIEKNKEIN 763
>LIM_HALRO (Q25132) LIM/homeobox protein LIM (HRLIM)
Length = 514
Score = 38.1 bits (87), Expect = 0.060
Identities = 45/180 (25%), Positives = 82/180 (45%), Gaps = 33/180 (18%)
Query: 294 KEHIEEEVDSILSVSAQT---NQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDD 350
+++ +E++ + SV+ ++ +++ + DY+ D+G + Y ++ + D D++ + D+DD
Sbjct: 74 RDYCGKELNKMASVTKESYVGGRLVDEDEDDYD-DEGDREVYEDDITFDDDDDDENDDDD 132
Query: 351 SQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMSRKN 410
S N L +V L DS T+ S T +N PMS
Sbjct: 133 SDVKMN-LRTNVNK---LRFDSGFEY-TLPTSSPENMTSFN-------------PMS--- 171
Query: 411 CDSSAVSLD-KEPDENIVKRHEFHESYTEAAPR------DKSNFCEEKKPIPTKYSGHKN 463
DSS + D + + I+ + SY + PR + N EE K IPT Y+G+ +
Sbjct: 172 -DSSHLFSDYSKHNSTILANGKSGTSYLTSTPRRIYDNTNMINIPEETKNIPTCYNGNNH 230
>DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contains:
Dentin phosphoprotein (Dentin phosphophoryn) (DPP);
Dentin sialoprotein (DSP)]
Length = 1253
Score = 38.1 bits (87), Expect = 0.060
Identities = 23/86 (26%), Positives = 42/86 (48%)
Query: 329 TDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNT 388
+D+ S DS + SD D +S S NS S + + +SDS + D+ + +S R+
Sbjct: 801 SDSSNSSDSSDSSDSSDSDSSNSSDSSNSSDSSDSSNSSDSSDSSDSSDSSDSDSSNRSD 860
Query: 389 PYNNQCQEEITEQFSTPMSRKNCDSS 414
N+ + ++ ++ S + DSS
Sbjct: 861 SSNSSDSSDSSDSSNSSDSSDSSDSS 886
Score = 37.7 bits (86), Expect = 0.078
Identities = 47/265 (17%), Positives = 95/265 (35%), Gaps = 6/265 (2%)
Query: 261 ECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLP 320
+ SD NS+ S D + S+ S+ + + + DS S + + D
Sbjct: 570 DSSDSNSSSDS-----DSSDSDSSDSSDSDSSDSSNSSDSSDSSDSSDSSDSSDSSDSKS 624
Query: 321 DY-EFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTI 379
D + + +D+ + S DS++ D DS S NS + S + +SDS + D+
Sbjct: 625 DSSKSESDSSDSDSKSDSSDSNSSDSSDNSDSSDSSNSSNSSDSSDSSDSSDSSSSSDSS 684
Query: 380 NLESKMRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEA 439
+ ++ ++ + + S + DSS S + + +S +
Sbjct: 685 SSSDSSNSSDSSDSSDSSNSSESSDSSDSSDSDSSDSSDSSNSNSSDSDSSNSSDSSDSS 744
Query: 440 APRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLD 499
D SN + + S + + D+ D ++ N + + +
Sbjct: 745 DSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNSSDSNDSSNSSDSSDSSNSSDSS 804
Query: 500 LSKRSYLNCDKQSEDIKETEEQSSS 524
S S + D D + + S+S
Sbjct: 805 NSSDSSDSSDSSDSDSSNSSDSSNS 829
Score = 33.9 bits (76), Expect = 1.1
Identities = 33/190 (17%), Positives = 67/190 (34%)
Query: 335 EQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQC 394
+ S+ SD+ + D DS S +S S + +SDS + D+ + ++ +
