

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137603.10 + phase: 0
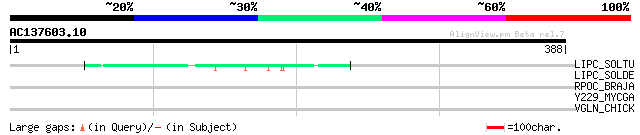
(388 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
LIPC_SOLTU (P80471) Light-induced protein, chloroplast precursor... 45 4e-04
LIPC_SOLDE (O99019) Light-induced protein, chloroplast precursor... 43 0.002
RPOC_BRAJA (Q89J75) DNA-directed RNA polymerase beta' chain (EC ... 31 4.7
Y229_MYCGA (P53660) Hypothetical protein MG447 homolog MYCGA2290 30 8.0
VGLN_CHICK (P81021) Vigilin 30 8.0
>LIPC_SOLTU (P80471) Light-induced protein, chloroplast precursor
(Chloroplastic drought-induced stress protein CDSP-34)
Length = 326
Score = 44.7 bits (104), Expect = 4e-04
Identities = 59/230 (25%), Positives = 90/230 (38%), Gaps = 51/230 (22%)
Query: 53 ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNS-SLIEGRWQLIFTTR 111
+ L +L G RG S+S + I I LE PT + +L+ G+W L +T+
Sbjct: 103 KKQLADSLYGTN-RGLSASSETRAEIVELITQLESKNPNPAPTEALTLLNGKWILAYTSF 161
Query: 112 PGTASPIQRTFVGVDFFSVFQEVYLQTNDPR---VTNIVSFSDAIGELKVEAAAS--IGD 166
G + R + + V E QT D V N V F+ + + A +
Sbjct: 162 SGLFPLLSRGNLPL----VRVEEISQTIDSESFTVQNSVVFAGPLATTSISTNAKFEVRS 217
Query: 167 GKRILFRFDRAAF-------------SFKFLPFKV---PY-------------------- 190
KR+ +F+ + +FL K+ P+
Sbjct: 218 PKRVQIKFEEGIIGTPQLTDSIVLPENVEFLGQKIDLSPFKGLITSVQDTASSVAKSISS 277
Query: 191 --PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQK 238
P+ F + + A+ WL TTYL LRISRG+ G+ FVL K+ P K
Sbjct: 278 QPPIKFPITNNNAQSWLLTTYLDDE--LRISRGDAGSVFVLIKEGSPLLK 325
>LIPC_SOLDE (O99019) Light-induced protein, chloroplast precursor
(C40.4)
Length = 326
Score = 42.7 bits (99), Expect = 0.002
Identities = 58/230 (25%), Positives = 89/230 (38%), Gaps = 51/230 (22%)
Query: 53 ENSLIQALVGIQGRGRSSSPQQLNAIERAIQVLEHIGGVSDPTNS-SLIEGRWQLIFTTR 111
+ L +L G RG S+S + I I LE PT + +L+ G+W L +T+
Sbjct: 103 KKQLADSLYGTN-RGLSASSETRAEIVELITQLESKNPNPAPTEALTLLNGKWILAYTSF 161
Query: 112 PGTASPIQRTFVGVDFFSVFQEVYLQTNDPR---VTNIVSFSDAIGELKVEAAAS--IGD 166
G + R + + V E QT D V N V F+ + + A +
Sbjct: 162 SGLFPLLSRGNLPL----VRVEEISQTIDSESFTVQNSVVFAGPLATTSISTNAKFEVRS 217
Query: 167 GKRILFRFDRAAF-------------SFKFLPFKV---PY-------------------- 190
KR+ +F+ + +FL K+ P+
Sbjct: 218 PKRVQIKFEEGIIGTPQLTDSIVLPENVEFLGQKIDVSPFKGLITSVQDTASSVVKSISS 277
Query: 191 --PVPFKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQK 238
P+ F + + A+ WL TTYL LRI RG+ G+ FVL K+ P K
Sbjct: 278 QPPIKFPITNNNAQSWLLTTYLDDE--LRIPRGDAGSVFVLIKEGSPLLK 325
>RPOC_BRAJA (Q89J75) DNA-directed RNA polymerase beta' chain (EC
2.7.7.6) (RNAP beta' subunit) (Transcriptase beta'
chain) (RNA polymerase beta' subunit)
Length = 1398
Score = 31.2 bits (69), Expect = 4.7
Identities = 26/110 (23%), Positives = 46/110 (41%), Gaps = 13/110 (11%)
Query: 275 WQMIWNSQTVTDSWLENAANGLMGKQIVEKNGRIKYVVNILLGLKFSMSGIFVKSSPKVY 334
WQ + + V+ W+E A +M ++ KN RI Y + L K +SG+ +VY
Sbjct: 554 WQGMDETGKVSTRWIETTAGRVMLGNLLPKNPRISYEIINKLMTKREISGVI----DQVY 609
Query: 335 EVTMDDAAIIGGPFGYPLEFGKKFILEILFNDGKVRISRGDNEIIFVHAR 384
+I F + + +N K IS G ++++ H +
Sbjct: 610 RHCGQKETVI---------FCDRIMALGFYNAFKAGISFGKDDMVVPHGK 650
>Y229_MYCGA (P53660) Hypothetical protein MG447 homolog MYCGA2290
Length = 596
Score = 30.4 bits (67), Expect = 8.0
Identities = 31/135 (22%), Positives = 55/135 (39%), Gaps = 17/135 (12%)
Query: 194 FKLLGDEAKGWLDTTYLSHSGNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAI 253
F + + W+ YL+ S NL+I N L + +T I + + + +
Sbjct: 92 FNFVDSNGQSWIYQDYLTASNNLKIPPFNIND---LIRAAISISAPITVIINALTLLITM 148
Query: 254 DKLISLNKNSGEEDPELEEGEW------QMIWNSQT------VTDSWLENAANGLMGKQI 301
+K G D E + W +I + T V +WL ++ANG M + +
Sbjct: 149 GLANQFSKALGRGDEEKIKEVWATGFITNVIVSIATSMIIVGVAKTWLTSSANGTMNRSL 208
Query: 302 --VEKNGRIKYVVNI 314
++ N R Y+V+I
Sbjct: 209 ESLDTNSRYGYIVSI 223
>VGLN_CHICK (P81021) Vigilin
Length = 1270
Score = 30.4 bits (67), Expect = 8.0
Identities = 16/49 (32%), Positives = 26/49 (52%)
Query: 214 GNLRISRGNKGTTFVLQKQTEPRQKLLTAISSGVGVREAIDKLISLNKN 262
GN+R R N G + + Q+L+T + + V+EA +L +L KN
Sbjct: 748 GNIRKVRDNTGARIIFPTSEDKDQELITIMGTEEAVKEAQKELEALIKN 796
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.136 0.390
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,381,006
Number of Sequences: 164201
Number of extensions: 1931842
Number of successful extensions: 4739
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 4732
Number of HSP's gapped (non-prelim): 7
length of query: 388
length of database: 59,974,054
effective HSP length: 112
effective length of query: 276
effective length of database: 41,583,542
effective search space: 11477057592
effective search space used: 11477057592
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137603.10


