

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.6 - phase: 0 /pseudo
(1211 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
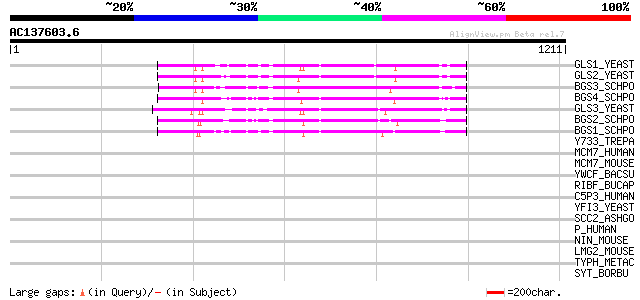
Score E
Sequences producing significant alignments: (bits) Value
GLS1_YEAST (P38631) 1,3-beta-glucan synthase component GLS1 (EC ... 329 2e-89
GLS2_YEAST (P40989) 1,3-beta-glucan synthase component GLS2 (EC ... 327 1e-88
BGS3_SCHPO (Q9P377) 1,3-beta-glucan synthase component bgs3 (EC ... 327 2e-88
BGS4_SCHPO (O74475) 1,3-beta-glucan synthase component bgs4 (EC ... 323 2e-87
GLS3_YEAST (Q04952) 1,3-beta-glucan synthase component FKS3 (EC ... 321 7e-87
BGS2_SCHPO (O13967) 1,3-beta-glucan synthase component bgs2 (EC ... 306 3e-82
BGS1_SCHPO (Q10287) 1,3-beta-glucan synthase component bgs1 (EC ... 304 1e-81
Y733_TREPA (O83715) Hypothetical protein TP0733 38 0.15
MCM7_HUMAN (P33993) DNA replication licensing factor MCM7 (CDC47... 37 0.25
MCM7_MOUSE (Q61881) DNA replication licensing factor MCM7 (CDC47... 36 0.72
YWCF_BACSU (P39604) Hypothetical protein ywcF 34 2.1
RIBF_BUCAP (Q8K9Z1) Riboflavin biosynthesis protein ribF [Includ... 33 3.6
C5P3_HUMAN (Q96JB5) CDK5 regulatory subunit-associated protein 3... 33 3.6
YFI3_YEAST (P43596) Hypothetical 90.9 kDa protein in GCN20-CMK1 ... 33 4.7
SCC2_ASHGO (Q750S2) Sister chromatid cohesion protein 2 33 6.1
P_HUMAN (Q04671) P protein (Melanocyte-specific transporter prot... 33 6.1
NIN_MOUSE (Q61043) Ninein 33 6.1
LMG2_MOUSE (Q61092) Laminin gamma-2 chain precursor (Kalinin/nic... 33 6.1
TYPH_METAC (Q8TL01) Putative thymidine phosphorylase (EC 2.4.2.4... 32 8.0
SYT_BORBU (O51662) Threonyl-tRNA synthetase (EC 6.1.1.3) (Threon... 32 8.0
>GLS1_YEAST (P38631) 1,3-beta-glucan synthase component GLS1 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
(CND1 protein) (CWN53 protein) (FKS1 protein)
(Papulacandin B sensitivity protein 1)
Length = 1876
Score = 329 bits (844), Expect = 2e-89
Identities = 239/740 (32%), Positives = 370/740 (49%), Gaps = 102/740 (13%)
Query: 322 PSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL--ESPNEDG 379
P + EA+RRISFF+ SL +P V NM +F+VLTP+Y E +L SLRE+ E
Sbjct: 810 PRDSEAERRISFFAQSLSTPIPEPLPVDNMPTFTVLTPHYAERILLSLREIIREDDQFSR 869
Query: 380 VSILFYLQKIFPDEWNNFLQRVNC---------SNEEELKEYDELEEEL----------- 419
V++L YL+++ P EW F++ NE E ++ D L+ ++
Sbjct: 870 VTLLEYLKQLHPVEWECFVKDTKILAEETAAYEGNENEAEKEDALKSQIDDLPFYCIGFK 929
Query: 420 ----------RRWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSD 469
R WAS R QTL RT+ G M Y +A++L +E + ++
Sbjct: 930 SAAPEYTLRTRIWASLRSQTLYRTISGFMNYSRAIKLLY---------RVENPEIVQMFG 980
Query: 470 DNSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYID 529
N+ G L + + +A KF ++VS Q+ K A+ +LR YP L++AY+D
Sbjct: 981 GNAEG---LERELEKMARRKFKFLVSMQRLAKFKPHELENAEFLLRA---YPDLQIAYLD 1034
Query: 530 EVEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKP 589
E ++ RI YS L+ + P+ ++++L G ILG+GK
Sbjct: 1035 EEPPLTEGEEPRI----YSALIDGHCEILDNGRRRPK-------FRVQLSGNPILGDGKS 1083
Query: 590 ENQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEFLKKH--------DGVRYP--- 638
+NQNHA++F RGE +Q ID NQDNY+EE LK+R++L EF + + G+RY
Sbjct: 1084 DNQNHALIFYRGEYIQLIDANQDNYLEECLKIRSVLAEFEELNVEQVNPYAPGLRYEEQT 1143
Query: 639 -----SILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRI 693
+I+G RE+IF+ + L + +E +F T+ R L+ + + HYGHPD +
Sbjct: 1144 TNHPVAIVGAREYIFSENSGVLGDVAAGKEQTFGTLFARTLSQ-IGGKLHYGHPDFINAT 1202
Query: 694 FHLTRGGVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKI 753
F TRGGVSKA K ++L+EDI+AG N+ LR G + H EY Q GKGRD+G I F KI
Sbjct: 1203 FMTTRGGVSKAQKGLHLNEDIYAGMNAMLRGGRIKHCEYYQCGKGRDLGFGTILNFTTKI 1262
Query: 754 ANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLS 813
