

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137825.12 - phase: 0 /pseudo
(451 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
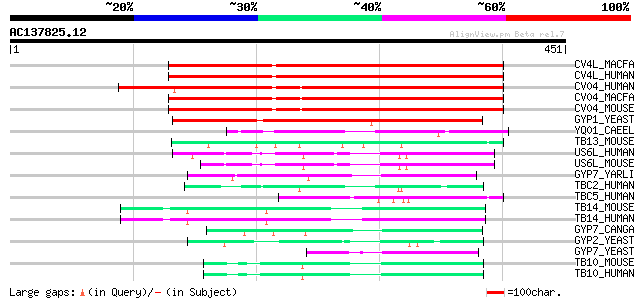
Score E
Sequences producing significant alignments: (bits) Value
CV4L_MACFA (Q95LL3) TBC1 domain family protein C6orf197 homolog ... 289 9e-78
CV4L_HUMAN (Q9NU19) TBC1 domain family protein C6orf197 289 9e-78
CV04_HUMAN (Q8WUA7) TBC1 domain family protein C22orf4 273 7e-73
CV04_MACFA (Q95KI1) TBC1 domain family protein C22orf4 homolog (... 271 2e-72
CV04_MOUSE (Q8R5A6) TBC1 domain family protein C22orf4 homolog (... 266 1e-70
GYP1_YEAST (Q08484) GTPase-activating protein GYP1 (GAP for YPT1) 249 1e-65
YQ01_CAEEL (Q09445) Hypothetical protein C04A2.1 in chromosome II 75 4e-13
TB13_MOUSE (Q8R3D1) TBC1 domain family member 13 72 2e-12
US6L_HUMAN (Q92738) USP6 N-terminal like protein (Related to the... 71 6e-12
US6L_MOUSE (Q80XC3) USP6 N-terminal like protein 68 5e-11
GYP7_YARLI (P09379) GTPase-activating protein GYP7 (GAP for YPT7) 67 9e-11
TBC2_HUMAN (Q9BYX2) TBC1 domain family member 2 (Prostate antige... 63 2e-09
TBC5_HUMAN (Q92609) TBC1 domain family member 5 59 3e-08
TB14_MOUSE (Q8CGA2) TBC1 domain family member 14 58 4e-08
TB14_HUMAN (Q9P2M4) TBC1 domain family member 14 58 4e-08
GYP7_CANGA (Q6FWI1) GTPase-activating protein GYP7 (GAP for YPT7) 57 7e-08
GYP2_YEAST (P53258) GTPase-activating protein GYP2 (MAC1-depende... 55 5e-07
GYP7_YEAST (P48365) GTPase-activating protein GYP7 (GAP for YPT7) 52 2e-06
TB10_MOUSE (P58802) TBC1 domain family member 10 (EBP50-PDX inte... 49 3e-05
TB10_HUMAN (Q9BXI6) TBC1 domain family member 10 (EBP50-PDX inte... 49 3e-05
>CV4L_MACFA (Q95LL3) TBC1 domain family protein C6orf197 homolog
(QtsA-20424)
Length = 505
Score = 289 bits (740), Expect = 9e-78
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 182 EKTRLEKFRQLLSSHNTDLDELRKCSWPGVPREVRPVTWRLLSGYLPANTERRKLTLQRK 241
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 242 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 298
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 299 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 358
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 359 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 418
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 419 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 450
>CV4L_HUMAN (Q9NU19) TBC1 domain family protein C6orf197
Length = 505
Score = 289 bits (740), Expect = 9e-78
Identities = 142/272 (52%), Positives = 188/272 (68%), Gaps = 3/272 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+ R+ KF ++LS LD LR+ +W GVP +RP WRLL GY P N++R++ L RK
Sbjct: 182 EKTRLEKFRQLLSSQNTDLDELRKCSWPGVPREVRPITWRLLSGYLPANTERRKLTLQRK 241
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
R EY I QYYD + E D RQI +D PRT P + FQQ VQ+ ERIL+ W
Sbjct: 242 REEYFGFIEQYYDSRNEEHHQDTY---RQIHIDIPRTNPLIPLFQQPLVQEIFERILFIW 298
