

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137821.7 - phase: 0
(821 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
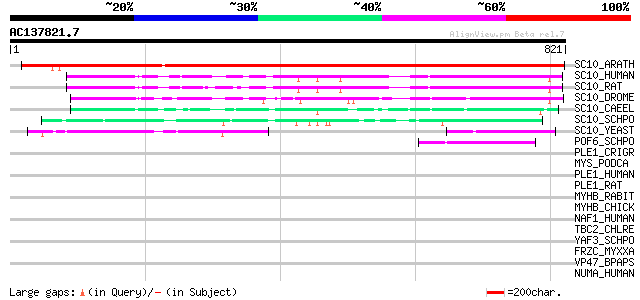
Score E
Sequences producing significant alignments: (bits) Value
SC10_ARATH (Q8RVQ5) Exocyst complex component Sec10 1285 0.0
SC10_HUMAN (O00471) Exocyst complex component Sec10 (hSec10) 172 3e-42
SC10_RAT (P97878) Exocyst complex component Sec10 (71 kDa compon... 170 2e-41
SC10_DROME (Q9XTM1) Exocyst complex component Sec10 159 3e-38
SC10_CAEEL (Q18406) Probable exocyst complex component Sec10 119 4e-26
SC10_SCHPO (O13705) Exocyst complex component sec10 92 7e-18
SC10_YEAST (Q06245) Exocyst complex component SEC10 86 5e-16
POF6_SCHPO (O74854) F-box protein pof6 50 3e-05
PLE1_CRIGR (Q9JI55) Plectin 1 (PLTN) (PCN) (300-kDa intermediate... 45 0.001
MYS_PODCA (Q05000) Myosin heavy chain (Fragment) 45 0.001
PLE1_HUMAN (Q15149) Plectin 1 (PLTN) (PCN) (Hemidesmosomal prote... 44 0.001
PLE1_RAT (P30427) Plectin 1 (PLTN) (PCN) 44 0.002
MYHB_RABIT (P35748) Myosin heavy chain, smooth muscle isoform (S... 41 0.015
MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle 40 0.019
NAF1_HUMAN (Q15025) Nef-associated factor 1 (Naf1) (HIV-1 Nef in... 39 0.043
TBC2_CHLRE (Q8VXP3) Tbc2 translation factor, chloroplast precursor 39 0.073
YAF3_SCHPO (Q09857) Hypothetical protein C29E6.03c in chromosome I 37 0.16
FRZC_MYXXA (P43500) Frizzy aggregation protein frzCD 37 0.16
VP47_BPAPS (Q9T1Q1) Putative protein p47 35 0.62
NUMA_HUMAN (Q14980) Nuclear mitotic apparatus protein 1 (NuMA pr... 35 0.62
>SC10_ARATH (Q8RVQ5) Exocyst complex component Sec10
Length = 829
Score = 1285 bits (3324), Expect = 0.0
Identities = 665/816 (81%), Positives = 743/816 (90%), Gaps = 15/816 (1%)
Query: 18 ASSAASFPLILDIDDFKGDFSFDALFGNLVNELLPSFKLEDLEA-------EGADAVQNK 70
+SS S PLILDI+DFKGDFSFDALFGNLVN+LLPSF E+ ++ G D + N
Sbjct: 12 SSSVNSVPLILDIEDFKGDFSFDALFGNLVNDLLPSFLDEEADSGDGHGNIAGVDGLTNG 71
Query: 71 Y-----SQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRT 125
+ + ++++P FPEV+ LLSLFKD+CKEL++LRKQ+DGRL+ LKK+VS QDSKHR+T
Sbjct: 72 HLRGQSAPLSSAPFFPEVDGLLSLFKDACKELVDLRKQVDGRLNTLKKEVSTQDSKHRKT 131
Query: 126 LAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFN 185
L E+EKGVDGLF SFARLD RISSVGQTAAKIGDHLQSADAQRETASQTI+LIKYLMEFN
Sbjct: 132 LTEIEKGVDGLFESFARLDGRISSVGQTAAKIGDHLQSADAQRETASQTIDLIKYLMEFN 191
Query: 186 SSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVA 245
SPGDLMELS LFSDDSRVAEAASIAQKLRSFAEEDIGR G A +A GNAT RGLEVA
Sbjct: 192 GSPGDLMELSALFSDDSRVAEAASIAQKLRSFAEEDIGRQG--ASTAAGNATPGRGLEVA 249
Query: 246 VANLQEYCNELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFID 305
VANLQ+YCNELENRLLSRFDAASQ+R+L+TM+ECAKILSQFNRGTSAMQHYVATRPMFID
Sbjct: 250 VANLQDYCNELENRLLSRFDAASQRRDLSTMSECAKILSQFNRGTSAMQHYVATRPMFID 309
Query: 306 VEVMNADTRLVLGDQAAQSSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSIL 365
VEVMN+D RLVLGD +Q SP+NVARGLS+L+KEITDTVRKEAATITAVFP+PNEVM+IL
Sbjct: 310 VEVMNSDIRLVLGDHGSQPSPSNVARGLSALFKEITDTVRKEAATITAVFPTPNEVMAIL 369
Query: 366 VQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG 425
VQRVLEQRVT +LDK+L KPSL++ P ++EGGLL YLRMLAV+YE+TQE+A+DLR VGCG
Sbjct: 370 VQRVLEQRVTGILDKILAKPSLMSPPPVQEGGLLLYLRMLAVAYERTQELAKDLRAVGCG 429
Query: 426 DLDVEGLTESLFSSHKDEYPEYEQASLRQLYKVKMEELRAESQ-ISDSSGTIGRSKGATV 484
DLDVE LTESLFSSHKDEYPE+E+ASL+QLY+ KMEELRAESQ +S+SSGTIGRSKGA++
Sbjct: 430 DLDVEDLTESLFSSHKDEYPEHERASLKQLYQAKMEELRAESQQVSESSGTIGRSKGASI 489
Query: 485 ASSQQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLE 544
+SS QQISVT VTEFVRWNEEAITRC LFSSQP+TLA VKA+FTCLLDQVS YI EGLE
Sbjct: 490 SSSLQQISVTFVTEFVRWNEEAITRCTLFSSQPATLAANVKAIFTCLLDQVSVYITEGLE 549
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
RARD L+EAA LRERFVLGTSVSRRVAAAAASAAEAAAAAGESSF+SFMVAVQR GSSVA
Sbjct: 550 RARDSLSEAAALRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFKSFMVAVQRCGSSVA 609
Query: 605 IIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSA 664
I+QQYF+NSISRLLLPVDGAHAA+CEEM+TA+S AEAAAYKGLQQCIETVMAEV+RLLS+
Sbjct: 610 IVQQYFANSISRLLLPVDGAHAASCEEMSTALSKAEAAAYKGLQQCIETVMAEVDRLLSS 669
Query: 665 EQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHK 724
EQK+TDY+SPDDG+A DHRPTNAC RVVAYLSRVLESAFTALEGLNKQAFL+ELGNRL K
Sbjct: 670 EQKSTDYRSPDDGIASDHRPTNACIRVVAYLSRVLESAFTALEGLNKQAFLTELGNRLEK 729
Query: 725 VLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLS 784
+LL HWQK+TFNPSGGLRLKRD+ EY FV+SF APSVDEKFELLGI+ANVFIVAP+SL
Sbjct: 730 LLLTHWQKFTFNPSGGLRLKRDLNEYVGFVKSFGAPSVDEKFELLGIIANVFIVAPDSLP 789
