

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.6 + phase: 0
(425 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
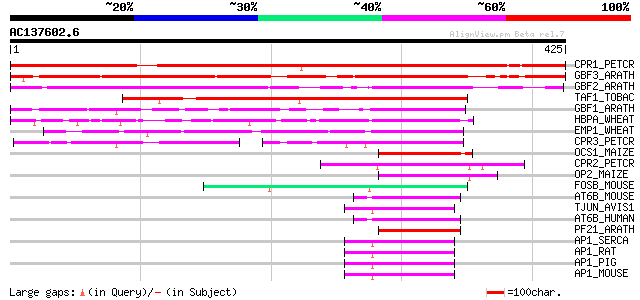
Score E
Sequences producing significant alignments: (bits) Value
CPR1_PETCR (Q99089) Common plant regulatory factor CPRF-1 432 e-121
GBF3_ARATH (P42776) G-box binding factor 3 (AtbZIP55) 393 e-109
GBF2_ARATH (P42775) G-box binding factor 2 (AtbZIP54) 311 2e-84
TAF1_TOBAC (Q99142) Transcriptional activator TAF-1 (Fragment) 265 2e-70
GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41) 177 6e-44
HBPA_WHEAT (P23922) Transcription factor HBP-1a (Histone-specifi... 164 4e-40
EMP1_WHEAT (P25032) DNA-binding EMBP-1 protein 118 3e-26
CPR3_PETCR (Q99091) Light-inducible protein CPRF-3 87 6e-17
OCS1_MAIZE (P24068) Ocs-element binding factor 1 (OCSBF-1) 58 5e-08
CPR2_PETCR (Q99090) Light-inducible protein CPRF-2 54 6e-07
OP2_MAIZE (P12959) Opaque-2 regulatory protein 50 1e-05
FOSB_MOUSE (P13346) Protein fosB 50 1e-05
AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor AT... 47 9e-05
TJUN_AVIS1 (P05411) Transforming protein jun 47 1e-04
AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor AT... 47 1e-04
PF21_ARATH (Q04088) Possible transcription factor PosF21 (AtbZIP59) 46 2e-04
AP1_SERCA (P54864) Transcription factor AP-1 (Proto-oncogene c-jun) 46 2e-04
AP1_RAT (P17325) Transcription factor AP-1 (Activator protein 1)... 46 2e-04
AP1_PIG (P56432) Transcription factor AP-1 (Activator protein 1)... 46 2e-04
AP1_MOUSE (P05627) Transcription factor AP-1 (Activator protein ... 46 2e-04
>CPR1_PETCR (Q99089) Common plant regulatory factor CPRF-1
Length = 411
Score = 432 bits (1111), Expect = e-121
Identities = 231/427 (54%), Positives = 295/427 (68%), Gaps = 18/427 (4%)
Query: 1 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVASGH 60
MGN+D+ K+ K EK SSP +Q+N HVYPDWAAMQAYYGPRVA+PPY+N VASG
Sbjct: 1 MGNTDDVKAVKPEKLSSPPPPAAPDQSNSHVYPDWAAMQAYYGPRVALPPYFNPAVASGQ 60
Query: 61 TPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE 120
+PHPYMWGPPQP+MPPYG PYAA+Y HGGVY HP VP+ +P+S++
Sbjct: 61 SPHPYMWGPPQPVMPPYGVPYAALYAHGGVYAHPGVPL--------------AASPMSMD 106
Query: 121 TPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNT 180
T KSSG + GL+KKLKG D LAMSIGNG A+S+E E SQS TEGSSDGS+ N+
Sbjct: 107 THAKSSGTNEHGLIKKLKGHDDLAMSIGNGKADSSEGEMERTLSQSKETEGSSDGSNENS 166
Query: 181 SGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMM--AVAPAGVTGQLVGPVA 238
A RKR R+ P GE K TQ S I A S+K++ VA V G++VG V