Sbjct: 1038 DSSDSSDSSNSSDSSDSSDSSDSSDSSGSSDSSDSSDSSDSSDSSDSSDSSDSSDSSESS 1097
Query: 395 QEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPI 454
+ S + S+ S D + + +S + D S+ +
Sbjct: 1098 DSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSDSSDSSDSS 1157
Query: 455 PTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRSYLNCDKQSED 514
+ S + + D+ D + N + ++ N S S + D S+
Sbjct: 1158 DSSDSSDSSDSSDSSDSSDSSDSSDSNESSDSSDSSDSSDSSNSSDSSDSSDSSDSTSDS 1217
Query: 515 IKETEEQSSS 524
E++ QS S
Sbjct: 1218 NDESDSQSKS 1227
Score = 33.5 bits (75), Expect = 1.5
Identities = 28/147 (19%), Positives = 55/147 (37%), Gaps = 1/147 (0%)
Query: 329 TDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNT 388
+D+ S+ SD+ + D DS S +S S + +SDS + D+ + ++
Sbjct: 898 SDSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSNSSDNSNSSDSSNSSDSSDSSDSSNSS 957
Query: 389 PYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFC 448
+N + S S + DSS S D + + +S + D S+
Sbjct: 958 DSSNSGDSSNSSDSSDSNSSDSSDSSN-SSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSS 1016
Query: 449 EEKKPIPTKYSGHKNQYLAAQDACDKT 475
+ + S + + D+ D +
Sbjct: 1017 DSSNSSDSSNSSDSSDSSDSSDSSDSS 1043
Score = 32.7 bits (73), Expect = 2.5
Identities = 29/142 (20%), Positives = 57/142 (39%), Gaps = 11/142 (7%)
Query: 335 EQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQC 394
+ S+ SD+ + D +DS S +S S + +SDS + D+ + +S + N+
Sbjct: 772 DSSDSSDSSNSSDSNDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSDSDSSNSSDSSNSSD 831
Query: 395 QEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPI 454
+ + + S + DSS D + R + S + D SN +
Sbjct: 832 SSDSSNSSDSSDSSDSSDSS--------DSDSSNRSDSSNSSDSSDSSDSSNSSDSS--- 880
Query: 455 PTKYSGHKNQYLAAQDACDKTS 476
+ S N+ + D+ D ++
Sbjct: 881 DSSDSSDSNESSNSSDSSDSSN 902
Score = 32.0 bits (71), Expect = 4.3
Identities = 25/147 (17%), Positives = 54/147 (36%)
Query: 329 TDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNT 388
+D+ S+ SD+ + D DS S +S S + + +SDS + D+ N ++
Sbjct: 969 SDSSDSNSSDSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSNSSDSSNSS 1028
Query: 389 PYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFC 448
++ + S + + S+ S D + +S + D S+
Sbjct: 1029 DSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSGSSDSSDSSDSSDSSDSSDSSDSS 1088
Query: 449 EEKKPIPTKYSGHKNQYLAAQDACDKT 475
+ + S + + D+ D +
Sbjct: 1089 DSSDSSESSDSSDSSDSSDSSDSSDSS 1115
Score = 31.6 bits (70), Expect = 5.6
Identities = 36/200 (18%), Positives = 77/200 (38%), Gaps = 20/200 (10%)
Query: 325 DQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESK 384