Query: 785 TLFEGTPSIRKDAQRFIQLREDYKSAKLASKLSSLW 820
TLFEG+PSIRKDAQRFIQLREDYKSAKLA+KLSSLW
Sbjct: 790 TLFEGSPSIRKDAQRFIQLREDYKSAKLATKLSSLW 825
>SC10_HUMAN (O00471) Exocyst complex component Sec10 (hSec10)
Length = 708
Score = 172 bits (436), Expect = 3e-42
Identities = 173/757 (22%), Positives = 317/757 (41%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNSEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA I QKL A+E R EV +Y ++LE +L+
Sbjct: 156 IKEAADIIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F + + Y+ Q
Sbjct: 200 FTSAQRRGEISRMREVAAVLLHFKGYSHCVDVYIKQC-------------------QEGA 240
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
N++ L + + V +F +P V++ L+Q V E ++ + + +
Sbjct: 241 YLRNDIFEDAGILCQRVNKQVGD-------IFSNPETVLAKLIQNVFEIKLQSFVKE--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLEECRKSDAEQYLKNLYDLYTRTTNLSSKLMEFNLGTDKQTFLSKLIKSIFISY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 346 LENYIEVETGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 465 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIGQMKHILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V AY+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
+++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N+ +VAP++L
Sbjct: 612 RLIYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 708
>SC10_RAT (P97878) Exocyst complex component Sec10 (71 kDa component
of rsec6/8 secretory complex) (p71)
Length = 708
Score = 170 bits (430), Expect = 2e-41
Identities = 176/757 (23%), Positives = 320/757 (42%), Gaps = 111/757 (14%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
++LL F + +EL + ++I ++ L++ + + + + EL+K F F L
Sbjct: 40 KRLLEEFVNHIQELQIMDERIQRKVEKLEQQCQKEAKEFAKKVQELQKSNQVAFQHFQEL 99
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
D IS V +GD L+ + R+ A + +L+KY EF G+L S +F++ +
Sbjct: 100 DEHISYVATKVCHLGDQLEGVNTPRQRAVEAQKLMKYFNEFLD--GELK--SDVFTNPEK 155
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+ EAA + QKL A+E R EV +Y ++LE +L+
Sbjct: 156 IKEAADVIQKLHLIAQE---------------LPFDRFSEVKSKIASKY-HDLECQLIQE 199
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQ 323
F +A ++ E++ M E A +L F +G S IDV + L + +
Sbjct: 200 FTSAQRRGEVSRMREVAAVLLHF-KGYSHC----------IDVYIKQCQEGAYLRNDIFE 248
Query: 324 SSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLV 383
+ R + K++ D +F +P V++ L+Q V E ++ + +
Sbjct: 249 DAAILCQR----VNKQVGD-----------IFSNPEAVLAKLIQNVFEVKLQSFVKD--- 290
Query: 384 KPSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCG---DLDVEGLTESLFSSH 440
L + YL+ L Y +T ++ L G + L +S+F S+
Sbjct: 291 -----QLEECRKSDAEQYLKSLYDLYTRTTSLSSKLMEFNLGTDKQTFLSKLIKSIFVSY 345
Query: 441 KDEYPEYEQASLR--------QLYKVKMEELRA--ESQISDSSGTIGRSKGATVASS--- 487
+ Y E E L+ + Y K + R+ I D I + + S
Sbjct: 346 LENYIEVEIGYLKSRSAMILQRYYDSKNHQKRSIGTGGIQDLKERIRQRTNLPLGPSIDT 405
Query: 488 --QQQISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVS-QYIAEGLE 544
+ +S VV ++ ++A RC+ S PS L +FT L++ + ++I LE
Sbjct: 406 HGETFLSQEVVVNLLQETKQAFERCHRLSD-PSDLPRNAFRIFTILVEFLCIEHIDYALE 464
Query: 545 RARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVA 604
G+ + + F+ VQ++ +
Sbjct: 465 TGLAGIPSSDSRNANLY------------------------------FLDVVQQANTIFH 494
Query: 605 IIQQYFSNSISRLLLPVDGAHAAAC-EEMATAMSSAEAAAYKGLQQCIETVMAEVERLLS 663
+ + F++ + L+ + C ++ + E G+ + + ++ +++ +L+
Sbjct: 495 LFDKQFNDHLMPLIS--SSPKLSECLQKKKEIIEQMEMKLDTGIDRTLNCMIGQMKHILA 552
Query: 664 AEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
AEQK TD+K P+D + TNAC +V AY+ + +E +++G N L ELG R H
Sbjct: 553 AEQKKTDFK-PEDENNVLIQYTNACVKVCAYVRKQVEKIKNSMDGKNVDTVLMELGVRFH 611
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL 783
++ H Q+Y+++ GG+ D+ EY + + F P V F+ L L N+ +VAP++L
Sbjct: 612 RLTYEHLQQYSYSCMGGMLAICDVAEYRKCAKDFKIPMVLHLFDTLHALCNLLVVAPDNL 671
Query: 784 STLFEGTPSIRKD---AQRFIQLREDYKSAKLASKLS 817
+ G D F+QLR DY+SA+LA S
Sbjct: 672 KQVCSGEQLANLDKNILHSFVQLRADYRSARLARHFS 708
>SC10_DROME (Q9XTM1) Exocyst complex component Sec10
Length = 710
Score = 159 bits (402), Expect = 3e-38
Identities = 170/756 (22%), Positives = 320/756 (41%), Gaps = 119/756 (15%)
Query: 90 FKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISS 149
F + K+L L+++ + L++ + + H + +A+L++ F +LD +I+S
Sbjct: 46 FIQTIKDLKILQEKQQSKCERLEESLRQEKESHAKKIAKLQERHQTAIDVFGQLDEKINS 105
Query: 150 VGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSRVAEAAS 209
V +G+ L++ + R + + +L+ ++ EF ++ G ++ ++ +F+D +R++EAA
Sbjct: 106 VAGKIMHLGEQLENVNTPRSRSVEAQKLLNFMSEFLAA-GPVI-VNDIFADAARLSEAAD 163