Sbjct: 167 KRAAVNGRKRGRDEAPNMIGEVKIETQSSVIPSPRAKSEKLLGITVATPMVAGKVVGTVV 226
Query: 239 SSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRS 298
S +MT++LEL++S H+ +P QPS ++P ++W+ N+R+LKRERRKQSNRESARRS
Sbjct: 227 SPSMTSSLELKDSPKEHAVNSPAGGQQPSTMMPNDSWLHNDRDLKRERRKQSNRESARRS 286
Query: 299 RLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPKE 358
RLRKQAEAEELA KV+SL AE+ +L++EINRL +E+L +N+ L E K A+ + +
Sbjct: 287 RLRKQAEAEELAIKVDSLTAENMALKAEINRLTLTAEKLTNDNSRLLEVMKNAQAERAAD 346
Query: 359 IILTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDASPR 418
+ L N + ++ + +ST NLLSRV +N+GS DR E E+ + SGAKLHQLLDA+PR
Sbjct: 347 VGLGNNNEKKASTLSTANLLSRV-DNAGSGDRD-EGESDVYEKTTKSGAKLHQLLDANPR 404
Query: 419 ADAVAAG 425
DAVAAG
Sbjct: 405 TDAVAAG 411
>GBF3_ARATH (P42776) G-box binding factor 3 (AtbZIP55)
Length = 382
Score = 393 bits (1009), Expect = e-109
Identities = 231/430 (53%), Positives = 281/430 (64%), Gaps = 53/430 (12%)
Query: 1 MGNSDEEKS--TKTEKPSSPVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPPYYNSPVA- 57
MGNS EE TK++KPSSP +QTNVHVYPDWAAMQAYYGPRVAMPPYYNS +A
Sbjct: 1 MGNSSEEPKPPTKSDKPSSP----PVDQTNVHVYPDWAAMQAYYGPRVAMPPYYNSAMAA 56
Query: 58 SGHTPHPYMWGPPQPMMPPYGHPYAAMYPHGG-VYTHPAVPIGPHPHSQGISSSPATGTP 116
SGH P PYMW P Q MM PYG PYAA+YPHGG VY HP +P+G P Q GT
Sbjct: 57 SGHPPPPYMWNP-QHMMSPYGAPYAAVYPHGGGVYAHPGIPMGSLPQGQKDPPLTTPGTL 115
Query: 117 LSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGS 176
LSI+TP KS+GNTD GLMKKLK FDGLAMS+GNG+ E+ R S T+GS+DGS
Sbjct: 116 LSIDTPTKSTGNTDNGLMKKLKEFDGLAMSLGNGNPENGAD-EHKRSRNSSETDGSTDGS 174
Query: 177 DGNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPA-GVTGQLVG 235
DGNT+GA++ + KRSREGTPT DG K++ + +V+P+ G TG +
Sbjct: 175 DGNTTGADEPKLKRSREGTPTKDG------------KQLVQASSFHSVSPSSGDTGVKLI 222
Query: 236 PVASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESA 295
+ + + S V + +NP + Q A++PPE W+QNERELKRERRKQSNRESA
Sbjct: 223 QGSGAIL--------SPGVSANSNPFMS-QSLAMVPPETWLQNERELKRERRKQSNRESA 273
Query: 296 RRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQ 355
RRSRLRKQAE EELARKVE+L AE+ +LRSE+N+L E S++LR NA L +K K +
Sbjct: 274 RRSRLRKQAETEELARKVEALTAENMALRSELNQLNEKSDKLRGANATLLDKLKCS---- 329
Query: 356 PKEIILTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDA 415
+ ++ P N+LSRV NSG+ D+ DN NS +KLHQLLD
Sbjct: 330 ---------EPEKRVPA---NMLSRV-KNSGAGDK----NKNQGDNDSNSTSKLHQLLDT 372
Query: 416 SPRADAVAAG 425
PRA AVAAG
Sbjct: 373 KPRAKAVAAG 382
>GBF2_ARATH (P42775) G-box binding factor 2 (AtbZIP54)