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKL 309
AIRHPASGYVQGINDLVTPF VVF+SE++E ++++ +++LS D + ++EAD +WC+SKL
Sbjct: 299 AIRHPASGYVQGINDLVTPFFVVFLSEYVEEDVENFDVTNLSQDMLRSIEADSFWCMSKL 358
Query: 310 LDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREI 369
LDG+QD+YTFAQPGIQ+ V L+ELV RIDE + H +E+LQFAFRW N LL+RE+
Sbjct: 359 LDGIQDNYTFAQPGIQKKVKALEELVSRIDEQVHNHFRRYEVEYLQFAFRWMNNLLMREL 418
Query: 370 PFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
P RLWDTY +E + F +Y+ A+FL+
Sbjct: 419 PLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLI 450
>CV04_HUMAN (Q8WUA7) TBC1 domain family protein C22orf4
Length = 517
Score = 273 bits (698), Expect = 7e-73
Identities = 146/321 (45%), Positives = 202/321 (62%), Gaps = 12/321 (3%)
Query: 89 EVQSTSKSVSAANVSKLKSSTSLGENLSEDIRKYTIGARATDSA-------RVTKFNKVL 141
E ++ + SA++ + L+ S SL + + + + + + SA R+ KF ++L
Sbjct: 146 ESHTSCPAESASDAAPLQRSQSLPHSATVTLGGTSDPSTLSSSALSEREASRLDKFKQLL 205
Query: 142 SGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYY 201
+G L+ LR L+WSG+P +RP W+LL GY P N DR+ L RK+ EY I YY
Sbjct: 206 AGPNTDLEELRRLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRKQKEYFAFIEHYY 265
Query: 202 DIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQG 261
D + E D RQI +D PR P+ Q + V + ERIL+ WAIRHPASGYVQG
Sbjct: 266 DSRNDEVHQDTY---RQIHIDIPRMSPEALILQPK-VTEIFERILFIWAIRHPASGYVQG 321
Query: 262 INDLVTPFLVVFISEHLEGG-IDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQDHYTFA 320
INDLVTPF VVFI E++E +D +S + ++ + N+EAD YWC+SKLLDG+QD+YTFA
Sbjct: 322 INDLVTPFFVVFICEYIEAEEVDTVDVSGVPAEVLCNIEADTYWCMSKLLDGIQDNYTFA 381
Query: 321 QPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDT 380
QPGIQ V L+ELV RIDE + +H++ + +LQFAFRW N LL+RE+P RLWDT
Sbjct: 382 QPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRWMNNLLMREVPLRCTIRLWDT 441
Query: 381 YLAEGDALPDFLVYIFASFLL 401
Y +E D F +Y+ A+FL+
Sbjct: 442 YQSEPDGFSHFHLYVCAAFLV 462
>CV04_MACFA (Q95KI1) TBC1 domain family protein C22orf4 homolog
(QtrA-11492) (Fragment)
Length = 497
Score = 271 bits (694), Expect = 2e-72
Identities = 136/273 (49%), Positives = 184/273 (66%), Gaps = 5/273 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+++R+ KF ++L+G L+ LR+L+WSG+P +RP W+LL GY P N DR+ L RK
Sbjct: 174 EASRLDKFEQLLAGPNTDLEELRKLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRK 233
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
+ EY I YYD + E D RQI +D PR P+ Q + V + ERIL+ W
Sbjct: 234 QKEYFAFIEHYYDSRNDEVHQDTY---RQIHIDIPRMSPEALILQPK-VTEIFERILFIW 289
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEGG-IDDWSMSDLSSDKISNVEADCYWCLSK 308
AIRHPASGYVQGINDLVTPF VVFI E++E +D +S + ++ + N+EAD YWC+SK
Sbjct: 290 AIRHPASGYVQGINDLVTPFFVVFICEYIEAEEVDTVDVSGVPAEVLRNIEADTYWCMSK 349
Query: 309 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIRE 368
LLDG+QD+YTFAQPGIQ V L+ELV RIDE + +H++ + +LQFAFRW N LL+RE
Sbjct: 350 LLDGIQDNYTFAQPGIQMKVKMLEELVSRIDEQVHRHLDQHEVRYLQFAFRWMNNLLMRE 409
Query: 369 IPFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