Query: 210 IAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAASQ 269
+ QKL + I+ GN S+ +++ +E+E RL+ F A +
Sbjct: 164 VIQKL----------YAISQDLPPGNFAESK------RKIEKKYDEVERRLIEEFATAQK 207
Query: 270 KRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNV 329
++ M A+ILSQF T + Y+ Q S ++
Sbjct: 208 SEDIERMKTLAQILSQFKGYTQCVDAYIEQ-------------------SQMQPYSGKDI 248
Query: 330 ARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRV-----TALLDKLLVK 384
G+ L K + ++K VF +P +VMS + + + ++ T L DK
Sbjct: 249 FIGIVPLCKHHYEIIQK-------VFANPQQVMSKFILNIYQLKLHQYAMTKLEDK---- 297
Query: 385 PSLVNLPSMEEGGLLFYLRMLAVSYEKTQEIARDLRTVGCGDLD---VEGLTESLFSSHK 441
EE YLR L Y +T +++ DL+ + +D ++ LT+ +F H
Sbjct: 298 -------KDEEK----YLRTLYELYSRTLKLSTDLQ-IYMSTIDDDLLQKLTQQIFIKHL 345
Query: 442 DEYPEYEQASLRQLYKVKMEELRAESQISDSSGTIG-RSKGATVASSQQQISVTVVTEF- 499
Y E E L ++E+ A + ++ T G R + +++ I++ + ++
Sbjct: 346 AGYAEMETKCLTAKCSTELEKFYASKKHQKTATTKGFRRNMEVLIATRANINIAAIEDYG 405
Query: 500 ----------VRWNEEA---ITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLERA 546
+ +EA + RC L S++ +K L + +++ LE
Sbjct: 406 GETFLSEELAINMLQEAKASLKRCRLLSNETELPGNAIKLNDILLRFLMHEHVDYALELG 465
Query: 547 RDGLTEAANLRERFVLGTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAII 606
+A L E V F VQ++ V ++
Sbjct: 466 ----LQAVPLAEGRVFPQLY-------------------------FFDVVQKTNIIVHLL 496
Query: 607 QQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSAEQ 666
+ S+ + ++ + M E +GL + I V+ V+ L EQ
Sbjct: 497 DKLCHTSVIPCVSNTP-KYSDYVFKKRILMEQIETKLDQGLDRSISAVIGWVKVYLQYEQ 555
Query: 667 KATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHKVL 726
K TDYK D D + AC +VV L V+ ++G N Q L+E G RLH+V+
Sbjct: 556 KKTDYKPETD---VDTISSAACLQVVQNLQPVIVQIKKCVDGENLQNVLTEFGTRLHRVI 612
Query: 727 LNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTL 786
+H Q FN +G + D+ EY + +R ++P V + F++L L N+ +V P++L +
Sbjct: 613 YDHLQTMQFNTAGAMCAICDVNEYRKCIRELDSPLVTQLFDILHALCNLLLVKPQNLQEV 672
Query: 787 FEGTPSIRKD---AQRFIQLREDYKSAKLASKLSSL 819
G D ++FIQLR D++ K + L +
Sbjct: 673 CTGDTLNYLDKSVVRQFIQLRTDFRIIKNTNYLKGI 708
>SC10_CAEEL (Q18406) Probable exocyst complex component Sec10
Length = 659
Score = 119 bits (297), Expect = 4e-26
Identities = 159/734 (21%), Positives = 289/734 (38%), Gaps = 93/734 (12%)
Query: 90 FKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISS 149
F++ L L Q +++ L+K ++ + + + L L + +LD + +
Sbjct: 3 FEEEIGSLQMLCDQFQNKINTLEKQMNEEKKDYVQKLHRLHEKNGEAIDKMKQLDHTMQA 62
Query: 150 VGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSRVAEAAS 209
V +GD L+S D R A +L+++ EF S D S +F+D ++ E+A
Sbjct: 63 VSTKVVHLGDQLESVDQPRSRAHDAHQLMQHFDEFLS---DQPLNSMIFTDPDKLLESAD 119
Query: 210 IAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAASQ 269
+ KL S ++E N A G V +E+ L+ F
Sbjct: 120 LVHKLYSISQE-------LNKDKFANVQARIGQRYKV---------VEDLLIEEF--VRS 161
Query: 270 KRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADTRLVLGDQAAQSSPNNV 329
+R+ MAE AKILS+F + + YV D QS
Sbjct: 162 QRDEKKMAEVAKILSEFKGYSGCVDRYV---------------------DFLCQSIHPRG 200
Query: 330 ARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVN 389
G + + R + I+A+FPSP+ VM LV + R+ + L +
Sbjct: 201 DGG--EILADCLQLCRTQQPRISAIFPSPHTVMQKLVLNIFTGRMKETITARLRE----- 253
Query: 390 LPSMEEGGLLFYLRMLAVSYEKTQEIARDLRT--VGCGDLDVEGLTESLFSSHKDEYPEY 447
+ YLR LA Y T ++ ++L + + LT+S+F + Y
Sbjct: 254 --CKDTDDQEHYLRDLAYLYSSTLKMCKELEKLHISPDSAFLSTLTDSIFQRYIATYCSE 311
Query: 448 EQASLRQ-----LYKVKMEELRAESQISDSSGTIGRSKGATVASSQQQISVTVVTEFVRW 502
E L L + + + QI + R A + + + T V E V
Sbjct: 312 ELKYLNDQCSNLLQRFYESKKHVKKQIGGGLHELKRDVAARLMNVETYGGETFVAEDVA- 370
Query: 503 NEEAITRCNLFSSQPSTLATLVKAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVL 562
S T +A C +++ +++ E D L +++
Sbjct: 371 ----------ISILQETKNAFGRANQLCDKEELPRHV----ENIVDVLL-------KYLY 409
Query: 563 GTSVSRRVAAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVD 622
G + V A + A + ++ F V + S + ++ + F + I ++
Sbjct: 410 GEHLDYAVETGLAGISLAESKTEPPAY--FFSVVAKCTSVILLMIKQFEDPIFPIIKETI 467
Query: 623 GAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDH 682
+ ++ ++ S E+ GL++ + +++ V+ L S EQK TD++ D D
Sbjct: 468 -VEPSVAKKWQQSLRSLESKMSLGLERQLNSIVGYVKFLFS-EQKKTDFRP--DSQQIDI 523
Query: 683 RPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLR 742
R + C +L+ + + +G N +A S+L RL K +L H Q++T+N +G +
Sbjct: 524 RVSPQCQLASRFLANQVAAMELGCDGENLEALQSDLATRLFKFMLTHIQQFTYNSTGAVL 583
Query: 743 LKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIVAPESL-----STLFEGTPSIRKDA 797