Length = 360
Score = 311 bits (798), Expect = 2e-84
Identities = 192/428 (44%), Positives = 247/428 (56%), Gaps = 73/428 (17%)
Query: 1 MGNSDEEKSTK-TEKPSSPVTVDQTNQTNVHVYP-DWAAMQAYYGPRVAMPPYYNSPVAS 58
MG+++E T ++KPS +Q+N VHVY DWAAMQAYYGPRV +P YYNS +A
Sbjct: 1 MGSNEEGNPTNNSDKPSQAAAPEQSN---VHVYHHDWAAMQAYYGPRVGIPQYYNSNLAP 57
Query: 59 GHTPHPYMWGPPQPMMPPYGHPYAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLS 118
GH P PYMW P PMM PYG PY P GGVY HP V +G P S+ TPL+
Sbjct: 58 GHAPPPYMWASPSPMMAPYGAPYPPFCPPGGVYAHPGVQMGSQPQGPVSQSASGVTTPLT 117
Query: 119 IETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAE-PGAESRQSQSVNTEGSSDGSD 177
I+ P S+GN+D G MKKLK FDGLAMSI N SAE +E R SQS +GSS+GSD
Sbjct: 118 IDAPANSAGNSDHGFMKKLKEFDGLAMSISNNKVGSAEHSSSEHRSSQSSENDGSSNGSD 177
Query: 178 GNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPV 237
GNT+G Q+RRKR ++ +P+T GE ++ + E D M
Sbjct: 178 GNTTGGEQSRRKRRQQRSPST-GERPSSQNSLPLRGENEKPDVTM--------------- 221
Query: 238 ASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARR 297
+ M TA+ +NS+ ++ PQP W NE+E+KRE+RKQSNRESARR
Sbjct: 222 GTPVMPTAMSFQNSAGMN------GVPQP--------W--NEKEVKREKRKQSNRESARR 265
Query: 298 SRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKIAKLGQPK 357
SRLRKQAE E+L+ KV++L AE+ SLRS++ +L SE+LR+EN A+ ++ K G+
Sbjct: 266 SRLRKQAETEQLSVKVDALVAENMSLRSKLGQLNNESEKLRLENEAILDQLKAQATGK-- 323
Query: 358 EIILTNIDSQRTTPVSTENLLSRVN-NNSGSNDRTVEDENGYCDNKPNSGAKLHQLLDAS 416
TENL+SRV+ NNS S +TV+ HQLL+AS
Sbjct: 324 ----------------TENLISRVDKNNSVSGSKTVQ----------------HQLLNAS 351
Query: 417 PRADAVAA 424
P D VAA
Sbjct: 352 PITDPVAA 359
>TAF1_TOBAC (Q99142) Transcriptional activator TAF-1 (Fragment)
Length = 265
Score = 265 bits (676), Expect = 2e-70
Identities = 144/271 (53%), Positives = 190/271 (69%), Gaps = 16/271 (5%)
Query: 87 HGGVYTHPAVPIGPHPHSQGISSSPAT-----GTPLSIETPPKSSGNTDQGLMKKLKGFD 141
HGGVY HP VPIG HP G+++SPA G LS++ KSS N+D+GL
Sbjct: 2 HGGVYAHPGVPIGSHPPGHGMATSPAVSQAMDGASLSLDASAKSSENSDRGL-------- 53
Query: 142 GLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNTSGANQTRRKRSREGTPTTDGE 201
LAMS+GNG A++ E GA+ SQS +TE S+DGSD N +G ++ +KRSRE TP G+
Sbjct: 54 -LAMSLGNGSADNIEGGADHGNSQSGDTEDSTDGSDTNGAGVSERSKKRSRETTPDNSGD 112
Query: 202 GKTNTQGSQISKEIAASDK--MMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKTN 259
K++ + Q + EI + ++AV P V +++G V S +MTT LE+RN +S H K +
Sbjct: 113 SKSHLRRCQPTGEINDDSEKAIVAVRPGKVGEKVMGTVLSPSMTTTLEMRNPASTHLKAS 172
Query: 260 PTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAE 319