+P RLWDTY +E + F +Y+ A+FL+
Sbjct: 410 VPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLV 442
>CV04_MOUSE (Q8R5A6) TBC1 domain family protein C22orf4 homolog
(Fragment)
Length = 353
Score = 266 bits (679), Expect = 1e-70
Identities = 134/273 (49%), Positives = 182/273 (66%), Gaps = 5/273 (1%)
Query: 130 DSARVTKFNKVLSGTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+++R+ KF ++L+G L+ LR+L+WSG+P +RP W+LL GY P N DR+ L RK
Sbjct: 30 ETSRLDKFKQLLAGPNTDLEELRKLSWSGIPKPVRPMTWKLLSGYLPANVDRRPATLQRK 89
Query: 190 RGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAW 249
+ EY I YY + E D RQI +D PR P+ Q + V + ERIL+ W
Sbjct: 90 QKEYFAFIEHYYSSRNDEVHQDTY---RQIHIDIPRMSPEALILQPK-VTEIFERILFIW 145
Query: 250 AIRHPASGYVQGINDLVTPFLVVFISEHLEG-GIDDWSMSDLSSDKISNVEADCYWCLSK 308
AIRHPASGYVQGINDLVTPF VVFI E+ + +D +S + ++ + N+EAD YWC+SK
Sbjct: 146 AIRHPASGYVQGINDLVTPFFVVFICEYTDREDVDKVDVSSVPAEVLRNIEADTYWCMSK 205
Query: 309 LLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIRE 368
LLDG+QD+YTFAQPGIQ V L+ELV RIDE + +H++ + +LQFAFRW N LL+RE
Sbjct: 206 LLDGIQDNYTFAQPGIQMKVKMLEELVSRIDERVHRHLDGHEVRYLQFAFRWMNNLLMRE 265
Query: 369 IPFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
+P RLWDTY +E + F +Y+ A+FL+
Sbjct: 266 LPLRCTIRLWDTYQSEPEGFSHFHLYVCAAFLV 298
>GYP1_YEAST (Q08484) GTPase-activating protein GYP1 (GAP for YPT1)
Length = 637
Score = 249 bits (636), Expect = 1e-65
Identities = 122/258 (47%), Positives = 176/258 (67%), Gaps = 10/258 (3%)
Query: 133 RVTKFNKVLSG-TVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRG 191
R++KF+ +L T++ +LR+++W+G+P RP VW+LL+GY P N+ R+EG L RKR
Sbjct: 254 RISKFDNILKDKTIINQQDLRQISWNGIPKIHRPVVWKLLIGYLPVNTKRQEGFLQRKRK 313
Query: 192 EYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAI 251
EY D + + S++ ++ QI +D PRT P + +Q + VQ SL+RILY WAI
Sbjct: 314 EYRDSLKHTF----SDQHSRDIPTWHQIEIDIPRTNPHIPLYQFKSVQNSLQRILYLWAI 369
Query: 252 RHPASGYVQGINDLVTPFLVVFISEHLEGG-IDDWSMSDLSS----DKISNVEADCYWCL 306
RHPASGYVQGINDLVTPF F++E+L IDD + D S+ ++I+++EAD +WCL
Sbjct: 370 RHPASGYVQGINDLVTPFFETFLTEYLPPSQIDDVEIKDPSTYMVDEQITDLEADTFWCL 429
Query: 307 SKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLI 366
+KLL+ + D+Y QPGI R V L +LV+RID + H +++ +EF+QFAFRW NCLL+
Sbjct: 430 TKLLEQITDNYIHGQPGILRQVKNLSQLVKRIDADLYNHFQNEHVEFIQFAFRWMNCLLM 489
Query: 367 REIPFDLITRLWDTYLAE 384
RE + R+WDTYL+E
Sbjct: 490 REFQMGTVIRMWDTYLSE 507
>YQ01_CAEEL (Q09445) Hypothetical protein C04A2.1 in chromosome II
Length = 330
Score = 74.7 bits (182), Expect = 4e-13
Identities = 65/232 (28%), Positives = 105/232 (45%), Gaps = 37/232 (15%)
Query: 177 PNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQ 236
P++ RKE L R D + Q +++PD +++QI +D PRT PD F + +
Sbjct: 80 PHTFRKELWL-RSCPSRADGVWQRHEVPDE--------VIKQIKLDLPRTFPDNKFLKTE 130
Query: 237 QVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKIS 296
+K+L R L+A A P+ GY QG+N + L+V
Sbjct: 131 GTRKTLGRALFAVAEHIPSVGYCQGLNFVAGVILLVV----------------------- 167