L D+ E V + + ++E L L N+ V P+ + S+L E R+
Sbjct: 584 LLCDVGELRTLVSKWRVQNALTQWESLQALTNLLAVLPDQVNETAHSSLLENVD--RQLI 641
Query: 798 QRFIQLREDYKSAK 811
F++LR D++S K
Sbjct: 642 HDFVRLRTDFRSIK 655
>SC10_SCHPO (O13705) Exocyst complex component sec10
Length = 811
Score = 91.7 bits (226), Expect = 7e-18
Identities = 162/818 (19%), Positives = 313/818 (37%), Gaps = 146/818 (17%)
Query: 48 NELLPSFKLEDLEAEGADAVQNKYSQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGR 107
N+ +PS + + AE QN S+ +S ++ + F EL L+ ++ R
Sbjct: 21 NKGIPSTEFVEKFAESLYETQNDGSKKLSS-----IDGSIKSFAACLHELNRLKSRVGDR 75
Query: 108 LHNLKK-DVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADA 166
+ + VQ+ H+++ EK L S L+ ++ + G L+ A+
Sbjct: 76 IRDYASASKQVQNEYHQKSNHLREKFAQVLELS-RHLEDNVNDMRSGLVDAGQELERAEN 134
Query: 167 QRETASQTIELIKYLMEFNSS-PGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRH 225
R+ + EL++Y +EF S P LM+ + D + A Q L E D+
Sbjct: 135 NRKRILSSAELLRYYLEFRSGKPQTLMDFFRTNNHDKMLLCAQRTRQLLALANEVDLPDS 194
Query: 226 GITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAASQKRELTTMAECAKILSQ 285
T +A ++++ LE L F+ +K MA IL +
Sbjct: 195 SET-----------------LARIEKFSEFLETNFLKFFNNEYRKPNWKGMASFGTILQE 237
Query: 286 FNRGTSAMQHYVATRPMFIDVEVMNADTRL-------VLGDQAAQSSPNNVARGLSSLYK 338
FN G S ++ +V FI + + + L +L D + P + + LSSL+
Sbjct: 238 FNGGASVVREFVNQHEFFIAADKVQSRQGLEQDPIWLILPDPTQKIPP--LIQTLSSLFS 295
Query: 339 EITDTVRKEAATITAVFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSMEEG-G 397
E+ + + A I VFP+P V+ QR+ Q + L++++ + +G
Sbjct: 296 ELCSVIEGDCAVIKRVFPNPELVLQTFFQRIFGQSIQNRLEEVM---------EIAKGKS 346
Query: 398 LLFYLRMLAVSYEKTQEIARDLRTV----GCGDLDVEGLTESLFSSHK---------DEY 444
L YLR L +++ DL+T+ G D L+ +L + D+Y
Sbjct: 347 NLAYLRTLQTVVSSLRKLVADLKTILENRGFSVSDNSPLSLALNQYMEDLLVPFIEVDDY 406
Query: 445 PEYEQASLR-----QLYKVKMEELRAES-------------------------------- 467
+ E+ SLR LYK ++R E+
Sbjct: 407 LKREEHSLRSLFRLSLYKYTSYKIRLETPEPGLLRSLMTPLQGNMVAPTGVHSQFNTKTE 466
Query: 468 ----QISD------SSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRCN-LFSSQ 516
+I+D SG+ T+ + ++ V F+ W+ EA+ R + L SSQ
Sbjct: 467 GFLLRIADIQENLIQSGSFITEDYITIEKNHSHLNSEKVYSFIGWHAEALNRASILISSQ 526
Query: 517 PSTLATLVKAVFTCLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAAS 576
+ V + L++ + + I E D + + + + ++ S+ +
Sbjct: 527 D------LSVVISSLVNLLDKLIRE------DYVFKELSSIQSYISSHDKSKNLDL---- 570
Query: 577 AAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEEMATAM 636
++V ++ I YFS + +++P G A++ E +
Sbjct: 571 --------------HYLVDIRECKK----IMGYFSAYLMSIVIPFTGVTASSRRETVNIL 612
Query: 637 SSA----EAAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVV 692
SS+ E A + + +E +LS ++ + Y ++ + + TR V
Sbjct: 613 SSSISVIECAVNDVFYATVHALGDHLEIILSPYRQIS-YAMTEEQIDSSTELRQSMTRNV 671
Query: 693 AYLSRVLESAFTALEGLNKQ--AFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEY 750
+++ + L + A + L +L++ + +G L L+ DI +
Sbjct: 672 QNYLDYIKNLYHRLGPYDPSLLALKQKTATMLAAMLISFAFRSKVTAAGALLLQNDINFF 731
Query: 751 GEFVRSFNAPSVDEKFELLGILANVFIVAPESLSTLFE 788
+ S+ +VD KF+L+ L ++ +V + L + +
Sbjct: 732 HSTLTSWGIDTVDAKFKLILQLLSLLMVKIDVLPAVMQ 769
>SC10_YEAST (Q06245) Exocyst complex component SEC10
Length = 871
Score = 85.5 bits (210), Expect = 5e-16
Identities = 75/372 (20%), Positives = 164/372 (43%), Gaps = 33/372 (8%)
Query: 27 ILDIDDFKGDFSFDALFGNLV----NELLPSFKLEDLEAEGADAVQNKYSQVATSPLFPE 82
+L D+F G + + L N++L ++L D Q+ + L P+
Sbjct: 14 LLKTDNFLGGLTVNEFVQELSKDHRNDVLIDANTKNLPTNEKD--QDAIREAIWKQLDPK 71
Query: 83 VEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFAR 142
+ F+ + KEL L ++ + + V+ Q+ H + +L K + +F +
Sbjct: 72 --PYIRTFESTLKELKNLNEETLNKRQYFSEQVATQEVIHSENVIKLSKDLHTTLLTFDK 129
Query: 143 LDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNS-SPGDLMELSPLFSDD 201
LD R+++V Q + +GD L++A +++ Q++ELI+ +F S D++E L +
Sbjct: 130 LDDRLTNVTQVVSPLGDKLETAIKKKQNYIQSVELIRRYNDFYSMGKSDIVEQLRLSKNW 189
Query: 202 SRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLL 261
++ + + L + S + ++ + + + +++Y +EN LL
Sbjct: 190 KLNLKSVKLMKNL------------LILSSKLETSSIPKTINTKLV-IEKYSEMMENELL 236
Query: 262 SRFDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVMNADT-------- 313
F++A ++ T + E A IL+ FN G + +Q ++ FID + ++ +
Sbjct: 237 ENFNSAYRENNFTKLNEIAIILNNFNGGVNVIQSFINQHDYFIDTKQIDLENEFENVFIK 296
Query: 314 RLVLGDQAAQSSPNNV--ARGLSSLYKEITDTVRKEAATITAVF-PSPNEVMSILVQRVL 370
+ +Q ++V + +L ++ ++ E+ + VF V+ + +QRV
Sbjct: 297 NVKFKEQLIDFENHSVIIETSMQNLINDVETVIKNESKIVKRVFEEKATHVIQLFIQRVF 356
Query: 371 EQRVTALLDKLL 382
Q++ + LL