PT+ Q S LP EAW+QNERELKRE+RKQSNRESARRSRLRKQAEAEELA +V+SL AE
Sbjct: 173 PTNVSQLSPALPNEAWLQNERELKREKRKQSNRESARRSRLRKQAEAEELAIRVQSLTAE 232
Query: 320 SASLRSEINRLAENSERLRMENAALKEKFKI 350
+ +L+SEIN+L ENSE+L++ENAAL E+ K+
Sbjct: 233 NMTLKSEINKLMENSEKLKLENAALMERLKM 263
>GBF1_ARATH (P42774) G-box binding factor 1 (AtbZIP41)
Length = 315
Score = 177 bits (448), Expect = 6e-44
Identities = 129/353 (36%), Positives = 179/353 (50%), Gaps = 67/353 (18%)
Query: 1 MGNSDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDWA-AMQAYYGPRVAMPPYYNSPVASG 59
MG S+++ KT KP+S + YPDW +MQAYYG P++ SPV S
Sbjct: 1 MGTSEDKMPFKTTKPTS-----SAQEVPPTPYPDWQNSMQAYYGGGGTPNPFFPSPVGSP 55
Query: 60 HTPHPYMWGPPQPMMPPYGHP--YAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPL 117
+PHPYMWG MMPPYG P Y AMYP G VY HP++P+ P+ S P P
Sbjct: 56 -SPHPYMWGAQHHMMPPYGTPVPYPAMYPPGAVYAHPSMPMPPN-------SGPTNKEPA 107
Query: 118 SIETP-PKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGS 176
+ KS GN+ KK +G D GN A S +SV T GSSD +
Sbjct: 108 KDQASGKKSKGNSK----KKAEGGDKALSGSGNDGA--------SHSDESV-TAGSSDEN 154
Query: 177 DGNTSGANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGP 236
D N + Q ++ G D ++ T EI S M VAP
Sbjct: 155 DENANQQEQGSIRKPSFGQMLADASSQSTTG------EIQGSVPMKPVAPG--------- 199
Query: 237 VASSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNERELKRERRKQSNRESAR 296
+ + ++L +S A +P +++ERELKR++RKQSNRESAR
Sbjct: 200 ---TNLNIGMDLWSSQ---------------AGVP----VKDERELKRQKRKQSNRESAR 237
Query: 297 RSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFK 349
RSRLRKQAE E+L ++VESL+ E+ SLR E+ RL+ ++L+ EN +++++ +
Sbjct: 238 RSRLRKQAECEQLQQRVESLSNENQSLRDELQRLSSECDKLKSENNSIQDELQ 290
>HBPA_WHEAT (P23922) Transcription factor HBP-1a (Histone-specific
transcription factor HBP1)
Length = 349
Score = 164 bits (415), Expect = 4e-40
Identities = 131/366 (35%), Positives = 178/366 (47%), Gaps = 56/366 (15%)
Query: 1 MGNSDEEKSTKTEKPSS----PVTVDQTNQTNVHVYPDWAAMQAYYGPRVAMPP--YYNS 54
MG++D +K KP P T T+ T VYP+W Q Y AMPP ++
Sbjct: 1 MGSNDPSTPSKASKPPEQEQPPAT---TSGTTAPVYPEWPGFQGY----PAMPPHGFFPP 53
Query: 55 PVASGHTPHPYMWGPPQPMMPPYGHPYAA--MYPHGGVYTHPAVPIGPHPHSQGISSSPA 112
PVA+G HPYMWGP Q M+PPYG P MYP G VY HP P G HP
Sbjct: 54 PVAAGQA-HPYMWGP-QHMVPPYGTPPPPYMMYPPGTVYAHPTAP-GVHP---------- 100
Query: 113 TGTPLSIETPPKSSGNTDQGLMKKLKGFDGLAMSIGNGHAESAEPGAESRQSQSVNTEGS 172
P +++GN + G G A + ++ EPG S S + T S
Sbjct: 101 ------FHYPMQTNGNLEPA------GAQGAAPGAAETNGKN-EPGKTSGPSANGVTSNS 147