Query: 297 NVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIE--DQGLEFL 354
N E+ L L+ Q++Y G++R + L L+R + +E D GL+ L
Sbjct: 168 NDESRAIDLLVHLVSQRQEYYGKNMIGLRRDMHVLHSLLREHCPRVVVTLEKLDVGLDML 227
Query: 355 QFAFRWFNCLLIREIPFDLITRLWDTYLAEGDA-LPDFLVYIFASFLLTTNS 405
+WF C + +P + + RLWD + EGD L V +F S ++ +S
Sbjct: 228 --VGKWFVCWFVESLPMETVLRLWDCLIYEGDEWLFRIAVALFRSNMIAISS 277
>TB13_MOUSE (Q8R3D1) TBC1 domain family member 13
Length = 400
Score = 72.4 bits (176), Expect = 2e-12
Identities = 76/355 (21%), Positives = 141/355 (39%), Gaps = 86/355 (24%)
Query: 132 ARVTKFNKVLSGTVVILDNLRELAWSGVP--DYMRPKVWRLLLGYEPPNSDRKEGVLGRK 189
+R+ F VL ++L+ LREL++SG+P +R W++LL Y P +L ++
Sbjct: 7 SRIADFQDVLKEPSIVLEKLRELSFSGIPCEGGLRCLCWKILLNYLPLERASWTSILAKQ 66
Query: 190 RGEYLDCISQ--------------------YYDIPDSERSDDEVN-------MLRQIAVD 222
RG Y + + + D P + D N +L QI D
Sbjct: 67 RGLYSQFLREMIIQPGIAKANMGVFREDVTFEDHPLNPNPDSRWNTYFKDNEVLLQIDKD 126
Query: 223 CPRTVPDVTFFQ-------------QQQVQKSLERILYAWAIRHPASGYVQGINDLVTPF 269
R PD++FFQ Q + + +R+ + G+ ++ +P
Sbjct: 127 VRRLCPDISFFQRATEYPCLLILDPQNEFETLRKRVEQTTLKSQTVARNRSGVTNMSSPH 186
Query: 270 --------------------------LVVFISEHLEGGIDDWS-------------MSDL 290
++FI L GI +D
Sbjct: 187 KNSAPSALNEYEVLPNGCEAHWEVVERILFIYAKLNPGIAYVQGMNEIVGPLYYTFATDP 246
Query: 291 SSDKISNVEADCYWCLSKLLDGMQDHY----TFAQPGIQRLVFKLKELVRRIDEPISQHI 346
+S+ + EAD ++C + L+ ++D++ +Q GI + K+ ++ D + +
Sbjct: 247 NSEWKEHAEADTFFCFTNLMAEIRDNFIKSLDDSQCGITYKMEKVYSTLKDKDVELYLKL 306
Query: 347 EDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
++Q ++ FAFRW LL +E + R+WD+ A+G+ DFL+ + + L+
Sbjct: 307 QEQSIKPQFFAFRWLTLLLSQEFLLPDVIRIWDSLFADGNRF-DFLLLVCCAMLI 360
>US6L_HUMAN (Q92738) USP6 N-terminal like protein (Related to the N
terminus of tre) (RN-tre)
Length = 828
Score = 70.9 bits (172), Expect = 6e-12
Identities = 65/270 (24%), Positives = 113/270 (41%), Gaps = 49/270 (18%)
Query: 133 RVTKFNKVLSGTVVI--LDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKR 190
R TK+ K+L G + + G+P +R +VW LLL P + + + +
Sbjct: 73 RTTKWLKMLKGWEKYKNTEKFHRRIYKGIPLQLRGEVWALLLEI-PKMKEETRDLYSKLK 131
Query: 191 GEYLDCISQYYDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQ--VQKSLERILYA 248
C PD +RQI +D RT D F+ + Q+SL +L A
Sbjct: 132 HRARGCS------PD----------IRQIDLDVNRTFRDHIMFRDRYGVKQQSLFHVLAA 175
Query: 249 WAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSK 308
++I + GY QG++ +T L+++++E D +W L K
Sbjct: 176 YSIYNTEVGYCQGMSQ-ITALLLMYMNEE-----------------------DAFWALVK 211
Query: 309 LLDGMQD--HYTFAQ--PGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCL 364
L G + H F Q P + R ++++ + + QH++ Q + + +WF
Sbjct: 212 LFSGPKHAMHGFFVQGFPKLLRFQEHHEKILNKFLSKLKQHLDSQEIYTSFYTMKWFFQC 271
Query: 365 LIREIPFDLITRLWDTYLAEGDALPDFLVY 394
+ PF L R+WD Y+ EG+ + + Y
Sbjct: 272 FLDRTPFTLNLRIWDIYIFEGERVLTAMSY 301
>US6L_MOUSE (Q80XC3) USP6 N-terminal like protein
Length = 819
Score = 67.8 bits (164), Expect = 5e-11
Identities = 59/245 (24%), Positives = 104/245 (42%), Gaps = 47/245 (19%)