G GEQ LSR+ Y LG + R L+ Y+ GF+ + L I L++ +F+ + L
Sbjct: 1263 GAGMGEQMLSREYYYLGTQLPVDRFLTFYYAHPGFHLNNLFIQ-LSLQMFMLTLVNLSSL 1321
Query: 814 GLEEGLSTQKAIRDNKPLQVALASQSF-------------VQIGFLMA-LPMLMEIGLER 859
E + + + V + +F + I F +A +P++++ +ER
Sbjct: 1322 AHESIMCIYDRNKPKTDVLVPIGCYNFQPAVDWVRRYTLSIFIVFWIAFVPIVVQELIER 1381
Query: 860 GFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADN 919
G A F L L+P+F F+ + L GGA+Y TGRGF F+
Sbjct: 1382 GLWKATQRFFCHLLSLSPMFEVFAGQIYSSALLSDLAIGGARYISTGRGFATSRIPFSIL 1441
Query: 920 YRLYSRSHFVKGIELMILLIVYQIFGN--GYRSGLSYLLITTPMWFMVGTWLYAPFLFNP 977
Y ++ S G M++L +FG +++ L + W + + ++APF+FNP
Sbjct: 1442 YSRFAGSAIYMGARSMLML----LFGTVAHWQAPLLW------FWASLSSLIFAPFVFNP 1491
Query: 978 SGFEWQKIVDDWTDWNKWIS 997
F W+ D+ D+ +W+S
Sbjct: 1492 HQFAWEDFFLDYRDYIRWLS 1511
>GLS2_YEAST (P40989) 1,3-beta-glucan synthase component GLS2 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1895
Score = 327 bits (838), Expect = 1e-88
Identities = 240/740 (32%), Positives = 370/740 (49%), Gaps = 102/740 (13%)
Query: 322 PSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL--ESPNEDG 379
P + EA+RRISFF+ SL +P V NM +F+VLTP+Y E +L SLRE+ E
Sbjct: 829 PRDSEAERRISFFAQSLSTPIPEPLPVDNMPTFTVLTPHYAERILLSLREIIREDDQFSR 888
Query: 380 VSILFYLQKIFPDEWNNFLQRVNC---------SNEEELKEYDELEEEL----------- 419
V++L YL+++ P EW+ F++ +NE+E ++ D L+ ++
Sbjct: 889 VTLLEYLKQLHPVEWDCFVKDTKILAEETAAYENNEDEPEKEDALKSQIDDLPFYCIGFK 948
Query: 420 ----------RRWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSD 469
R WAS R QTL RT+ G M Y +A++L ++ + + G N+D
Sbjct: 949 SAAPEYTLRTRIWASLRSQTLYRTISGFMNYSRAIKLLYRVENPEIVQMFGG-----NAD 1003
Query: 470 DNSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYID 529
R + + +A KF ++VS Q+ K A+ +LR YP L++AY+D
Sbjct: 1004 GLER-------ELEKMARRKFKFLVSMQRLAKFKPHELENAEFLLRA---YPDLQIAYLD 1053
Query: 530 EVEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKP 589
E ++ RI YS L+ + P+ ++++L G ILG+GK
Sbjct: 1054 EEPPLNEGEEPRI----YSALIDGHCEILENGRRRPK-------FRVQLSGNPILGDGKS 1102
Query: 590 ENQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEF--------------LKKHD-G 634
+NQNHA++F RGE +Q ID NQDNY+EE LK+R++L EF LK D
Sbjct: 1103 DNQNHALIFYRGEYIQLIDANQDNYLEECLKIRSVLAEFEELGIEQIHPYTPGLKYEDQS 1162
Query: 635 VRYP-SILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRI 693
+P +I+G RE+IF+ + L + +E +F T+ R LA + + HYGHPD +
Sbjct: 1163 TNHPVAIVGAREYIFSENSGVLGDVAAGKEQTFGTLFARTLAQ-IGGKLHYGHPDFINAT 1221
Query: 694 FHLTRGGVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKI 753
F TRGGVSKA K ++L+EDI+AG N+ LR G + H EY Q GKGRD+G I F KI
Sbjct: 1222 FMTTRGGVSKAQKGLHLNEDIYAGMNAVLRGGRIKHCEYYQCGKGRDLGFGTILNFTTKI 1281
Query: 754 ANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLS 813
G GEQ LSR+ Y LG + R L+ Y+ GF+ + L I L++ +F+ + L
Sbjct: 1282 GAGMGEQMLSREYYYLGTQLPIDRFLTFYYAHPGFHLNNLFIQ-LSLQMFMLTLVNLHAL 1340
Query: 814 GLEEGLSTQKAIRDNKPLQVALASQSF-------------VQIGFLMA-LPMLMEIGLER 859
E L + + + +F + I F +A +P++++ +ER
Sbjct: 1341 AHESILCVYDRDKPITDVLYPIGCYNFHPAIDWVRRYTLSIFIVFWIAFVPIVVQELIER 1400
Query: 860 GFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADN 919
G A F L L+P+F F+ + + GGA+Y TGRGF F+
Sbjct: 1401 GLWKATQRFFRHILSLSPMFEVFAGQIYSSALLSDIAVGGARYISTGRGFATSRIPFSIL 1460
Query: 920 YRLYSRSHFVKGIELMILLIVYQIFGN--GYRSGLSYLLITTPMWFMVGTWLYAPFLFNP 977
Y ++ S G M++L +FG +++ L + W + ++APF+FNP
Sbjct: 1461 YSRFAGSAIYMGSRSMLML----LFGTVAHWQAPLLW------FWASLSALIFAPFIFNP 1510
Query: 978 SGFEWQKIVDDWTDWNKWIS 997
F W+ D+ D+ +W+S
Sbjct: 1511 HQFAWEDFFLDYRDYIRWLS 1530
>BGS3_SCHPO (Q9P377) 1,3-beta-glucan synthase component bgs3 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1826
Score = 327 bits (837), Expect = 2e-88
Identities = 239/731 (32%), Positives = 365/731 (49%), Gaps = 94/731 (12%)