Query: 173 SDGSDGNTSG--ANQTRRKRSREGTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVT 230
GSD + G AN S+E +G + G S +K M + P +
Sbjct: 148 ESGSDSESEGSDANSQNDSHSKENDVNENGSAQN---GVSHSSSHGTFNKPMPLVPVQ-S 203
Query: 231 GQLVGPVASSAMTTALELRNSSSVHSKTNPTSTPQ-PSAVLPPEAWIQNERELKRERRKQ 289
G ++G VA A + + + S P + PS E W +ERELK+++RK
Sbjct: 204 GAVIG-VAGPATNLNIGMDYWGATGSSPVPAMRGKVPSGSARGEQW--DERELKKQKRKL 260
Query: 290 SNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFK 349
SNRESARRSRLRKQAE EEL ++ E+L +E++SLR E++R+ + E L +N +LK
Sbjct: 261 SNRESARRSRLRKQAECEELGQRAEALKSENSSLRIELDRIKKEYEELLSKNTSLK---- 316
Query: 350 IAKLGQ 355
AKLG+
Sbjct: 317 -AKLGE 321
>EMP1_WHEAT (P25032) DNA-binding EMBP-1 protein
Length = 354
Score = 118 bits (295), Expect = 3e-26
Identities = 98/324 (30%), Positives = 145/324 (44%), Gaps = 31/324 (9%)
Query: 27 TNVHVYPDWAA-MQAYYGPRVAMPPYYNSPVASGHTPHPYMWGPPQPMMPPYGHPYAAMY 85
T + +WAA M AYY A+ HPY P P +G A
Sbjct: 21 TPAQAHAEWAASMHAYYA------------AAASAAGHPYAAWPLPPQAQQHGLVAAGA- 67
Query: 86 PHGGVYTHPAVPIGPHPHS--QGISSSPATGTPLSIETPPKSSGNTDQGLMKKLKGFDGL 143
G Y AVP P P + + +S A G P ++G + + K +
Sbjct: 68 --GAAYGAGAVPHVPPPPAGTRHAHASMAAGVPYMAGESASAAGKGKR--VGKTQRVPSG 123
Query: 144 AMSIGNGHAESAEPGAESRQSQSVNTEGSSDGSDGNTSGANQTRRKRSREGTPTTDGEGK 203
++ +G ++ G+ + N +GSS + SGA +T EG P+ +
Sbjct: 124 EINSSSGSGDAGSQGSSEKGDAGANQKGSSSSAKRRKSGAAKT------EGEPSQAATVQ 177
Query: 204 TNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSSVHSKTNPTST 263
+ + ++ K++ +AP P + M L SS V + N ++
Sbjct: 178 NAVTEPPLEDKERSASKLLVLAPGRAALTSAAPNLNIGMDP-LSASPSSLVQGEVNAAAS 236
Query: 264 PQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASL 323
Q +A L +ERELKRERRKQSNRESARRSRLRKQ E EELA+KV L A + +L
Sbjct: 237 SQSNASLSQ----MDERELKRERRKQSNRESARRSRLRKQQECEELAQKVSELTAANGTL 292
Query: 324 RSEINRLAENSERLRMENAALKEK 347
RSE+++L ++ + + EN L K
Sbjct: 293 RSELDQLKKDCKTMETENKKLMGK 316
>CPR3_PETCR (Q99091) Light-inducible protein CPRF-3
Length = 296
Score = 87.4 bits (215), Expect = 6e-17
Identities = 56/163 (34%), Positives = 88/163 (53%), Gaps = 17/163 (10%)
Query: 194 GTPTTDGEGKTNTQGSQISKEIAASDKMMAVAPAGVTGQLVGPVASSAMTTALELRNSSS 253
G +D +G+T+ + K I S G Q ASS+ L + +
Sbjct: 108 GRKISDEKGRTSAK-----KSIGVSGSTSFAVDKGAENQ---KAASSSDNDCPSLSSENG 159
Query: 254 VHS----KTNPTSTPQPSAVLP-----PEAWIQNERELKRERRKQSNRESARRSRLRKQA 304
V ++NP P A++ P+ + +ERELKR+RRKQSNRESARRSRLRKQA
Sbjct: 160 VDGSLEVRSNPLDVAAPGAIVVHDGMLPDQRVNDERELKRQRRKQSNRESARRSRLRKQA 219
Query: 305 EAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEK 347