Sbjct: 357 AQKIEPRFEVLL 368
Score = 68.6 bits (166), Expect = 7e-11
Identities = 49/164 (29%), Positives = 81/164 (48%), Gaps = 6/164 (3%)
Query: 647 LQQCIETVMAEVERLLSAEQKATDYKSPDDGMAPDHRPTNACTRVVAYLSRVLESAFTAL 706
LQ+ I + + +L ++K + + D P +V L+ + E + L
Sbjct: 697 LQETITVISTKFSAILCKQKKKDFVPKSQELLDQDTLPA---IEIVNILNLIFEQSSKFL 753
Query: 707 EGLNKQAFLSELGNRLHKVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKF 766
+G N Q FL+ +G L+ +LL+H+ + N GG+ + +DI Y + + S+ +KF
Sbjct: 754 KGKNLQTFLTLIGEELYGLLLSHYSHFQVNSIGGVVVTKDIIGYQTAIEDWGVASLIDKF 813
Query: 767 ELLGILANVFIVAPESLSTLF-EG-TPSIRKD-AQRFIQLREDY 807
L LAN+F V PE L +L EG I +D Q +I RED+
Sbjct: 814 ATLRELANLFTVQPELLESLTKEGHLADIGRDIIQSYISNREDF 857
>POF6_SCHPO (O74854) F-box protein pof6
Length = 872
Score = 49.7 bits (117), Expect = 3e-05
Identities = 42/175 (24%), Positives = 75/175 (42%), Gaps = 2/175 (1%)
Query: 605 IIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEVERLLSA 664
++ +F+ +S + + D A E+ E AA+ GL + I ++ + LL
Sbjct: 643 MMDSFFNEEMSPICVKDDMFDPAIAEKKKFEQLLDERAAF-GLHKGINVLIEHADFLLET 701
Query: 665 EQKATDYKSPDDGMAPDH-RPTNACTRVVAYLSRVLESAFTALEGLNKQAFLSELGNRLH 723
+ + G + PT A VV +L + + F E+G RL
Sbjct: 702 KTPMNLFSDQTIGSITNTIEPTAAAKNVVQFLGFHMRILVGRADHEILDVFYKEVGMRLF 761
Query: 724 KVLLNHWQKYTFNPSGGLRLKRDITEYGEFVRSFNAPSVDEKFELLGILANVFIV 778
L + + + F+ GGL+L D Y EF+ S + S+ F+ L +A++FI+
Sbjct: 762 DSLTRYIKSHKFSVDGGLKLLSDCNLYYEFIHSLHQSSLLPYFKTLKEIAHLFII 816
>PLE1_CRIGR (Q9JI55) Plectin 1 (PLTN) (PCN) (300-kDa intermediate
filament-associated protein) (IFAP300) (Fragment)
Length = 4473
Score = 44.7 bits (104), Expect = 0.001
Identities = 111/531 (20%), Positives = 207/531 (38%), Gaps = 74/531 (13%)
Query: 60 EAEGADAVQNKYSQVATSPLFP--EVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSV 117
+ + ADA K+ + A L +VE+ L+ + +E + +D L LK +V+
Sbjct: 2085 QKQAADAEMEKHKKFAEQTLRQKAQVEQELTTLRLQLEETDHQKSILDEELQRLKAEVT- 2143
Query: 118 QDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIEL 177
+ ++ R + E V +L +RI + + L+ D + + E
Sbjct: 2144 EAARQRSQVEEELFSVRVQMEELGKLKARIEAENRALI-----LRDKDNTQRFLEEEAEK 2198
Query: 178 IKYLMEFNSSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNAT 237
+K + E + +R++ AA A +LR AEED+ + A
Sbjct: 2199 MKQVAE----------------EAARLSVAAQEAARLRQLAEEDLAQQ---------RAL 2233
Query: 238 ASRGLEVAVANLQEYCN-ELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHY 296
A + L+ + +QE + E LL + +Q++ + ++ Q T Q
Sbjct: 2234 AEKMLKEKMQAVQEATRLKAEAELLQQQKELAQEQARRLQEDKEQMAQQLVEETQGFQRT 2293
Query: 297 V-ATRPMFIDVEVMNADTRLVLGDQA-AQSSPNNVARGLSSLYKEITDTV-RKEAATITA 353
+ R +++ +L + + + AQ+ A+ +EI + + R E AT
Sbjct: 2294 LEVERQRQLEMSAEAERLKLRMAEMSRAQARAEEDAQRFRKQAEEIGEKLHRTELATQEK 2353
Query: 354 VFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQ 413
V LVQ + QR + D ++ ++ L E+ L ++L + E+ Q
Sbjct: 2354 V---------TLVQTLEIQRQQSDHDAERLREAIAEL-EREKEKLKQEAKLLQLKSEEMQ 2403
Query: 414 EIARDLRTVGCGDLDVEGLTESLFSSHKDEYPEYEQASLRQLYK---VKMEELRAESQIS 470
+ ++ L L+E ++ + E E+A L QL++ K ++LR E Q
Sbjct: 2404 TVQQEQILQETQALQKSFLSEKDSLLQRERFIEQEKAKLEQLFQDEVAKAQQLREEQQRQ 2463
Query: 471 DSSGTIGRSKGATVASSQQ-QISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFT 529
+ + K VAS ++ + E VR +E + L Q
Sbjct: 2464 QRQ--MEQEKQELVASMEEARRRQCEAEEAVRRKQEELQHLELQRQQQEK---------- 2511
Query: 530 CLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEA 580
LL + +Q + E L+R L E + S +AA A+AA+A
Sbjct: 2512 -LLAEENQRLRERLQR----------LEEEHRAALAHSEEIAATQAAAAKA 2551
>MYS_PODCA (Q05000) Myosin heavy chain (Fragment)
Length = 692
Score = 44.7 bits (104), Expect = 0.001
Identities = 65/299 (21%), Positives = 119/299 (39%), Gaps = 44/299 (14%)
Query: 96 ELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDG--------------LFASFA 141
E +LR+ D +L +++ H+R + L+ +D A
Sbjct: 303 EYTQLRQSSDRKLSEKDEELEGLRKNHQRQMESLQNTIDSESRSKAEQQKLRKKYDADMM 362
Query: 142 RLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFS-- 199
L+S++ S + AA+ ++ AQ I+ ++ +++ S D M S S
Sbjct: 363 ELESQLESSNRVAAESQKQMKKLQAQ-------IKELQSMIDDESRGRDDMRDSASRSER 415
Query: 200 ---------DDSRVA-EAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANL 249
D++RVA E A A+KL E+ + A+ N A+ E +L
Sbjct: 416 RANDLAVQLDEARVALEQAERARKLAE-NEKSENSDRVAELQALYNNVANAKAEGDYHSL 474
Query: 250 QEYCNELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHYVATRPMFIDVEVM 309
QE +LEN AS+ + MAE A+++S+ N A +R + + +V
Sbjct: 475 QEEIEDLENEA-----KASEDKAQRAMAEVARLMSELNSAQEATSTAEKSRQL-VSKQVA 528
Query: 310 NADTRL----VLGDQAAQSSPNNVARGLSSLYKEITDTVRKEAATITAVFPSPNEVMSI 364