Query: 156 WSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEVNM 215
+ G+P +R +VW LLL P + + + + C PD
Sbjct: 98 YKGIPLQLRGEVWALLLEI-PKMKEETRDLYSKLKHRARGCS------PD---------- 140
Query: 216 LRQIAVDCPRTVPDVTFFQQQQ--VQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVF 273
+RQI +D RT D F+ + Q+SL +L A++I + GY QG++ +T L+++
Sbjct: 141 IRQIDLDVNRTFRDHIMFRDRYGVKQQSLFHVLAAYSIYNTEVGYCQGMSQ-ITALLLMY 199
Query: 274 ISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQD--HYTFAQ--PGIQRLVF 329
++E D +W L KL G + H F Q P + R
Sbjct: 200 MNEE-----------------------DAFWALVKLFSGPKHAMHGFFVQGFPKLLRFQE 236
Query: 330 KLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEGDALP 389
++++ + + QH++ Q + + +WF + PF L R+WD Y+ EG+ +
Sbjct: 237 HHEKILNKFLSKLKQHLDSQEIYTSFYTMKWFFQCFLDRTPFRLNLRIWDIYIFEGERVL 296
Query: 390 DFLVY 394
+ Y
Sbjct: 297 TAMSY 301
>GYP7_YARLI (P09379) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 730
Score = 67.0 bits (162), Expect = 9e-11
Identities = 63/264 (23%), Positives = 112/264 (41%), Gaps = 53/264 (20%)
Query: 145 VVILDNLRELAW-SGVPDYMRPKVWRLLLGYEPPNS---DRKEGVLGRKRGEYLDCISQY 200
+V ++ ++E + G+ +RP+ W LLG P +S +RKE ++ + R +Y ++
Sbjct: 374 IVTVNEVKERIFHGGLAPAVRPEGWLFLLGVYPWDSTAAERKE-LVSKLRVDYNRLKKEW 432
Query: 201 YDIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKS------------------- 241
+ D ER D + L +I D RT ++TFF + +K
Sbjct: 433 WVQEDKERDDFWRDQLSRIEKDVHRTDRNITFFAECDAKKDGDDDNYDKDEFGFSSQINS 492
Query: 242 ------LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKI 295
L +L + + GYVQG++DL++P VV + L
Sbjct: 493 NIHLIQLRDMLITYNQHNKNLGYVQGMSDLLSPLYVVLQDDTL----------------- 535
Query: 296 SNVEADCYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQ 355
+W S ++ M+ +Y Q G++ + L LV+ + + +H+E L
Sbjct: 536 ------AFWAFSAFMERMERNYLRDQSGMRNQLLCLDHLVQFMLPSLYKHLEKTESTNLF 589
Query: 356 FAFRWFNCLLIREIPFDLITRLWD 379
F FR RE+ +D + RLW+
Sbjct: 590 FFFRMLLVWFKRELLWDDVLRLWE 613
>TBC2_HUMAN (Q9BYX2) TBC1 domain family member 2 (Prostate antigen
recognized and indentified by SEREX) (PARIS-1)
Length = 917
Score = 62.8 bits (151), Expect = 2e-09
Identities = 64/251 (25%), Positives = 101/251 (39%), Gaps = 51/251 (20%)
Query: 143 GTVVILDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYD 202
G +V L++L +GVP RP+VWR L+ R ++L Y +
Sbjct: 610 GDLVPSAELKQLLRAGVPREHRPRVWRWLV---------------HLRVQHLHTPGCYQE 654
Query: 203 IPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQ--QQQVQKSLERILYAWAIRHPASGYVQ 260
+ S E RQI +D RT P+ F L R+L A++ ++P GY Q
Sbjct: 655 LL-SRGQAREHPAARQIELDLNRTFPNNKHFTCPTSSFPDKLRRVLLAFSWQNPTIGYCQ 713
Query: 261 GINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDGMQ--DHY- 317
G+N L L+V E +WCL +++ + D+Y
Sbjct: 714 GLNRLAAIALLVL-----------------------EEEESAFWCLVAIVETIMPADYYC 750
Query: 318 ---TFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLI 374
T +Q + L L E + R+ + QH D L F WF + + +++
Sbjct: 751 NTLTASQVDQRVLQDLLSEKLPRLMAHLGQHHVDLSL----VTFNWFLVVFADSLISNIL 806