+++EL ++++L+ E+ LR + R++E + EN ++KE+
Sbjct: 220 KSDELQERLDNLSKENRILRKNLQRISEACAEVTSENHSIKEE 262
Score = 69.7 bits (169), Expect = 1e-11
Identities = 52/177 (29%), Positives = 77/177 (43%), Gaps = 19/177 (10%)
Query: 4 SDEEKSTKTEKPSSPVTVDQTNQTNVHVYPDW-AAMQAYYGPRVAMPPYYNSPVASGHTP 62
SD E+ T + P +V++ T +PD ++MQAYYG A P + + +P
Sbjct: 2 SDGEEGTPMKHPKPASSVEEAPITTTP-FPDLLSSMQAYYGG--AAPAAFYASTVGSPSP 58
Query: 63 HPYMWGPPQPMMPPYGHP--YAAMYPHGGVYTHPAVPIGPHPHSQGISSSPATGTPLSIE 120
HPYMW + PYG P Y A++ GG++THP VP P+ P S E
Sbjct: 59 HPYMWRNQHRFILPYGIPMQYPALFLPGGIFTHPIVPTDPNL------------APTSGE 106
Query: 121 TPPKSSGNTDQGLMKKLKGFDG-LAMSIGNGHAESAEPGAESRQSQSVNTEGSSDGS 176
K S + KK G G + ++ G + S+++E DGS
Sbjct: 107 VGRKISDEKGRTSAKKSIGVSGSTSFAVDKGAENQKAASSSDNDCPSLSSENGVDGS 163
>OCS1_MAIZE (P24068) Ocs-element binding factor 1 (OCSBF-1)
Length = 151
Score = 57.8 bits (138), Expect = 5e-08
Identities = 31/72 (43%), Positives = 46/72 (63%), Gaps = 2/72 (2%)
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
+RE+R+ SNRESARRSRLRKQ +EL ++V L A++A + + +A R+ EN
Sbjct: 26 RREKRRLSNRESARRSRLRKQQHLDELVQEVARLQADNARVAARARDIASQYTRVEQENT 85
Query: 343 ALKEKFKIAKLG 354
L+ + A+LG
Sbjct: 86 VLRA--RAAELG 95
>CPR2_PETCR (Q99090) Light-inducible protein CPRF-2
Length = 401
Score = 54.3 bits (129), Expect = 6e-07
Identities = 46/168 (27%), Positives = 79/168 (46%), Gaps = 12/168 (7%)
Query: 239 SSAMTTALELRNSSSVHSKTNPTSTPQPSAVLPPEAWIQNER-----ELKRERRKQSNRE 293
++ + AL+ +++ V S T+ +S E + R + KR RR SNRE
Sbjct: 151 ATPLLPALQKKSAIQVKSTTSGSSRDHSDDDDELEGETETTRNGDPSDAKRVRRMLSNRE 210
Query: 294 SARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENAALKEKFKI--A 351
SARRSR RKQA EL +V L E++SL + +++ ++N LK + A
Sbjct: 211 SARRSRRRKQAHMTELETQVSQLRVENSSLLKRLTDISQRYNDAAVDNRVLKADIETMRA 270
Query: 352 KLGQPKEII-----LTNIDSQRTTPVSTENLLSRVNNNSGSNDRTVED 394
K+ +E + L + ++ +ST + S + S ++ T +D
Sbjct: 271 KVKMAEETVKRVTGLNPMFQSMSSEISTIGMQSFSGSPSDTSADTTQD 318
>OP2_MAIZE (P12959) Opaque-2 regulatory protein
Length = 453
Score = 50.1 bits (118), Expect = 1e-05
Identities = 32/93 (34%), Positives = 50/93 (53%), Gaps = 2/93 (2%)
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
+R R+K+SNRESARRSR RK A +EL +V L AE++ L I L + ++N
Sbjct: 227 ERVRKKESNRESARRSRYRKAAHLKELEDQVAQLKAENSCLLRRIAALNQKYNDANVDNR 286
Query: 343 ALKEKFKI--AKLGQPKEIILTNIDSQRTTPVS 373
L+ + AK+ ++ + I+ + P S
Sbjct: 287 VLRADMETLRAKVKMGEDSLKRVIEMSSSVPSS 319
>FOSB_MOUSE (P13346) Protein fosB
Length = 338
Score = 49.7 bits (117), Expect = 1e-05