Query: 324 NLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL--ESPNEDGVS 381
N EA+RRISFF+ SL +P A V M SF+VL P+Y E++L SLRE+ E ++
Sbjct: 788 NSEAERRISFFAQSLGGKIPDAVPVPKMPSFTVLIPHYGEKILLSLREIIREQDPMSRIT 847
Query: 382 ILFYLQKIFPDEWNNFLQRVNCS-------------NEEELKEYDELEEEL--------- 419
+L YL++++P++W+NF+Q E+ K+ ++E+L
Sbjct: 848 LLEYLKQLYPNDWDNFVQDTKLMAGDVGVEETKSDVKSEKGKKQGTVKEDLPFYCIGFKS 907
Query: 420 ---------RRWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSDD 470
R WAS R QTL RT GMM Y +AL+L L + +L+ DD
Sbjct: 908 TAPEYTLRTRIWASLRSQTLYRTASGMMNYSRALKL---LYRVEQPNLL---------DD 955
Query: 471 NSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYIDE 530
L Q + +A KF +S Q+Y R A+ +LR +P L++AY+D+
Sbjct: 956 CDGNFERLEHQLEQMAYRKFRLCISMQRYAKFNRDEYENAEFLLR---AHPELQIAYLDQ 1012
Query: 531 VEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKPE 590
E P KVY + + P + + Y+I+L G ILG+GK +
Sbjct: 1013 DPSEDGEEP----KVYATLINGFCPFENGRRLPK---------YRIRLSGNPILGDGKAD 1059
Query: 591 NQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEF----------LKKHDGVRYP-S 639
NQN A+ F RGE LQ ID NQDNY+EE +K+RN+L EF K R+P +
Sbjct: 1060 NQNMALPFVRGEYLQLIDANQDNYIEECMKIRNVLSEFEEMDCATLTPYTKKGNARHPVA 1119
Query: 640 ILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLTRG 699
+LG RE++F+ + L + +E +F T+ R LA + + HYGHPD + IF TRG
Sbjct: 1120 MLGAREYVFSENSGILGDVAAGKEQTFGTLFSRSLA-LIGGKLHYGHPDFLNTIFMTTRG 1178
Query: 700 GVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGE 759
GVSKA K ++++EDI+AG + R G + H +Y Q GKGRD+G I F KI G GE
Sbjct: 1179 GVSKAQKGLHVNEDIYAGMTALQRGGRIKHCDYFQCGKGRDLGFGTIINFTTKIGTGMGE 1238
Query: 760 QTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQITV---LTVYVFL-YGRLY--LVLS 813
Q+LSR+ + LG + FFRMLS Y+ GF+ + + I + L + VF+ G +Y + +
Sbjct: 1239 QSLSREYFYLGTQLPFFRMLSFYYAHAGFHLNNVFIMISMQLLMLVFVNLGAMYHTVEIC 1298
Query: 814 GLEEGLSTQKAIRDN-----KPL--QVALASQSFVQIGFLMALPMLMEIGLERGFRTALS 866
+ G + ++ KP+ + S + F+ LP+++ LE+G A++
Sbjct: 1299 DYQAGAAINASLYPPGCYMLKPVLDWIRRCIISIFIVFFISFLPLVVHDLLEKGVIRAVA 1358
Query: 867 EFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADNYRLYSRS 926
L+P+F F + L +GGA+Y TGRG F+ Y LY+ S
Sbjct: 1359 RLCKQIFSLSPMFEVFVTQNYANSIFTNLTYGGARYIATGRGLATTRVPFSVLYSLYTGS 1418
Query: 927 HFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKIV 986
G L+++L +FG Y+ M+ +V PF++NP F +
Sbjct: 1419 SIYLGSRLIMML----LFGTMTVWTTHYVYFWVTMFALV----ICPFIYNPHQFSFVDFF 1470
Query: 987 DDWTDWNKWIS 997
D+ ++ +W+S
Sbjct: 1471 VDYREFLRWLS 1481
>BGS4_SCHPO (O74475) 1,3-beta-glucan synthase component bgs4 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1955
Score = 323 bits (828), Expect = 2e-87
Identities = 239/735 (32%), Positives = 363/735 (48%), Gaps = 94/735 (12%)
Query: 322 PSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL--ESPNEDG 379
P+N EA+RR+SFF+ SL +P V NM +F+VL P+Y E++L SLRE+ E
Sbjct: 874 PANSEAERRLSFFAQSLATPIPEPVPVDNMPTFTVLIPHYAEKILLSLREIIREEDQLSR 933
Query: 380 VSILFYLQKIFPDEWNNFLQRVNCSNEEELK-EYDELEEE-------------------- 418
V++L YL+++ P EW+ F++ EE E D + E+
Sbjct: 934 VTLLEYLKQLHPVEWDCFVKDTKILVEENAPYENDSVSEKEGTYKSKVDDLPFYCIGFKS 993
Query: 419 --------LRRWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSDD 470
R WAS R QTL RT+ G M Y +A++L ++ + + G N+D
Sbjct: 994 AMPEYTLRTRIWASLRSQTLYRTISGFMNYSRAIKLLYRVENPEIVQMFGG-----NTD- 1047
Query: 471 NSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYIDE 530
R ER L +A KF VVS Q+Y + A+ +LR YP L++AY+DE
Sbjct: 1048 --RLEREL----DRMARRKFKLVVSMQRYAKFTKEEYENAEFLLRA---YPDLQIAYLDE 1098
Query: 531 VEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKPE 590
+P +E + ++ L+ + P+ Y+I+L G ILG+GK +
Sbjct: 1099 --DPPEE--EGAEPQLFAALIDGHSEIMENERRRPK-------YRIRLSGNPILGDGKSD 1147