Query: 375 TRLWDTYLAEG 385
R+WD +L EG
Sbjct: 807 LRVWDAFLYEG 817
>TBC5_HUMAN (Q92609) TBC1 domain family member 5
Length = 795
Score = 58.9 bits (141), Expect = 3e-08
Identities = 57/215 (26%), Positives = 95/215 (43%), Gaps = 35/215 (16%)
Query: 219 IAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHL 278
I D RT P++ FFQQ+ V+K L +L+ +A + Y QG+++L+ P + V +H
Sbjct: 163 IEQDVKRTFPEMQFFQQENVRKILTDVLFCYARENEQLLYKQGMHELLAPIVFVLHCDH- 221
Query: 279 EGGIDDWSMSDLSSDKISNV------EADCYWCLSKLL------------DGMQDHYT-- 318
S S S+++ V E D Y S+L+ DG + T
Sbjct: 222 -QAFLHASESAQPSEEMKTVLNPEYLEHDAYAVFSQLMETAEPWFSTFEHDGQKGKETLM 280
Query: 319 ----FAQP-------GIQRLVFKLKE-LVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLI 366
FA+P I V ++++ L+++ D + H+ + + RW L
Sbjct: 281 TPIPFARPQDLGPTIAIVTKVNQIQDHLLKKHDIELYMHLNRLEIAPQIYGLRWVRLLFG 340
Query: 367 REIPFDLITRLWDTYLAEGDALPDFLVYIFASFLL 401
RE P + +WD A+G +L + YIF + LL
Sbjct: 341 REFPLQDLLVVWDALFADGLSL-GLVDYIFVAMLL 374
>TB14_MOUSE (Q8CGA2) TBC1 domain family member 14
Length = 679
Score = 58.2 bits (139), Expect = 4e-08
Identities = 73/307 (23%), Positives = 124/307 (39%), Gaps = 41/307 (13%)
Query: 91 QSTSKSVSAANVSKLKSSTSLGENLSEDIR-KYTIGARATDSARVTKFNKVLSG--TVVI 147
Q + V A +LK + + L E + + +IG +A +T N++L T+
Sbjct: 322 QQYEEMVLQAKKRELKEAQRRRKQLEERCKVEESIG-----NAVLTWNNEILPNWETMWC 376
Query: 148 LDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGV-LGRKRGEYLDCISQYYDIPDS 206
+R+L W G+P +R KVW L +G E + + L R + + + ++ +
Sbjct: 377 SKKVRDLWWQGIPPSVRGKVWSLAIGNELNITHELFDICLARAKERWRSLSTGGSEVENE 436
Query: 207 E---RSDDEVNMLRQIAVDCPRTVPDVTFFQQ-QQVQKSLERILYAWAIRHPASGYVQGI 262
+ + D L I +D RT P++ FQQ L IL A+ P GYVQG
Sbjct: 437 DAGFSAADREASLELIKLDISRTFPNLCIFQQGGPYHDMLHSILGAYTCYRPDVGYVQG- 495
Query: 263 NDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVE-ADCYWCLSKLLD-GMQDHYTFA 320
MS +++ I N++ AD + S LL+ Q +
Sbjct: 496 ------------------------MSFIAAVLILNLDTADAFIAFSNLLNKPCQMAFFRV 531
Query: 321 QPGIQRLVFKLKELVRRIDEP-ISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
G+ F E+ + P + H + L + W L + +P DL R+WD
Sbjct: 532 DHGLMLTYFAAFEVFFEENLPKLFAHFKKNNLTADIYLIDWIFTLYSKSLPLDLACRIWD 591
Query: 380 TYLAEGD 386
+ +G+
Sbjct: 592 VFCRDGE 598
>TB14_HUMAN (Q9P2M4) TBC1 domain family member 14
Length = 678
Score = 58.2 bits (139), Expect = 4e-08
Identities = 74/307 (24%), Positives = 124/307 (40%), Gaps = 41/307 (13%)
Query: 91 QSTSKSVSAANVSKLKSSTSLGENLSEDIR-KYTIGARATDSARVTKFNKVLSG--TVVI 147
Q + V A +LK + + L E R + +IG +A +T N++L T+
Sbjct: 321 QQYEEMVVQAKKRELKEAQRRKKQLEERCRVEESIG-----NAVLTWNNEILPNWETMWC 375
Query: 148 LDNLRELAWSGVPDYMRPKVWRLLLGYEPPNSDRKEGV-LGRKRGEYLDCISQYYDIPDS 206
+R+L W G+P +R KVW L +G E + + L R + + + ++ +
Sbjct: 376 SRKVRDLWWQGIPPSVRGKVWSLAIGNELNITHELFDICLARAKERWRSLSTGGSEVENE 435
Query: 207 E---RSDDEVNMLRQIAVDCPRTVPDVTFFQQ-QQVQKSLERILYAWAIRHPASGYVQGI 262
+ + D L I +D RT P++ FQQ L IL A+ P GYVQG