D +D+ S DS + SD D DS S + S+ +SDS + D+ + +S
Sbjct: 548 DSSDSDSSDSSNSSDSSDSSDSDSSDSNSSSD--------SDSSDSDSSDSSDSDSSDSS 599
Query: 385 MRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDK 444
N+ + ++ + S + DS + S E D + +S ++++ +
Sbjct: 600 ------NSSDSSDSSDSSDSSDSSDSSDSKSDSSKSESDSS------DSDSKSDSSDSNS 647
Query: 445 SNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSMLAYNLIGRMLEEFAIAEDLNLDLSKRS 504
S+ + + S + + + D+ D +S + ++ + S S
Sbjct: 648 SDSSDNSDSSDSSNSSNSSDSSDSSDSSDSSSSSDSSSSSDSSNSSDSSDSSDSSNSSES 707
Query: 505 YLNCDKQSEDIKETEEQSSS 524
+ D D ++ + S+S
Sbjct: 708 SDSSDSSDSDSSDSSDSSNS 727
Score = 30.8 bits (68), Expect = 9.6
Identities = 28/147 (19%), Positives = 54/147 (36%), Gaps = 7/147 (4%)
Query: 329 TDAYMEEQSEDSDNESDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNT 388
+D+ S DSD+ + DS S NS S + + +SD+ + D+ N ++
Sbjct: 895 SDSSDSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSN---SSDNSNSSDSSNSSDSSDSS 951
Query: 389 PYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFC 448
+N + S + +SS D N + +S + D SN
Sbjct: 952 DSSNSSDSSNSGDSSNSSDSSDSNSS----DSSDSSNSSDSSDSSDSSDSSDSSDSSNSS 1007
Query: 449 EEKKPIPTKYSGHKNQYLAAQDACDKT 475
+ + S + + + D+ D +
Sbjct: 1008 DSSDSSDSSDSSNSSDSSNSSDSSDSS 1034
Score = 30.8 bits (68), Expect = 9.6
Identities = 28/151 (18%), Positives = 55/151 (35%), Gaps = 4/151 (2%)
Query: 329 TDAYMEEQSEDSDNESDK----DEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESK 384
+D+ S DS N SD D DS S +S + S + +SDS + D+ N
Sbjct: 992 SDSSDSSDSSDSSNSSDSSDSSDSSDSSNSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDS 1051
Query: 385 MRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDK 444
++ ++ + S + S+ S D + + + +S + D
Sbjct: 1052 SDSSDSSDSSDSSGSSDSSDSSDSSDSSDSSDSSDSSDSSDSSESSDSSDSSDSSDSSDS 1111
Query: 445 SNFCEEKKPIPTKYSGHKNQYLAAQDACDKT 475
S+ + + S + + D+ D +
Sbjct: 1112 SDSSDSSDSSDSSDSSDSSNSSDSSDSSDSS 1142
>YG3R_YEAST (P53288) Hypothetical 22.2 kDa protein in NSR1-TIF4631
intergenic region
Length = 203
Score = 37.4 bits (85), Expect = 0.10
Identities = 25/78 (32%), Positives = 40/78 (51%), Gaps = 1/78 (1%)
Query: 278 VINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQS 337
+++ S+ S+ +E EE +DS VSA + D D E + D + S
Sbjct: 87 LVSSFSSSSSSSEEESEEEEEESLDSSFLVSASLSLSEDDEEEDSESEDEDEDEDSDSDS 146
Query: 338 EDSDNESDKDEDDSQCSE 355
DSD++SD+DED+ + SE
Sbjct: 147 -DSDSDSDEDEDEDEDSE 163
>GCN2_YEAST (P15442) Serine/threonine-protein kinase GCN2 (EC
2.7.1.37)
Length = 1590
Score = 37.4 bits (85), Expect = 0.10
Identities = 41/183 (22%), Positives = 76/183 (41%), Gaps = 21/183 (11%)