+ +RL G + ++ + + + L ++ RK A I A S +V +
Sbjct: 529 DLQSRLEDAEAQGGKGLKNQLRKLEQRIMELESDVDTEARKGADAIKAARKSEKKVKEL 587
>PLE1_HUMAN (Q15149) Plectin 1 (PLTN) (PCN) (Hemidesmosomal protein 1)
(HD1)
Length = 4684
Score = 44.3 bits (103), Expect = 0.001
Identities = 134/672 (19%), Positives = 248/672 (35%), Gaps = 101/672 (15%)
Query: 60 EAEGADAVQNKYSQVATSPLFP--EVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSV 117
+ + ADA K+ + A L +VE+ L+ + +E + +D L LK + +
Sbjct: 2296 QKQAADAEMEKHKKFAEQTLRQKAQVEQELTTLRLQLEETDHQKNLLDEELQRLKAEAT- 2354
Query: 118 QDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIEL 177
+ ++ R + E V ++L +RI + + L+ D + + E
Sbjct: 2355 EAARQRSQVEEELFSVRVQMEELSKLKARIEAENRALI-----LRDKDNTQRFLQEEAEK 2409
Query: 178 IKYLMEFNSSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNAT 237
+K + E + +R++ AA A +LR AEED+ + A
Sbjct: 2410 MKQVAE----------------EAARLSVAAQEAARLRQLAEEDLAQQ---------RAL 2444
Query: 238 ASRGLEVAVANLQEYCN-ELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHY 296
A + L+ + +QE + E LL + +Q++ + ++ Q T Q
Sbjct: 2445 AEKMLKEKMQAVQEATRLKAEAELLQQQKELAQEQARRLQEDKEQMAQQLAEETQGFQRT 2504
Query: 297 V-ATRPMFIDVEVMNADTRLVLGDQA-AQSSPNNVARGLSSLYKEITDTV-RKEAATITA 353
+ A R +++ +L + + + AQ+ A+ +EI + + R E AT
Sbjct: 2505 LEAERQRQLEMSAEAERLKLRVAEMSRAQARAEEDAQRFRKQAEEIGEKLHRTELATQEK 2564
Query: 354 VFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQ 413
V LVQ + QR + D ++ ++ L E+ L ++L + E+ Q
Sbjct: 2565 V---------TLVQTLEIQRQQSDHDAERLREAIAEL-EREKEKLQQEAKLLQLKSEEMQ 2614
Query: 414 EIARDLRTVGCGDLDVEGLTESLFSSHKDEYPEYEQASLRQLYK---VKMEELRAESQ-- 468
+ ++ L L+E ++ + E E+A L QL++ K ++LR E Q
Sbjct: 2615 TVQQEQLLQETQALQQSFLSEKDSLLQRERFIEQEKAKLEQLFQDEVAKAQQLREEQQRQ 2674
Query: 469 -----------ISDSSGTIGRSKGATVASSQQQISVTVVTEFVRWNEEAITRCN-LFSSQ 516
++ R A ++Q + + + R EE + N Q
Sbjct: 2675 QQQMEQERQRLVASMEEARRRQHEAEEGVRRKQEELQQLEQQRRQQEELLAEENQRLREQ 2734
Query: 517 PSTLATLVKAVFTCLLDQVSQYIA--EGLERARDGL-------------------TEAAN 555
L +A + + +A + L RD L A
Sbjct: 2735 LQLLEEQHRAALAHSEEVTASQVAATKTLPNGRDALDGPAAEAEPEHSFDGLRRKVSAQR 2794
Query: 556 LRERFVLGTSVSRRVAAAAASAAEAAA-------AAGESSFRSFMVAVQRSGSSV--AII 606
L+E +L +R+A + E A G SS ++ SV A+
Sbjct: 2795 LQEAGILSAEELQRLAQGHTTVDELARREDVRHYLQGRSSIAGLLLKATNEKLSVYAALQ 2854
Query: 607 QQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCIETVMAEV-ERLLSAE 665
+Q S + +LL A + + + A +G+ V E+ +LLSAE
Sbjct: 2855 RQLLSPGTALILLEAQAASGFLLDPVRNRRLTVNEAVKEGV------VGPELHHKLLSAE 2908
Query: 666 QKATDYKSPDDG 677
+ T YK P G
Sbjct: 2909 RAVTGYKDPYTG 2920
>PLE1_RAT (P30427) Plectin 1 (PLTN) (PCN)
Length = 4687
Score = 43.5 bits (101), Expect = 0.002
Identities = 110/531 (20%), Positives = 208/531 (38%), Gaps = 74/531 (13%)
Query: 60 EAEGADAVQNKYSQVATSPLFP--EVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSV 117
+ + ADA K+ + A L +VE+ L+ + +E + +D L LK +V+
Sbjct: 2299 QKQAADAEMEKHKKFAEQTLRQKAQVEQELTTLRLQLEETDHQKSILDEELQRLKAEVT- 2357
Query: 118 QDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIEL 177
+ ++ R + E V +L +RI + + L+ D + + E
Sbjct: 2358 EAARQRSQVEEELFSVRVQMEELGKLKARIEAENRALI-----LRDKDNTQRFLEEEAEK 2412
Query: 178 IKYLMEFNSSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAVGNAT 237
+K + E + +R++ AA A +LR AEED+ + A
Sbjct: 2413 MKQVAE----------------EAARLSVAAQEAARLRQLAEEDLAQQ---------RAL 2447
Query: 238 ASRGLEVAVANLQEYCN-ELENRLLSRFDAASQKRELTTMAECAKILSQFNRGTSAMQHY 296
A + L+ + +QE + E LL + +Q++ A+ ++ Q T Q
Sbjct: 2448 AEKMLKEKMQAVQEATRLKAEAELLQQQKELAQEQARRLQADKEQMAQQLVEETQGFQRT 2507
Query: 297 V-ATRPMFIDVEVMNADTRLVLGDQA-AQSSPNNVARGLSSLYKEITDTV-RKEAATITA 353
+ A R +++ +L + + + AQ+ A+ +EI + + R E AT
Sbjct: 2508 LEAERQRQLEMSAEAERLKLRMAEMSRAQARAEEDAQRFRKQAEEIGEKLHRTELATQEK 2567
Query: 354 VFPSPNEVMSILVQRVLEQRVTALLDKLLVKPSLVNLPSMEEGGLLFYLRMLAVSYEKTQ 413
V LVQ + QR + D ++ ++ L E+ L ++L + E+ Q
Sbjct: 2568 V---------TLVQTLEIQRQQSDQDAERLREAIAEL-EREKEKLKQEAKLLQLKSEEMQ 2617
Query: 414 EIARDLRTVGCGDLDVEGLTESLFSSHKDEYPEYEQASLRQLYK---VKMEELRAESQIS 470
+ ++ L L+E ++ + E E+A L QL++ K ++L+ E Q
Sbjct: 2618 TVQQEQILQETQALQKSFLSEKDSLLQRERFIEQEKAKLEQLFQDEVAKAKQLQEEQQRQ 2677
Query: 471 DSSGTIGRSKGATVASSQQ-QISVTVVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFT 529
+ + K VAS ++ + E VR +E + R Q
Sbjct: 2678 QQQ--MEQEKQELVASMEEARRRQREAEEGVRRKQEELQRLEQQRQQQEK---------- 2725
Query: 530 CLLDQVSQYIAEGLERARDGLTEAANLRERFVLGTSVSRRVAAAAASAAEA 580
LL + +Q + E L+R L E + S +A + A+A +A
Sbjct: 2726 -LLAEENQRLRERLQR----------LEEEHRAALAHSEEIATSQAAATKA 2765