Identities = 55/214 (25%), Positives = 87/214 (39%), Gaps = 12/214 (5%)
Query: 149 NGHAESAEPGAESRQSQSVNTEGSSDGSDGNTSGANQTRRKRSREGTPT---TDGEGKTN 205
+G S+ P AES+ SV++ GS + + A S T T T + +
Sbjct: 11 SGSRCSSSPSAESQYLSSVDSFGSPPTAAASQECAGLGEMPGSFVPTVTAITTSQDLQWL 70
Query: 206 TQGSQISKEIAASDKMMAVAPAGVTG-QLVGPVASSAMTTALELRNSSSVHSKTNPTSTP 264
Q + IS + + +A P V + G S+ +A +S + T+T
Sbjct: 71 VQPTLISSMAQSQGQPLASQPPAVDPYDMPGTSYSTPGLSAYSTGGASGSGGPSTSTTTS 130
Query: 265 QPSAVLPPEA--------WIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESL 316
P + P A + E E KR R++ N+ +A + R R++ + L + + L
Sbjct: 131 GPVSARPARARPRRPREETLTPEEEEKRRVRRERNKLAAAKCRNRRRELTDRLQAETDQL 190
Query: 317 NAESASLRSEINRLAENSERLRMENAALKEKFKI 350
E A L SEI L + ERL A K KI
Sbjct: 191 EEEKAELESEIAELQKEKERLEFVLVAHKPGCKI 224
>AT6B_MOUSE (O35451) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp)
Length = 699
Score = 47.0 bits (110), Expect = 9e-05
Identities = 29/82 (35%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query: 264 PQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASL 323
P P PPE + + LKR++R NRESA +SR +K+ + L +++++ A++ L
Sbjct: 308 PMPGNSCPPEV---DAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQL 364
Query: 324 RSEINRLAENSERLRMENAALK 345
R E L E L EN+ LK
Sbjct: 365 RRENAALRRRLEALLAENSGLK 386
>TJUN_AVIS1 (P05411) Transforming protein jun
Length = 296
Score = 46.6 bits (109), Expect = 1e-04
Identities = 27/86 (31%), Positives = 45/86 (51%), Gaps = 2/86 (2%)
Query: 257 KTNPTSTPQPSAVLPPEAWI--QNERELKRERRKQSNRESARRSRLRKQAEAEELARKVE 314
K P + P+ PP I +++ +K ER++ NR +A +SR RK L KV+
Sbjct: 191 KEEPQTVPEMPGETPPLFPIDMESQERIKAERKRMRNRIAASKSRKRKLERIARLEEKVK 250
Query: 315 SLNAESASLRSEINRLAENSERLRME 340
+L A+++ L S N L E +L+ +
Sbjct: 251 TLKAQNSELASTANMLREQVAQLKQK 276
>AT6B_HUMAN (Q99941) Cyclic-AMP-dependent transcription factor ATF-6
beta (Activating transcription factor 6 beta)
(ATF6-beta) (cAMP responsive element binding
protein-like 1) (cAMP response element binding
protein-related protein) (Creb-rp) (G13 protein)
Length = 703
Score = 46.6 bits (109), Expect = 1e-04
Identities = 29/82 (35%), Positives = 45/82 (54%), Gaps = 3/82 (3%)
Query: 264 PQPSAVLPPEAWIQNERELKRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASL 323
P P PPE + + LKR++R NRESA +SR +K+ + L +++++ A++ L
Sbjct: 311 PMPGNSCPPEV---DAKLLKRQQRMIKNRESACQSRRKKKEYLQGLEARLQAVLADNQQL 367
Query: 324 RSEINRLAENSERLRMENAALK 345
R E L E L EN+ LK
Sbjct: 368 RRENAALRRRLEALLAENSELK 389