Query: 296 HIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQS------EDSDNESDKDED 349
H EE++ +ILS +++ ++++ + A++EE S E +D ESD E
Sbjct: 563 HTEEKLSTILS------EVMLLASLNHQYVVRYYAAWLEEDSMDENVFESTDEESDLSES 616
Query: 350 DSQCSENSL--HCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPMS 407
S EN L S+ + + + N D I+ S + + N +++ E S
Sbjct: 617 SSDFEENDLLDQSSIFKNRTNHDLDNSNWDFIS-GSGYPDIVFENSSRDDENEDLDHDTS 675
Query: 408 RKNCDSSAVSLDKEPD--ENIVKRHEFHESYTEAAPRD----KSNFCEEKKPIPTKYSGH 461
+ S DKE +N+ +R F + T + + +CE + +S +
Sbjct: 676 STSSSESQDDTDKESKSIQNVPRRRNFVKPMTAVKKKSTLFIQMEYCENRTLYDLIHSEN 735
Query: 462 KNQ 464
NQ
Sbjct: 736 LNQ 738
>BRC1_GORGO (Q6J6I8) Breast cancer type 1 susceptibility protein
homolog
Length = 1863
Score = 37.0 bits (84), Expect = 0.13
Identities = 38/156 (24%), Positives = 66/156 (41%), Gaps = 19/156 (12%)
Query: 261 ECSDMNSTPKSLLQILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLP 320
EC N T ++LL + + +N CSN+ T Q+ H+ EE S+ + ++DL
Sbjct: 1250 ECLSKN-TEENLLSLKNSLNDCSNQVILAKTSQEHHLSEETKCSASLFSSQCSELEDLTA 1308
Query: 321 DYEFDQGF---TDAYMEEQSED-----SDNE--SDKDEDDSQCSENS-------LHCSVT 363
+ F + M QSE SD E SD +E + EN+ +
Sbjct: 1309 NTNTQDPFLIGSSKQMRHQSESQGVGLSDKELVSDDEERGTGLEENNQEEQSMDSNLGEA 1368
Query: 364 PSECLNSDS-DKNCDTINLESKMRNTPYNNQCQEEI 398
S C + S ++C ++ +S + T + Q+ +
Sbjct: 1369 ASGCESETSVSEDCSGLSSQSDILTTQQRDTMQDNL 1404
>NAP1_YEAST (P25293) Nucleosome assembly protein
Length = 417
Score = 36.6 bits (83), Expect = 0.17
Identities = 19/66 (28%), Positives = 38/66 (56%), Gaps = 1/66 (1%)
Query: 293 QKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQ 352
Q E +EE+++ L++ + ++D L D FT A +E + E+ + E+D+DED+ +
Sbjct: 326 QDEELEEDLEERLALDYSIGEQLKDKLIPRAVDW-FTGAALEFEFEEDEEEADEDEDEEE 384
Query: 353 CSENSL 358
++ L
Sbjct: 385 DDDHGL 390
>MDN1_YEAST (Q12019) Midasin (MIDAS-containing protein)
Length = 4910
Score = 36.6 bits (83), Expect = 0.17
Identities = 33/148 (22%), Positives = 58/148 (38%), Gaps = 6/148 (4%)
Query: 284 NKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNE 343
NK D + E +E D + + ++DL E D TD+ EE E+ D
Sbjct: 4091 NKEQQD---KDERDDENEDDAVEMEGDMAGELEDLSNGEENDDEDTDSEEEELDEEID-- 4145
Query: 344 SDKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFS 403
D +EDD ++ + D+D+N D N E ++ + Q ++
Sbjct: 4146 -DLNEDDPNAIDDKMWDDKASDNSKEKDTDQNLDGKNQEEDVQAAENDEQQRDNKEGGDE 4204
Query: 404 TPMSRKNCDSSAVSLDKEPDENIVKRHE 431
P + ++ D + + +EN V E
Sbjct: 4205 DPNAPEDGDEEIENDENAEEENDVGEQE 4232
>ECX1_METMA (Q8PTT8) Probable exosome complex exonuclease 1 (EC
3.1.13.-)
Length = 493