Query: 591 NQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEFLKKH-DGV-----------RYP 638
NQN ++ F RGE +Q ID NQDNY+EE LK+R++L EF + D V ++P
Sbjct: 1148 NQNMSLPFYRGEYIQLIDANQDNYLEECLKIRSVLAEFEEMETDNVNPYSESARERNKHP 1207
Query: 639 -SILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLT 697
+ILG RE+IF+ ++ L + +E +F T+ R LA + + HYGHPD + IF T
Sbjct: 1208 VAILGAREYIFSENIGILGDVAAGKEQTFGTLFSRTLAQ-IGGKLHYGHPDFLNGIFMTT 1266
Query: 698 RGGVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGN 757
RGGVSKA K ++++EDI+AG N+ LR G + H EY Q GKGRD+G I F K+ G
Sbjct: 1267 RGGVSKAQKGLHVNEDIYAGMNAMLRGGRIKHCEYFQCGKGRDLGFGSILNFNTKVGTGM 1326
Query: 758 GEQTLSRDVYRLGHRFDFFRMLSCYFTTIGFY----FSTLQITVLTVYVFLYGRLYLVLS 813
GEQ LSR+ Y LG + R LS YF GF+ F L + + V + G +Y V++
Sbjct: 1327 GEQMLSREYYYLGTQLQLDRFLSFYFAHPGFHLNNMFIMLSVQLFMVVLINLGAIYHVVT 1386
Query: 814 ----GLEEGLSTQKAIRDNKPLQVALASQ-------SFVQIGFLMALPMLMEIGLERGFR 862
+ LS +I Q+ S + ++ +P+ + +ERG
Sbjct: 1387 VCYYNGNQKLSYDTSIVPRGCYQLGPVLSWLKRCVISIFIVFWISFIPLTVHELIERGVW 1446
Query: 863 TALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADNYRL 922
A F +P+F F+ + L +GGA+Y TGRGF F+ Y
Sbjct: 1447 RATKRFFKQIGSFSPLFEVFTCQVYSQAITSDLAYGGARYIGTGRGFATARLPFSILYSR 1506
Query: 923 YSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEW 982
++ G +++L +FG +++L+ W + APFLFNP F+W
Sbjct: 1507 FAVPSIYIGARFLMML----LFGT-MTVWVAHLIY---WWVSIMALCVAPFLFNPHQFDW 1558
Query: 983 QKIVDDWTDWNKWIS 997
D+ ++ +W+S
Sbjct: 1559 NDFFVDYREFIRWLS 1573
>GLS3_YEAST (Q04952) 1,3-beta-glucan synthase component FKS3 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1785
Score = 321 bits (823), Expect = 7e-87
Identities = 245/778 (31%), Positives = 372/778 (47%), Gaps = 135/778 (17%)
Query: 313 TTKESAMDVPSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL 372
+T +S PSN EAKRRISFF+ SL + V M +F+VL P+Y+E++L L+E+
Sbjct: 687 STFKSMEFFPSNSEAKRRISFFAQSLATPISEPVPVDCMPTFTVLVPHYSEKILLGLKEI 746
Query: 373 ---ESPNEDGVSILFYLQKIFPDEWN-------------NFLQRVNCSNEEELKEY---- 412
ESP +++L YL+ + P EW +FL+ S++E+ E
Sbjct: 747 IREESPKSK-ITVLEYLKHLHPTEWECFVKDTKLLSMEKSFLKEAESSHDEDRLEIPDAL 805
Query: 413 ---------------------DELEEEL---------------------RRWASYRGQTL 430
D ++E++ R WAS R QTL
Sbjct: 806 YDPRSSPLSDHTESRKLPTEDDLIKEKINDLPFSYFGFNSSEPSYTLRTRIWASLRTQTL 865
Query: 431 TRTVRGMMYYRKALELQAFLDMAKDEDLMEGY-KAIENSDDNSRGERSLWTQCQAVADMK 489
RT+ G M Y KA++L ++ L G +A+EN +N +A K
Sbjct: 866 YRTLSGFMNYSKAIKLLYRIENPSLVSLYRGNNEALENDLEN-------------MASRK 912
Query: 490 FSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYI-DEVEEPSKERPKRISKVYYS 548
F VV+ Q+Y + + +LR YP++ ++Y+ +E+E+ E K YYS
Sbjct: 913 FRMVVAMQRYAKFNKDEVEATELLLRA---YPNMFISYLLEELEQNESE------KTYYS 963
Query: 549 CLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKPENQNHAIMFTRGEGLQTID 608
CL +E + E L + I+KI+L G ILG+GK +NQNH+I+F RGE +Q ID
Sbjct: 964 CLTNGY------AEFDEESGLRKPIFKIRLSGNPILGDGKSDNQNHSIIFYRGEYIQVID 1017
Query: 609 MNQDNYMEEALKMRNLLQEFLKKH--------DGVRYP------SILGLREHIFTGSVSS 654
NQDNY+EE LK+R++L EF + G+ Y +I+G RE+IF+ ++
Sbjct: 1018 ANQDNYLEECLKIRSVLSEFEELELNPTIPYIPGIEYEEEPPPIAIVGSREYIFSENIGV 1077
Query: 655 LAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLTRGGVSKASKVINLSEDI 714
L + +E +F T+ R LA + + HYGHPD + IF TRGG+SKA + ++L+EDI
Sbjct: 1078 LGDIAAGKEQTFGTLFARTLAE-IGGKLHYGHPDFLNGIFMTTRGGLSKAQRGLHLNEDI 1136
Query: 715 FAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGEQTLSRDVYRLGHRFD 774
+AG N+ R G + H +Y Q GKGRD+G I F KI G GEQ LSR+ Y LG +
Sbjct: 1137 YAGMNAICRGGKIKHSDYYQCGKGRDLGFGSILNFTTKIGAGMGEQLLSREYYYLGTQLP 1196
Query: 775 FFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEE--------------GLS 820
R LS ++ GF+ + L I+ F+ L L L L L
Sbjct: 1197 MDRFLSFFYAHPGFHLNNLFISFSVQLFFV---LLLNLGALNHEIIACFYDKDAPITNLE 1253