>MYHB_RABIT (P35748) Myosin heavy chain, smooth muscle isoform (SMMHC)
Length = 1972
Score = 40.8 bits (94), Expect = 0.015
Identities = 49/231 (21%), Positives = 101/231 (43%), Gaps = 25/231 (10%)
Query: 3 EPRDATKTDLKTAKSASSAASFPL---ILDIDDFKGDFSFDA-LFGNLVNELLPSFKLED 58
E +A + L+ K + A L IL +DD S + L +++L + E+
Sbjct: 960 EEEEAARQKLQLEKVTAEAKIKKLEDDILVMDDQNNKLSKERKLLEERISDLTTNLAEEE 1019
Query: 59 LEAEGADAVQNKYSQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQ 118
+A+ ++NK+ + + E+E L + S +EL +L++++DG +L + ++
Sbjct: 1020 EKAKNLTKLKNKHESMIS-----ELEVRLKKEEKSRQELEKLKRKMDGEASDLHEQIADL 1074
Query: 119 DSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAK-------IGDHLQSADAQRETA 171
++ +L K + L A+ ARL+ S K I D + D++R
Sbjct: 1075 QAQIAELKMQLAKKEEELQAALARLEDETSQKNNALKKIRELEGHISDLQEDLDSERAAR 1134
Query: 172 SQTIELIKYLMEFNSSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDI 222
++ + + L E EL L ++ + + Q+LR+ E+++
Sbjct: 1135 NKAEKQKRDLGE---------ELEALKTELEDTLDTTATQQELRAKREQEV 1176
Score = 32.0 bits (71), Expect = 6.8
Identities = 42/195 (21%), Positives = 83/195 (42%), Gaps = 22/195 (11%)
Query: 82 EVEKLLSLFKDSCKELLELRKQIDGRLHNLKKD---VSVQDSKHRRTLAELEKGVDGLFA 138
E+E+ + K ++L+ + + +H L+K + Q + + L ELE +
Sbjct: 1501 ELERTNKMLKAEMEDLVSSKDDVGKNVHELEKSKRALETQMEEMKTQLEELEDELQATED 1560
Query: 139 SFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLF 198
+ RL+ + ++ + LQ+ D Q E + +L + L E+ +
Sbjct: 1561 AKLRLEVNMQAL---KVQFERDLQARDEQNEEKRR--QLQRQLHEYETE----------- 1604
Query: 199 SDDSRVAEAASIAQKLRSFAEEDIGRHGITAPSAV-GNATASRGLEVAVANLQEYCNELE 257
+D R A + A K + E D+ + A SA+ G A + L A ++++ ELE
Sbjct: 1605 LEDERKQRALAAAAKKK--LEGDLKDLELQADSAIKGREEAIKQLLKLQAQMKDFQRELE 1662
Query: 258 NRLLSRFDAASQKRE 272
+ SR + + +E
Sbjct: 1663 DARASRDEIFATAKE 1677
>MYHB_CHICK (P10587) Myosin heavy chain, gizzard smooth muscle
Length = 1978
Score = 40.4 bits (93), Expect = 0.019
Identities = 47/211 (22%), Positives = 93/211 (43%), Gaps = 14/211 (6%)
Query: 91 KDSCKELLELRK-QIDGRLHNLKKDVSVQDSKHRRTLAE---LEKGVDGLFASFARLDSR 146
+++ ++ L+L K DG++ ++ D+ + + ++ + E LE+ V L + A + +
Sbjct: 967 EEAARQKLQLEKVTADGKIKKMEDDILIMEDQNNKLTKERKLLEERVSDLTTNLAEEEEK 1026
Query: 147 ISSVGQTAAKIGDHLQSADAQ---RETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
++ + K + + + E + Q +E IK +E SS DL E + ++
Sbjct: 1027 AKNLTKLKNKHESMISELEVRLKKEEKSRQELEKIKRKLEGESS--DLHE--QIAELQAQ 1082
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
+AE + K + + R R LE +++LQE +LE+ +R
Sbjct: 1083 IAELKAQLAKKEEELQAALARLEDETSQKNNALKKIRELESHISDLQE---DLESEKAAR 1139
Query: 264 FDAASQKRELTTMAECAKILSQFNRGTSAMQ 294
A QKR+L+ E K + T+A Q
Sbjct: 1140 NKAEKQKRDLSEELEALKTELEDTLDTTATQ 1170
>NAF1_HUMAN (Q15025) Nef-associated factor 1 (Naf1) (HIV-1 Nef
interacting protein) (Virion-associated nuclear
shuttling protein) (VAN) (hVAN) (TNFAIP3 interacting
protein 1)
Length = 636
Score = 39.3 bits (90), Expect = 0.043
Identities = 58/222 (26%), Positives = 92/222 (41%), Gaps = 25/222 (11%)
Query: 59 LEAEGADAVQNKYSQVATSPLFPEV------EKLLSLFKDSCKELLELRKQIDGRLHNLK 112
+E G AV + T+ PEV EK + + + ELLE+ KQ D ++K
Sbjct: 269 MEGTGKKAVAGQQQASVTAGKVPEVVALGAAEKKVKMLEQQRSELLEVNKQWDQHFRSMK 328
Query: 113 KDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRET-- 170
+ + ++ R+ LA+L+K V L A R AK ++ D ++ T
Sbjct: 329 QQYEQKITELRQKLADLQKQVTDLEAE-REQKQRDFDRKLLLAKSKIEMEETDKEQLTAE 387
Query: 171 ASQTIELIKYLMEFNSSPGDLMELSPLFSDDSRVAEAASIAQKLRSFAEEDIGRHGITAP 230
A + + +KYL + +LSPL R + I Q+L EE + T P
Sbjct: 388 AKELRQKVKYLQD---------QLSPL--TRQREYQEKEI-QRLNKALEEALSIQ--TPP 433
Query: 231 SAVGNATASRGLEVAVANLQEYC--NELENRLLSRFDAASQK 270
S+ A S A+ QE NEL + + F+ Q+
Sbjct: 434 SSPPTAFGSPEGAGALLRKQELVTQNELLKQQVKIFEEDFQR 475
>TBC2_CHLRE (Q8VXP3) Tbc2 translation factor, chloroplast precursor
Length = 1115
Score = 38.5 bits (88), Expect = 0.073
Identities = 30/105 (28%), Positives = 49/105 (46%), Gaps = 11/105 (10%)
Query: 571 AAAAASAAEAAAAAGESSFRSFMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACE 630
AAAA + A +AAAAG+S + + AV + A++ + S +D AA
Sbjct: 931 AAAAPAGASSAAAAGDSP--AALSAVPAAAGDGALVPSFMS---------IDDDGTAAVA 979
Query: 631 EMATAMSSAEAAAYKGLQQCIETVMAEVERLLSAEQKATDYKSPD 675
ATA+++AE AA+ T +A + L + +A P+
Sbjct: 980 AAATALAAAEPAAHAATSTTTATAVAHPQPQLLPQAQALPQPGPE 1024
>YAF3_SCHPO (Q09857) Hypothetical protein C29E6.03c in chromosome I
Length = 1044
Score = 37.4 bits (85), Expect = 0.16
Identities = 37/159 (23%), Positives = 73/159 (45%), Gaps = 10/159 (6%)
Query: 67 VQNKYSQVATSPLFPEVEKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTL 126