Score = 36.6 bits (83), Expect = 0.17
Identities = 41/173 (23%), Positives = 72/173 (40%), Gaps = 8/173 (4%)
Query: 285 KSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNES 344
+S D+ ++E EEE L A+ + DL D E D G EE+ E+ + E
Sbjct: 313 ESEEDLETEEEEFEEEA---LEEEAEPEE---DLEEDLEEDLGEELEEEEEELEEEEFEE 366
Query: 345 DKDEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFST 404
+ E++++ E SL C+ E ++ + ++E++ P + EE E+ +
Sbjct: 367 EALEEETEL-EASLECAPELKEFDEIEARLEKEDASIEAEEEIEPEAEEATEEGLEEEAE 425
Query: 405 PMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEEKKPIPTK 457
+ + + E +E E E TEA ++ EEK P K
Sbjct: 426 IEETAASEEENIEAEAEAEEEAEPEVEAEEISTEAEEAEEEPE-EEKSEGPWK 477
Score = 35.4 bits (80), Expect = 0.39
Identities = 32/146 (21%), Positives = 63/146 (42%), Gaps = 11/146 (7%)
Query: 299 EEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCSENSL 358
EE + +S + + +++PD+E + + +EE+ E+S+ + + +E++ + E +L
Sbjct: 276 EEPEVEISEEVEAEILASEVIPDFEDE---LEEEIEEELEESEEDLETEEEEFE--EEAL 330
Query: 359 HCSVTPSECLNSDSDKNC--DTINLESKMRNTPYNNQCQEEITE---QFSTPMSRKNCDS 413
P E L D +++ + E ++ + + EE TE K D
Sbjct: 331 EEEAEPEEDLEEDLEEDLGEELEEEEEELEEEEFEEEALEEETELEASLECAPELKEFDE 390
Query: 414 SAVSLDKEPDENIVKRHEFHESYTEA 439
L+KE D +I E EA
Sbjct: 391 IEARLEKE-DASIEAEEEIEPEAEEA 415
>Z407_HUMAN (Q9C0G0) Zinc finger protein 407 (Fragment)
Length = 1165
Score = 36.2 bits (82), Expect = 0.23
Identities = 26/104 (25%), Positives = 44/104 (42%), Gaps = 19/104 (18%)
Query: 347 DEDDSQCSENSLHCSVTPSECLNSDSDKNCDTINLESKMRNTPYNNQCQEEITEQFSTPM 406
+E+ Q SE S PS+ L S + +C +N E + M
Sbjct: 15 EEEHQQNSEEFQIISGQPSDTLKSRNAADCSILN-------------------ENTNLDM 55
Query: 407 SRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAAPRDKSNFCEE 450
S+ C + +V ++ E + N + H F E++ +A +DK EE
Sbjct: 56 SKVLCAADSVEVETEEESNFNEDHSFCETFQQAPVKDKVRKPEE 99
>KIN4_YEAST (Q01919) Serine/threonine-protein kinase KIN4 (EC
2.7.1.37)
Length = 800
Score = 36.2 bits (82), Expect = 0.23
Identities = 51/235 (21%), Positives = 95/235 (39%), Gaps = 38/235 (16%)
Query: 219 EKKEVVPLPVLRELQLLIEPKATVVNKSLR-----KAFVNLLTEFLFECSDMNSTPKSLL 273
+ K+ VPL + + KAT+ N S+ A ++ + D+N K +
Sbjct: 538 DAKDPVPLNAIHDTN-----KATISNNSIMLLSEGPAAKTSPVDYHYAIGDLNHGDKPIT 592
Query: 274 QILDVINKCSNKSTHDVTLQKEHIEEEVDSILSVSAQTNQIIQDLLPDYEFDQGFTDAYM 333
+++D INK + +E I+ E S + V+ + P D+ ++ +
Sbjct: 593 EVIDKINKDLTHKAAENGFPRESIDPESTSTILVTKE---------PTNSTDEDHVESQL 643
Query: 334 E---EQSEDSDNESDKDE----DDSQCSENSLHCSVTPSECL--NSDSDKNCDTINLESK 384
E S SD SDKD + + S SL+ S+ S + S N + S
Sbjct: 644 ENVGHSSNKSDASSDKDSKKIYEKKRFSFMSLYSSLNGSRSTVESRTSKGNAPPV---SS 700