Query: 821 TQKAIRDNKPL--QVALASQSFVQIGFLMALPMLMEIGLERGFRTALSEFILMQLQLAPV 878
T + +P V++ S + F+ P+L++ LE+G A S F+ L +AP+
Sbjct: 1254 TPVGCYNIQPALHWVSIFVLSIFIVFFIAFAPLLIQEVLEKGIWRAASRFLHHLLSMAPL 1313
Query: 879 FFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADNYRLYSRSHFVKGIELMILL 938
F F ++ L GGAKY TGRGF + F Y + G ++ +L
Sbjct: 1314 FEVFVCQVYSNSLLMDLTFGGAKYISTGRGFAITRLDFFTLYSRFVNISIYSGFQVFFML 1373
Query: 939 IVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEWQKIVDDWTDWNKWI 996
+ I + ++ L + W V + +APF+FNP F + D+ + W+
Sbjct: 1374 LFAII--SMWQPALLW------FWITVISMCFAPFIFNPHQFAFMDFFIDYKTFIHWL 1423
>BGS2_SCHPO (O13967) 1,3-beta-glucan synthase component bgs2 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
(Meiotic expression up-regulated protein 21)
Length = 1894
Score = 306 bits (783), Expect = 3e-82
Identities = 231/743 (31%), Positives = 358/743 (48%), Gaps = 108/743 (14%)
Query: 322 PSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL--ESPNEDG 379
P++ EA+RR+SFF+ SL +P V M +F+VL P+Y E++L SL+E+ E
Sbjct: 846 PAHSEAERRLSFFAQSLATPIPEPIPVDAMPTFTVLVPHYGEKILLSLKEIIREQDKLSR 905
Query: 380 VSILFYLQKIFPDEWNNFLQRVNCSNEEEL-------------------KEYDEL----- 415
V++L YL+++ +EW F++ EE+ K++D+L
Sbjct: 906 VTLLEYLKQLHANEWKCFVRDTKILAEEDALSNQDLNSQDESMKAEQLHKKFDDLPFYCI 965
Query: 416 -------EEELRR--WASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIE 466
E LR WAS R QTL RTV G M Y +A++L ++ L EG +
Sbjct: 966 GFKNATPEYTLRTRIWASLRSQTLYRTVSGFMNYSRAIKLLYRVENPDVAQLFEGQMDV- 1024
Query: 467 NSDDNSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVA 526
L + +A KF VS Q+Y + ILR YP L +A
Sbjct: 1025 -----------LEYELDRMASRKFKMCVSMQRYAKFTADEIENTEFILRA---YPDLLIA 1070
Query: 527 YIDEVEEPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGE 586
Y+DE +P KE + Y+ L+ + + +P+ Y+IKL G ILG+
Sbjct: 1071 YLDE--DPPKEG--ETTPQLYAALIDGYSELDENKKRKPK-------YRIKLSGNPILGD 1119
Query: 587 GKPENQNHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEF----LKKHDGVRYPS--- 639
GK +NQN ++ F RGE +Q ID NQDNY+EE LK+R++L EF LK +D +
Sbjct: 1120 GKSDNQNLSLPFYRGEYIQLIDANQDNYLEECLKIRSILAEFEAFDLKTNDPYAETNALY 1179
Query: 640 ------ILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRI 693
I+G RE+IF+ ++ L + +E +F T+ R +A + + HYGHPD + I
Sbjct: 1180 QNNPVAIMGAREYIFSENIGILGDVAAGKEQTFGTLFARTMAQ-IGGKLHYGHPDFLNAI 1238
Query: 694 FHLTRGGVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKI 753
+ TRGGVSKA K ++++EDI+AG + R G + H EY Q GKGRD+G I F KI
Sbjct: 1239 YMTTRGGVSKAQKGLHVNEDIYAGMTALQRGGRIKHCEYYQCGKGRDLGFGSILNFTTKI 1298
Query: 754 ANGNGEQTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLS 813
G GEQ +SR+ Y LG + F R LS Y+ GF+ + + I +L+V +F+ + + L
Sbjct: 1299 GTGMGEQMVSREYYYLGTQLPFDRFLSFYYAHPGFHINNIFI-MLSVQLFMV--VLVNLG 1355
Query: 814 GLEEGLSTQKAIRDNKPLQVALASQSFVQIG-----------------FLMALPMLMEIG 856
G+ ++ D K L V + + Q+ F+ +P+ ++
Sbjct: 1356 GMYHVVTVCDYDHDQK-LTVPMRPEGCYQLNPVVNWLKRCIISIFIVFFISFVPLTVQEL 1414
Query: 857 LERGFRTALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKF 916
ERG AL+ +P+F F+ T L GGA+Y TGRGF F
Sbjct: 1415 TERGAWRALTRLGKHFASFSPMFEVFACQTYAQSVIANLSFGGARYIGTGRGFATARLSF 1474
Query: 917 ADNYRLYSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGT--WLYAPFL 974
+ + ++ G +++L+ G + I ++F + T +PF+
Sbjct: 1475 SLLFSRFAGPSIYLGSRTLLMLLF----------GTMTVWIPHLIYFWISTLAMCISPFI 1524
Query: 975 FNPSGFEWQKIVDDWTDWNKWIS 997
FNP F W D+ ++ +W+S
Sbjct: 1525 FNPHQFSWTDFFVDYREFIRWLS 1547
>BGS1_SCHPO (Q10287) 1,3-beta-glucan synthase component bgs1 (EC
2.4.1.34) (1,3-beta-D-glucan-UDP glucosyltransferase)
Length = 1729
Score = 304 bits (778), Expect = 1e-81
Identities = 227/734 (30%), Positives = 349/734 (46%), Gaps = 98/734 (13%)