Sbjct: 436 DAGFSAADREASLELIKLDISRTFPNLCIFQQGGPYHDMLHSILGAYTCYRPDVGYVQG- 494
Query: 263 NDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVE-ADCYWCLSKLLD-GMQDHYTFA 320
MS +++ I N++ AD + S LL+ Q +
Sbjct: 495 ------------------------MSFIAAVLILNLDTADAFIAFSNLLNKPCQMAFFRV 530
Query: 321 QPGIQRLVFKLKELVRRIDEP-ISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWD 379
G+ F E+ + P + H + L + W L + +P DL R+WD
Sbjct: 531 DHGLMLTYFAAFEVFFEENLPKLFAHFKKNNLTPDIYLIDWIFTLYSKSLPLDLACRIWD 590
Query: 380 TYLAEGD 386
+ +G+
Sbjct: 591 VFCRDGE 597
>GYP7_CANGA (Q6FWI1) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 745
Score = 57.4 bits (137), Expect = 7e-08
Identities = 55/252 (21%), Positives = 96/252 (37%), Gaps = 51/252 (20%)
Query: 161 DYMRPKVWRLLLGYEPPNSDRKEGVLGRK--RGEYLDCISQYYDIPDSERSDDEV---NM 215
D R +VW LLG P +S E RK EY++ ++ D + +D+E +
Sbjct: 394 DATRKEVWPFLLGVYPWDSSEDERKQLRKALHDEYMELKQKWVDREVNLDNDEEEYWKDQ 453
Query: 216 LRQIAVDCPRTVPDVTFFQQQQVQ-----------------------KSLERILYAWAIR 252
L +I D R ++ ++ K L IL + I
Sbjct: 454 LFRIEKDVKRNDRNIDIYKYNTSDNLPFPEDTAPTTDDDDSIKNPNLKKLADILTTYNIF 513
Query: 253 HPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDG 312
+P GYVQG+ DL++P + E +WC + ++
Sbjct: 514 NPNLGYVQGMTDLLSPLYYIIRDEETT-----------------------FWCFTNFMER 550
Query: 313 MQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFD 372
M+ ++ Q GI+ + L +L + + +S H++ L F FR RE +D
Sbjct: 551 MERNFLRDQSGIRDQMLALTDLCQLMLPRLSAHLQKCDSSDLFFCFRMLLVWFKREFNYD 610
Query: 373 LITRLWDTYLAE 384
I +W+ + +
Sbjct: 611 DIFNIWEVFFTD 622
>GYP2_YEAST (P53258) GTPase-activating protein GYP2 (MAC1-dependent
regulator) (MIC1 protein)
Length = 950
Score = 54.7 bits (130), Expect = 5e-07
Identities = 57/249 (22%), Positives = 100/249 (39%), Gaps = 55/249 (22%)
Query: 145 VVILDNLRELAWSGVPDYMRPKVWRLLLG---YEPPNSDRKEGVLGRKRGEYLDCISQYY 201
VV R+L GVP+ MR ++W L G NS E +L G+ I +
Sbjct: 231 VVQTPMFRKLIRIGVPNRMRGEIWELCSGAMYMRYANSGEYERILNENAGKTSQAIDE-- 288
Query: 202 DIPDSERSDDEVNMLRQIAVDCPRTVPDVTFFQQQQVQKSLERILYAWAIRHPASGYVQG 261
I D R++P+ + +Q ++ + L +L A++ ++P GY Q
Sbjct: 289 -----------------IEKDLKRSLPEYSAYQTEEGIQRLRNVLTAYSWKNPDVGYCQA 331
Query: 262 INDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLD-GMQDHYTFA 320
+N +V FL +F+SE +WCL L D + +Y+
Sbjct: 332 MNIVVAGFL-IFMSEE-----------------------QAFWCLCNLCDIYVPGYYSKT 367
Query: 321 QPG--IQRLVFK--LKELVRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITR 376
G + + VF+ +++ + + E I QH ++ + WF L +P + R
Sbjct: 368 MYGTLLDQRVFESFVEDRMPVLWEYILQH----DIQLSVVSLPWFLSLFFTSMPLEYAVR 423
Query: 377 LWDTYLAEG 385
+ D + G
Sbjct: 424 IMDIFFMNG 432
>GYP7_YEAST (P48365) GTPase-activating protein GYP7 (GAP for YPT7)
Length = 746
Score = 52.4 bits (124), Expect = 2e-06
Identities = 35/140 (25%), Positives = 59/140 (42%), Gaps = 23/140 (16%)
Query: 242 LERILYAWAIRHPASGYVQGINDLVTPFLVVFISEHLEGGIDDWSMSDLSSDKISNVEAD 301
L+ IL + + + GYVQG+ DL++P V+ E W
Sbjct: 513 LQNILITYNVYNTNLGYVQGMTDLLSPIYVIMKEE--------WKT-------------- 550