Query: 385 MRNTPYNNQCQEEITEQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEA 439
+ +N+ +IT+Q +N + DK+ ++N ++ + SY E+
Sbjct: 701 RNPSGQSNRSNIKITQQ-----QPRNLSDRVPNPDKKINDNRIRDNA--PSYAES 748
>DNL1_HUMAN (P18858) DNA ligase I (EC 6.5.1.1)
(Polydeoxyribonucleotide synthase [ATP])
Length = 919
Score = 36.2 bits (82), Expect = 0.23
Identities = 42/170 (24%), Positives = 66/170 (38%), Gaps = 21/170 (12%)
Query: 333 MEEQSEDSDNES--DKDEDDSQCSENSLHCSVTPSECLNSDSDK-NCDTINLESKMRNTP 389
+EEQSED D E+ K+E++ + + SL + +E D D+ L++ TP
Sbjct: 137 LEEQSEDEDREAKRKKEEEEEETPKESLTEAEVATEKEGEDGDQPTTPPKPLKTSKAETP 196
Query: 390 YNNQCQEEIT---------EQFSTPMSRKNCDSSAVSLDKEPDENIVKRHEFHESYTEAA 440
+ + E+ EQ P SS + K + VK E E A
Sbjct: 197 TESVSEPEVATKQELQEEEEQTKPPRRAPKTLSSFFTPRKPAVKKEVKEEEPGAPGKEGA 256
Query: 441 PRDKSNFCEEKKPIPTKYSGHKNQYLAAQDACDKTSM-LAYNLIGRMLEE 489
+ P+ Y+ KN Y +DAC K + Y + R E+
Sbjct: 257 AEGPLD--------PSGYNPAKNNYHPVEDACWKPGQKVPYLAVARTFEK 298
>DMD_CHICK (P11533) Dystrophin
Length = 3660
Score = 36.2 bits (82), Expect = 0.23
Identities = 46/192 (23%), Positives = 77/192 (39%), Gaps = 29/192 (15%)
Query: 195 EECYQFSPEVKSLQSDIMQAILSVEKKEVVPLPVLRELQLLIEPKATVVNKSLRKAFVNL 254
EE + E+K + D MQ KEV + + I N++L+K L
Sbjct: 1193 EELQKAVEELKRAKEDAMQ-------KEVKVKLITDSVNNFIAKAPPAANEALKKELDVL 1245
Query: 255 LTEFLFECSDMNSTPKS----------LLQILDVINKCSN------KSTHDVTLQKEHIE 298
+T + CS +N K+ LL LD NK N K+T ++ E I
Sbjct: 1246 ITSYQRLCSRLNGKCKTLEEVWACWHELLSYLDAENKWLNEVELKLKATENIQGGAEEIS 1305
Query: 299 EEVDSILSV----SAQTNQIIQDLLPDYEFDQGFTDAYMEEQSEDSDNESDKDEDDSQCS 354
E +DS+ + NQI + L D G D + E+ E + ++ + ++
Sbjct: 1306 ESLDSLERLMRHPEDNRNQIRE--LAQTLTDGGILDELINEKLEKFNTRWEELQQEAVRR 1363
Query: 355 ENSLHCSVTPSE 366
+ SL S+ ++
Sbjct: 1364 QKSLEQSIQSAQ 1375
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.131 0.366
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 63,231,340
Number of Sequences: 164201
Number of extensions: 2693147
Number of successful extensions: 10483
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 30
Number of HSP's successfully gapped in prelim test: 142
Number of HSP's that attempted gapping in prelim test: 9673
Number of HSP's gapped (non-prelim): 459
length of query: 552
length of database: 59,974,054
effective HSP length: 115
effective length of query: 437
effective length of database: 41,090,939
effective search space: 17956740343
effective search space used: 17956740343
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 68 (30.8 bits)
Medicago: description of AC137825.14