Query: 322 PSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLREL--ESPNEDG 379
P+N EA RRISFF+ SL +P + M +F+VL P+Y+E++L SLRE+ E
Sbjct: 694 PANSEAARRISFFAQSLAESIPKTSSIDAMPTFTVLVPHYSEKILLSLREIIREEDQLSR 753
Query: 380 VSILFYLQKIFPDEWNNFLQRVNCSNEEE--------------LKEYD------------ 413
V++L YL++++P EW NF+ +E K YD
Sbjct: 754 VTLLEYLKQLYPVEWRNFVDDTKLLADENDSVIGSIDNEKNGVNKAYDLPFYCVGFKSAT 813
Query: 414 -ELEEELRRWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSDDNS 472
E R WAS R QTL RT+ G Y +A++L L + +L+E + D
Sbjct: 814 PEYTLRTRIWASLRTQTLYRTINGFSNYSRAIKL---LYRTETPELVEW-----TNGDPV 865
Query: 473 RGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYIDEVE 532
R + L +A+ KF + VS Q+Y + A A+ +LR YP L++AY+DE
Sbjct: 866 RLDEEL----DLMANRKFRFCVSMQRYAKFTKEEAENAEFLLRA---YPDLQIAYMDEDP 918
Query: 533 EPSKERPKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPGPAILGEGKPENQ 592
+ + + YS L+ + P+ Y+I+L G ILG+GK +NQ
Sbjct: 919 QSRHNDERHL----YSVLIDGHCPIMENGKRRPK-------YRIRLSGNPILGDGKSDNQ 967
Query: 593 NHAIMFTRGEGLQTIDMNQDNYMEEALKMRNLLQEFLKKHDGVRYP-------------S 639
N +I + RGE +Q ID NQDNY+EE LK+R++L EF + + P +
Sbjct: 968 NMSIPYIRGEYVQMIDANQDNYLEECLKIRSILAEFEQLTPPLHSPYSVNAKAADNHPVA 1027
Query: 640 ILGLREHIFTGSVSSLAWFMSNQETSFVTIGQRLLANPLRVRFHYGHPDVFDRIFHLTRG 699
ILG RE+IF+ + L + +E +F T+ R+L+ + + HYGHPD + +F +TRG
Sbjct: 1028 ILGAREYIFSENTGMLGDVAAGKEQTFGTLFARILSL-IGGKLHYGHPDFINVLFMITRG 1086
Query: 700 GVSKASKVINLSEDIFAGFNSTLREGNVTHHEYIQVGKGRDVGLNQISMFEAKIANGNGE 759
GVSKA K ++++EDI+AG + R G + H +Y Q GKGRD+G I F KI G E
Sbjct: 1087 GVSKAQKGLHVNEDIYAGMIALQRGGRIKHCDYYQCGKGRDLGFGSILNFTTKIGTGMAE 1146
Query: 760 QTLSRDVYRLGHRFDFFRMLSCYFTTIGFYFSTL----QITVLTVYVFLYGRLYLVL--- 812
Q LSR+ + LG + F R LS ++ GF+ + + + +L + + G +Y V+
Sbjct: 1147 QMLSREYFNLGTQLPFDRFLSFFYAHAGFHVNNMVIMFSLQLLMLVIINLGAMYTVVPVC 1206
Query: 813 ----------SGLEEGLSTQKAIRDNKPLQVALASQSFVQIGFLMALPMLMEIGLERGFR 862
S EG K + + L+ + S F+ G + E+G ERG
Sbjct: 1207 RYRQFDSLTASLYPEGCYQLKPVLE--WLKRCILS-IFIVFGIAFVPLAVCELG-ERGAI 1262
Query: 863 TALSEFILMQLQLAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADNYRL 922
+ L+P+F F+ L GGA+Y T RGF F+ Y
Sbjct: 1263 RMVIRLAKQIFSLSPIFEIFTCQIYAQSLIANLTFGGARYIGTSRGFATVRVPFSLLYSR 1322
Query: 923 YSRSHFVKGIELMILLIVYQIFGNGYRSGLSYLLITTPMWFMVGTWLYAPFLFNPSGFEW 982
+S G LM +L+ + S ++L W + +PFL+NP F W
Sbjct: 1323 FSGPSLYFGSRLMYMLL--------FGSITAWLPHYIYFWITLTALCISPFLYNPHQFAW 1374
Query: 983 QKIVDDWTDWNKWI 996
D+ ++ +W+
Sbjct: 1375 TDFFVDYREFMRWL 1388
>Y733_TREPA (O83715) Hypothetical protein TP0733
Length = 219
Score = 38.1 bits (87), Expect = 0.15
Identities = 31/91 (34%), Positives = 43/91 (47%), Gaps = 10/91 (10%)
Query: 755 NGNGEQTLSRDVYRLGHRFDFFRMLS--------CYFTTIGFYFSTLQITVLTVYVFLYG 806
NGNG+ L + LG+ + F + S CY TT Y+ ++ ITV Y F G
Sbjct: 83 NGNGK-LLGGGGFHLGYEYFFTKNFSLGGQVSFECYRTTGSNYYFSVPITVNPTYTFAVG 141
Query: 807 RLYLVLSGLEEGLSTQKAIRDNKPLQVALAS 837
R + LS L GL+ Q + P +A AS
Sbjct: 142 RWRIPLS-LGVGLNIQSYLSKKAPGLIAEAS 171
>MCM7_HUMAN (P33993) DNA replication licensing factor MCM7 (CDC47
homolog) (P1.1-MCM3)
Length = 719
Score = 37.4 bits (85), Expect = 0.25
Identities = 34/119 (28%), Positives = 54/119 (44%), Gaps = 5/119 (4%)
Query: 418 ELRR--WASYRGQ-TLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSDDNSRG 474
E+RR WAS T RT+ ++ AL +D+ + ED+ E + +E S D+ G
Sbjct: 587 EMRREAWASKDATYTSARTLLAILRLSTALARLRMVDVVEKEDVNEAIRLMEMSKDSLLG 646
Query: 475 ERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYIDEVEE 533
++ + Q AD+ F+ V G R A + + R P+ A +DE EE
Sbjct: 647 DKGQTARTQRPADVIFATVRELVSGGRSVRFSEAEQRCVSRGFT--PAQFQAALDEYEE 703
>MCM7_MOUSE (Q61881) DNA replication licensing factor MCM7 (CDC47
homolog)
Length = 719
Score = 35.8 bits (81), Expect = 0.72