Query: 302 CYWCLSKLLDGMQDHYTFAQPGIQRLVFKLKELVRRIDEPISQHIEDQGLEFLQFAFRWF 361
+WC + +D M+ ++ Q GI + L ELV+ + +S+H+ L F FR
Sbjct: 551 -FWCFTHFMDIMERNFLRDQSGIHEQMLTLVELVQLMLPELSEHLNKCDSGNLFFCFRML 609
Query: 362 NCLLIREIPFDLITRLWDTY 381
RE + I +W+ +
Sbjct: 610 LVWFKREFEMEDIMHIWENF 629
>TB10_MOUSE (P58802) TBC1 domain family member 10 (EBP50-PDX
interactor of 64 kDa) (EPI64 protein)
Length = 500
Score = 48.9 bits (115), Expect = 3e-05
Identities = 47/231 (20%), Positives = 87/231 (37%), Gaps = 45/231 (19%)
Query: 158 GVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEVNMLR 217
G+P +R + W+ L G + L + G++ + + S + L
Sbjct: 111 GIPPSLRGRAWQYLSGGKVK--------LQQNPGKF----------DELDMSPGDPKWLD 152
Query: 218 QIAVDCPRTVPDVTFFQQQ--QVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
I D R P F + Q+ L R+L A+ + P GY Q + L+ +
Sbjct: 153 VIERDLHRQFPFHEMFVSRGGHGQQDLFRVLKAYTLYRPEEGYCQAQAPIAAVLLMHMPA 212
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDG-MQDHYTFAQPGIQRLVFKLKEL 334
E +WCL ++ + + +Y+ IQ L L
Sbjct: 213 EQ------------------------AFWCLVQVCEKYLPGYYSEKLEAIQLDGEILFSL 248
Query: 335 VRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEG 385
++++ +H+ Q ++ L + WF C R +P+ + R+WD + EG
Sbjct: 249 LQKVSPVAHKHLSRQKIDPLLYMTEWFMCAFARTLPWSSVLRVWDMFFCEG 299
>TB10_HUMAN (Q9BXI6) TBC1 domain family member 10 (EBP50-PDX
interactor of 64 kDa) (EPI64 protein)
Length = 508
Score = 48.9 bits (115), Expect = 3e-05
Identities = 47/231 (20%), Positives = 87/231 (37%), Gaps = 45/231 (19%)
Query: 158 GVPDYMRPKVWRLLLGYEPPNSDRKEGVLGRKRGEYLDCISQYYDIPDSERSDDEVNMLR 217
G+P +R + W+ L G + L + G++ + + S + L
Sbjct: 111 GIPPSLRGRAWQYLSGGKVK--------LQQNPGKF----------DELDMSPGDPKWLD 152
Query: 218 QIAVDCPRTVPDVTFFQQQ--QVQKSLERILYAWAIRHPASGYVQGINDLVTPFLVVFIS 275
I D R P F + Q+ L R+L A+ + P GY Q + L+ +
Sbjct: 153 VIERDLHRQFPFHEMFVSRGGHGQQDLFRVLKAYTLYRPEEGYCQAQAPIAAVLLMHMPA 212
Query: 276 EHLEGGIDDWSMSDLSSDKISNVEADCYWCLSKLLDG-MQDHYTFAQPGIQRLVFKLKEL 334
E +WCL ++ + + +Y+ IQ L L
Sbjct: 213 EQ------------------------AFWCLVQICEKYLPGYYSEKLEAIQLDGEILFSL 248
Query: 335 VRRIDEPISQHIEDQGLEFLQFAFRWFNCLLIREIPFDLITRLWDTYLAEG 385
++++ +H+ Q ++ L + WF C R +P+ + R+WD + EG
Sbjct: 249 LQKVSPVAHKHLSRQKIDPLLYMTEWFMCAFSRTLPWSSVLRVWDMFFCEG 299
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.135 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 52,069,064
Number of Sequences: 164201
Number of extensions: 2144592
Number of successful extensions: 6753
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 32
Number of HSP's successfully gapped in prelim test: 47
Number of HSP's that attempted gapping in prelim test: 6506
Number of HSP's gapped (non-prelim): 130
length of query: 451
length of database: 59,974,054
effective HSP length: 114
effective length of query: 337
effective length of database: 41,255,140
effective search space: 13902982180
effective search space used: 13902982180
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137825.12