++ Y++ + E++ L + K+ +E ++ +L ++K + SK+R L
Sbjct: 673 IELDYTKSNCKQMEEEMQVLREGHESEIKDFIEEHSKLTKQLDDIKNQFGIISSKNRDLL 732
Query: 127 AELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSADAQRETASQTI---ELIKYLME 183
+ELEK L S A L+S+ + + + L +A E+ TI EL K +
Sbjct: 733 SELEKS-KSLNNSLAALESKNKKLENDLNLLTEKLNKKNADTESFKNTIREAELSKKALN 791
Query: 184 FNSSPGDLMELSPLFSD-DSRVAEAASIAQKLRSFAEED 221
N L + SD ++++E ++ Q+L+S +D
Sbjct: 792 DN-----LGNKENIISDLKNKLSEESTRLQELQSQLNQD 825
>FRZC_MYXXA (P43500) Frizzy aggregation protein frzCD
Length = 417
Score = 37.4 bits (85), Expect = 0.16
Identities = 55/248 (22%), Positives = 107/248 (42%), Gaps = 28/248 (11%)
Query: 436 LFSSHKDEYPEYEQASLRQLYKVKMEELR-AESQISDSSGTIGRSKGATVASSQQQISVT 494
L +S + E EQA+ MEEL+ A +QI++++G++ R T+ +++
Sbjct: 156 LAASTQHETSSTEQAAAIHETTATMEELKHASAQIAENAGSVARVAEETLGAARAGRGA- 214
Query: 495 VVTEFVRWNEEAITRCNLFSSQPSTLATLVKAVFTCL--LDQVSQY-----IAEGLERAR 547
+ EF++ ++ + + + L+ V+ + T + +D+++ + LE +R
Sbjct: 215 -IGEFIQAMQQIRSDGVAVADSIAKLSKRVERIGTVVEVIDEIADRSDLLALNAALEGSR 273
Query: 548 DG---------LTEAANLRERFVLGT-------SVSRRVAAAAASAAEAAAAAGESSFRS 591
G E L E + T + R AAAA AAEA+ +A ES +
Sbjct: 274 AGEAGKGFSIVAAEMRRLAENVLDSTKEIKNLITEIREATAAAAGAAEASKSATESGEKL 333
Query: 592 FMVAVQRSGSSVAIIQQYFSNSISRLLLPVDGAHAAACEEMATAMSSAEAAAYKGLQQCI 651
VA Q +A +Q+ ++ +R++ A E++ +M+ E + Q
Sbjct: 334 GAVAAQAVEGILAGVQE--TSDAARVINLATQQQRTATEQVVASMAEIEDVTRQTTQASK 391
Query: 652 ETVMAEVE 659
+ A E
Sbjct: 392 QATGAAAE 399
>VP47_BPAPS (Q9T1Q1) Putative protein p47
Length = 190
Score = 35.4 bits (80), Expect = 0.62
Identities = 27/99 (27%), Positives = 54/99 (54%), Gaps = 14/99 (14%)
Query: 57 EDLEAEGADAVQNKYSQVATSPLFPEVEKLLS-----LFKDSCKELLELRKQIDGRLHNL 111
+DL AE AD ++ +++A +V K LS + KD E+ ++RK + + ++
Sbjct: 59 KDLSAEIADVRKDLSAEIA------DVRKDLSAEIADVRKDLSAEIADVRKDLSAEIADV 112
Query: 112 KKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISSV 150
+KD++++ K +A++ K + L F + D++IS V
Sbjct: 113 RKDMAIRFEKTDAQIADVRKDMVNL---FDKTDAQISLV 148
>NUMA_HUMAN (Q14980) Nuclear mitotic apparatus protein 1 (NuMA
protein) (SP-H antigen)
Length = 2115
Score = 35.4 bits (80), Expect = 0.62
Identities = 60/249 (24%), Positives = 100/249 (40%), Gaps = 35/249 (14%)
Query: 55 KLEDLEAEGADAVQNKYSQVATSPLFPEVEKL---LSLFKDSCKELLELRKQIDGRLHNL 111
K E A G+ A + T P P++E L +S + C++ E ++ L
Sbjct: 1096 KKEKEHASGSGAQSEAAGR--TEPTGPKLEALRAEVSKLEQQCQKQQEQADSLERSLEAE 1153
Query: 112 KKDVSVQDSKHRRTLAELEKGVDGLFASFARLDSRISSVGQTAAKIGDHLQSAD---AQ- 167
+ + +DS +LE+ L S + L S + K+ DH ++ D AQ
Sbjct: 1154 RASRAERDSALETLQGQLEEKAQELGHSQSALASAQRELAAFRTKVQDHSKAEDEWKAQV 1213
Query: 168 ---RETASQTIELIKYLMEFNS--------SPGDLMELSPLFSDDSRVAEAASIAQKLRS 216
R+ A + LI L E S G+ EL L +S ++ + ++LR
Sbjct: 1214 ARGRQEAERKNSLISSLEEEVSILNRQVLEKEGESKELKRLVMAESE--KSQKLEERLRL 1271
Query: 217 FAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSRFDAASQKRELTTM 276
E TA ++ A S L V +L+E + R + + ++ELT+
Sbjct: 1272 LQAE-------TASNSARAAERSSALREEVQSLREEAEK------QRVASENLRQELTSQ 1318
Query: 277 AECAKILSQ 285
AE A+ L Q
Sbjct: 1319 AERAEELGQ 1327
Score = 31.6 bits (70), Expect = 8.9
Identities = 37/189 (19%), Positives = 78/189 (40%), Gaps = 24/189 (12%)
Query: 84 EKLLSLFKDSCKELLELRKQIDGRLHNLKKDVSVQDSKHRRTLAELEKGVDGLFASFARL 143
E+L++L K+ C++ + ++ ++ ++ +Q S+ + LAEL A+ AR
Sbjct: 833 EQLMTL-KEECEKARQELQEAKEKVAGIESHSELQISRQQNELAELH-------ANLARA 884
Query: 144 DSRISSVGQTAAKIGDHLQSADAQRETASQTIELIKYLMEFNSSPGDLMELSPLFSDDSR 203
++ A K+ D L + + S+ + ++ L+ G+ E +
Sbjct: 885 LQQVQEKEVRAQKLADDLSTLQEKMAATSKEVARLETLVR---KAGEQQET----ASREL 937
Query: 204 VAEAASIAQKLRSFAEEDIGRHGITAPSAVGNATASRGLEVAVANLQEYCNELENRLLSR 263
V E A + + EE GR + +A L+ ++ NELE +
Sbjct: 938 VKEPARAGDRQPEWLEEQQGRQFCSTQAA---------LQAMEREAEQMGNELERLRAAL 988
Query: 264 FDAASQKRE 272
++ Q++E
Sbjct: 989 MESQGQQQE 997
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.315 0.129 0.348
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 81,377,639
Number of Sequences: 164201
Number of extensions: 3138862
Number of successful extensions: 12371
Number of sequences better than 10.0: 83
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 12257
Number of HSP's gapped (non-prelim): 180
length of query: 821
length of database: 59,974,054
effective HSP length: 119
effective length of query: 702
effective length of database: 40,434,135
effective search space: 28384762770
effective search space used: 28384762770
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC137821.7