Identities = 34/124 (27%), Positives = 55/124 (43%), Gaps = 7/124 (5%)
Query: 412 YDELEEELR--RWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKDEDLMEGYKAIENSD 469
Y E+ E R + A+Y T RT+ ++ AL +D+ + ED+ E + +E S
Sbjct: 585 YVEMRREARASKDATY---TSARTLLAILRLSTALARLRMVDIVEKEDVNEAIRLMEMSK 641
Query: 470 DNSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSLRVAYID 529
D+ GE+ + Q AD+ F+ + G A + I R P+ A +D
Sbjct: 642 DSLLGEKGQTARTQRPADVIFATIRELVSRGRSVHFSEAEQRCISRGFT--PAQFQAALD 699
Query: 530 EVEE 533
E EE
Sbjct: 700 EYEE 703
>YWCF_BACSU (P39604) Hypothetical protein ywcF
Length = 393
Score = 34.3 bits (77), Expect = 2.1
Identities = 27/107 (25%), Positives = 52/107 (48%), Gaps = 6/107 (5%)
Query: 776 FRMLSCYFTTIGFYFSTLQITVLTVYVFLYGRLYLVLSGLEEGLSTQKAIRDNKPL---- 831
F +L T+ YF Q+ L++Y+F+ G L L++ + S I+ K
Sbjct: 51 FYLLGAVAITVLLYFDLEQLEKLSLYIFIIGILSLIILKISPE-SIAPVIKGAKSWFRIG 109
Query: 832 QVALASQSFVQIGFLMALPMLMEIGLERGFRTALSEFILMQLQLAPV 878
++ + F+++G +M L ++ +G RT L + I + L++A V
Sbjct: 110 RITIQPSEFMKVGLIMMLASVIGKANPKGVRT-LRDDIHLLLKIAGV 155
>RIBF_BUCAP (Q8K9Z1) Riboflavin biosynthesis protein ribF [Includes:
Riboflavin kinase (EC 2.7.1.26) (Flavokinase); FMN
adenylyltransferase (EC 2.7.7.2) (FAD pyrophosphorylase)
(FAD synthetase)]
Length = 312
Score = 33.5 bits (75), Expect = 3.6
Identities = 36/137 (26%), Positives = 58/137 (42%), Gaps = 10/137 (7%)
Query: 815 LEEGLSTQKAIRDNKPLQVALASQSFVQIGFLMALPMLMEIGLERGFRTALSEFILMQLQ 874
L G+ K I N V++ + V +G L L +IG + T L IL + Q
Sbjct: 3 LIRGIHNLKKINSNSA--VSIGNFDGVHLGHQKLLSNLYKIGKKNNILTVL---ILFEPQ 57
Query: 875 LAPVFFTFSLGTKTHYYGRTLLHGGAKYRPTGRGFVVFHAKFADNYRLYSRSHFVKGIEL 934
P+ F L K T + KY + + ++ KF +++ S F+K I +
Sbjct: 58 --PLEF---LNNKNSPKRLTTIQNKIKYIQSWKIDIILCIKFNESFSSLSAEKFIKNILI 112
Query: 935 MILLIVYQIFGNGYRSG 951
L I + I G+ +R G
Sbjct: 113 TKLNIKFIIIGDDFRFG 129
>C5P3_HUMAN (Q96JB5) CDK5 regulatory subunit-associated protein 3
(CDK5 activator-binding protein C53) (HSF-27 protein)
(MSTP016) (PP1553)
Length = 506
Score = 33.5 bits (75), Expect = 3.6
Identities = 41/188 (21%), Positives = 78/188 (40%), Gaps = 42/188 (22%)
Query: 336 NSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLRELESPNEDGVSILFYLQKIFPDEWN 395
N+ DMP + ++ +LS S + ++ +L L+ E+ +
Sbjct: 42 NAAIQDMPESEEIAQLLSGSYIHYFHCLRILDLLKGTEASTK------------------ 83
Query: 396 NFLQRVNCSNEEELKEYDELEEELRRWASYRGQTLTRTVRGMMYYRKALELQAFLDMAKD 455
N R + + +K++ E+ + +Y + + VR + Y +L+ Q +AK
Sbjct: 84 NIFGRYS---SQRMKDWQEIIALYEKDNTYLVELSSLLVRNVNYEIPSLKKQ----IAKC 136
Query: 456 EDLMEGYKAIENSDDNSRGERSLWTQCQA-VADMKFSYVVSCQQYGIDKRSGAARAQDIL 514
+ L + Y E +CQA A+M+ + SC+QYGI +G ++L
Sbjct: 137 QQLQQEYSRKEE-------------ECQAGAAEMREQFYHSCKQYGI---TGENVRGELL 180
Query: 515 RLMARYPS 522
L+ PS
Sbjct: 181 ALVKDLPS 188
>YFI3_YEAST (P43596) Hypothetical 90.9 kDa protein in GCN20-CMK1
intergenic region
Length = 787
Score = 33.1 bits (74), Expect = 4.7
Identities = 37/129 (28%), Positives = 53/129 (40%), Gaps = 20/129 (15%)
Query: 464 AIENSDDNSRGERSLWTQCQAVADMKFSYVVSCQQYGIDKRSGAARAQDILRLMARYPSL 523
+I+N ++G S +AD V D+ +++ D LR R P
Sbjct: 7 SIQNLQQEAQGSSSA-----QLADHDHDRVSMAMPLQTDQSVSVSQSSDNLRRSRRVPKP 61
Query: 524 RVAYIDEVEEPSKER---PKRISKVYYSCLVKAMPKSSSPSETEPEQCLDQVIYKIKLPG 580
R + DE EE KER PKR A PK +PS T+ + D+V K
Sbjct: 62 RTSIYDEYEEELKERANKPKRKR--------PAPPKKKAPS-TQNSKSNDKV---EKKKT 109
Query: 581 PAILGEGKP 589
+I +GKP
Sbjct: 110 TSIAKDGKP 118
>SCC2_ASHGO (Q750S2) Sister chromatid cohesion protein 2
Length = 1479
Score = 32.7 bits (73), Expect = 6.1
Identities = 28/122 (22%), Positives = 56/122 (44%), Gaps = 13/122 (10%)
Query: 163 VRECYASFKSIIRYLVQGDR-----EKQVIEYILSEVDKHIEAGDLISEFKLSALPSLYG 217
++ C S + I + LV GD+ E +I Y+L++ + LIS S+L L
Sbjct: 838 IKNCRVSIRVIAQLLVGGDKICDLFEHFLIFYVLNKNAHTEDQNKLIS----SSLCLLTD 893
Query: 218 QFVALI----KYLLDNKHEDRDQVVILFQDMLEVVTRDIMMEDHLLSLVDSIHGGSGQEG 273
Q + ++ +DN+ E+ + ++ F +L + +DH+ +L +H + +
Sbjct: 894 QVIEMVIENEAADVDNQREEEGRNIMKFLSVLSSCQDSFITKDHITALYPYLHSDTKSDF 953
Query: 274 ML 275
L
Sbjct: 954 QL 955
>P_HUMAN (Q04671) P protein (Melanocyte-specific transporter protein)
Length = 838
Score = 32.7 bits (73), Expect = 6.1
Identities = 27/113 (23%), Positives = 59/113 (51%), Gaps = 13/113 (11%)
Query: 1007 EKSWESWWEE-EQEHLKYSGMRGIIAEILLSLRFFIYQYGL---VYHLNFTKSTKSVLPT 1062
+K+WE+ +E +++H G+ ++A+ L L F I+ + L V ++ ++L
Sbjct: 601 DKNWETNIQELQKKHRISDGI--LLAKCLTVLGFVIFMFFLNSFVPGIHLDLGWIAILGA 658
Query: 1063 GLWHIMVGDLSNISYLEDFQLVFRLMKGLVFVTFVSILVTMIALAHMTLQDIV 1115
+W +++ D+ DF+++ ++ + F ++ V M ALAH+ L + V
Sbjct: 659 -IWLLILADI------HDFEIILHRVEWATLLFFAALFVLMEALAHLHLIEYV 704
>NIN_MOUSE (Q61043) Ninein
Length = 2035
Score = 32.7 bits (73), Expect = 6.1
Identities = 40/153 (26%), Positives = 69/153 (44%), Gaps = 23/153 (15%)
Query: 135 ALDMAKDSNGKDRELRKRIEFDNYMSC--AVRECYASFKSIIRYLVQG------------ 180
ALD + D N EL +E + ++ + ASFK+ IR+L++
Sbjct: 328 ALDFSLDGNINLTELTLALENELLVTKNGIHQAALASFKAEIRHLLERVDQVVREKEKLR 387
Query: 181 ---DREKQVIEYILSEVDKHIEAGDLISEFKLSALPSLYGQFVALIKYLLDNKHEDR--- 234
D+ +++ + SEVD H A + +E+ L L Y + +A +K L + E
Sbjct: 388 SDLDKAEKLKSLMASEVDDHHAAIERRNEYNLRKLDEEYKERIAALKNELRQEREQMLQQ 447
Query: 235 --DQVVILFQDMLEVVTRDIMMEDHL-LSLVDS 264
Q V L Q++ + T + + D L LSL ++
Sbjct: 448 VGKQRVELEQEIQKAKTEENYIRDRLALSLKEN 480
>LMG2_MOUSE (Q61092) Laminin gamma-2 chain precursor
(Kalinin/nicein/epiligrin 100 kDa subunit) (Laminin B2t
chain)
Length = 1191
Score = 32.7 bits (73), Expect = 6.1
Identities = 25/75 (33%), Positives = 40/75 (53%), Gaps = 7/75 (9%)
Query: 256 DHLLSLVDSIHGGSGQEGMLLLEQQHQLFASEGAIRFPIEPVTEAWTEKIKR----LYLL 311
D +L L+D G +EGM+LLEQ LF ++ I + P+ E+++R L+LL
Sbjct: 1101 DGILHLIDQ-PGSVDEEGMMLLEQ--GLFQAKTQINSRLRPLMSDLEERVRRQRNHLHLL 1157
Query: 312 LTTKESAMDVPSNLE 326
T+ + + NLE
Sbjct: 1158 ETSIDGILADVKNLE 1172
>TYPH_METAC (Q8TL01) Putative thymidine phosphorylase (EC 2.4.2.4)
(TdRPase)
Length = 506
Score = 32.3 bits (72), Expect = 8.0
Identities = 23/83 (27%), Positives = 41/83 (48%), Gaps = 6/83 (7%)
Query: 190 ILSEVDKHIEAGDLISEFKLSALPSLYGQFVALIKYLLDNKHEDRDQVVILFQDMLEVVT 249
+ SEV +H E D E +P+L + +IK +LD K +D++ +L D++E
Sbjct: 61 VFSEVYEHFEDWDKPVE----VVPALRSKSSGVIKKMLDKKSVLQDEIKLLVNDIVEENL 116
Query: 250 RDIMMEDHLLSLVDSIHGGSGQE 272
D+ + + + IHG + E
Sbjct: 117 SDVELSAFITA--SYIHGMTDDE 137
>SYT_BORBU (O51662) Threonyl-tRNA synthetase (EC 6.1.1.3)
(Threonine--tRNA ligase) (ThrRS)
Length = 581
Score = 32.3 bits (72), Expect = 8.0
Identities = 24/103 (23%), Positives = 42/103 (40%), Gaps = 17/103 (16%)
Query: 319 MDVPSNLEAKRRISFFSNSLFMDMPTAPKVRNMLSFSVLTPYYTEEVLFSLRELESPNED 378
+D+ N + + +S + + F+D+ P V NM + F L +
Sbjct: 98 IDLIKNFDLQNEVSIYKSHNFVDLCRGPHVENMNKI--------DPKAFKLTSIAGAYWR 149
Query: 379 GVSILFYLQKIFPDEWNNFLQRVNCSNEEELKEYDELEEELRR 421
G L +I+ WN NE+EL+ Y L EE+++
Sbjct: 150 GSEKNPMLTRIYGTLWN---------NEKELRSYLNLREEIKK 183
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.324 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 141,003,278
Number of Sequences: 164201
Number of extensions: 6188737
Number of successful extensions: 14450
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 14362
Number of HSP's gapped (non-prelim): 43
length of query: 1211
length of database: 59,974,054
effective HSP length: 122
effective length of query: 1089
effective length of database: 39,941,532
effective search space: 43496328348
effective search space used: 43496328348
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 72 (32.3 bits)
Medicago: description of AC137603.6