>PF21_ARATH (Q04088) Possible transcription factor PosF21 (AtbZIP59)
Length = 398
Score = 46.2 bits (108), Expect = 2e-04
Identities = 25/63 (39%), Positives = 39/63 (61%)
Query: 283 KRERRKQSNRESARRSRLRKQAEAEELARKVESLNAESASLRSEINRLAENSERLRMENA 342
KR +R +NR+SA RS+ RK EL RKV++L E+ +L +++ L ++ L +EN
Sbjct: 203 KRAKRIWANRQSAARSKERKTRYIFELERKVQTLQTEATTLSAQLTLLQRDTNGLTVENN 262
Query: 343 ALK 345
LK
Sbjct: 263 ELK 265
>AP1_SERCA (P54864) Transcription factor AP-1 (Proto-oncogene c-jun)
Length = 314
Score = 45.8 bits (107), Expect = 2e-04
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 2/86 (2%)
Query: 257 KTNPTSTPQPSAVLPPEAWI--QNERELKRERRKQSNRESARRSRLRKQAEAEELARKVE 314
K P + P+ PP + I +++ +K ER++ NR +A + R RK L KV+
Sbjct: 209 KEEPQTVPEMPGETPPLSPIDMESQERIKAERKRMRNRIAASKCRKRKLERIARLEEKVK 268
Query: 315 SLNAESASLRSEINRLAENSERLRME 340
+L A+++ L S N L E +L+ +
Sbjct: 269 TLKAQNSELASTANMLREQVAQLKQK 294
>AP1_RAT (P17325) Transcription factor AP-1 (Activator protein 1)
(AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus
17 oncogene homolog)
Length = 334
Score = 45.8 bits (107), Expect = 2e-04
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 2/86 (2%)
Query: 257 KTNPTSTPQPSAVLPPEAWI--QNERELKRERRKQSNRESARRSRLRKQAEAEELARKVE 314
K P + P+ PP + I +++ +K ER++ NR +A + R RK L KV+
Sbjct: 229 KEEPQTVPEMPGETPPLSPIDMESQERIKAERKRMRNRIAASKCRKRKLERIARLEEKVK 288
Query: 315 SLNAESASLRSEINRLAENSERLRME 340
+L A+++ L S N L E +L+ +
Sbjct: 289 TLKAQNSELASTANMLREQVAQLKQK 314
>AP1_PIG (P56432) Transcription factor AP-1 (Activator protein 1)
(AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus
17 oncogene homolog)
Length = 331
Score = 45.8 bits (107), Expect = 2e-04
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 2/86 (2%)
Query: 257 KTNPTSTPQPSAVLPPEAWI--QNERELKRERRKQSNRESARRSRLRKQAEAEELARKVE 314
K P + P+ PP + I +++ +K ER++ NR +A + R RK L KV+
Sbjct: 226 KEEPQTVPEMPGETPPLSPIDMESQERIKAERKRMRNRIAASKCRKRKLERIARLEEKVK 285
Query: 315 SLNAESASLRSEINRLAENSERLRME 340
+L A+++ L S N L E +L+ +
Sbjct: 286 TLKAQNSELASTANMLREQVAQLKQK 311
>AP1_MOUSE (P05627) Transcription factor AP-1 (Activator protein 1)
(AP1) (Proto-oncogene c-jun) (V-jun avian sarcoma virus
17 oncogene homolog) (Jun A) (AH119)
Length = 334
Score = 45.8 bits (107), Expect = 2e-04
Identities = 26/86 (30%), Positives = 45/86 (52%), Gaps = 2/86 (2%)
Query: 257 KTNPTSTPQPSAVLPPEAWI--QNERELKRERRKQSNRESARRSRLRKQAEAEELARKVE 314
K P + P+ PP + I +++ +K ER++ NR +A + R RK L KV+
Sbjct: 229 KEEPQTVPEMPGETPPLSPIDMESQERIKAERKRMRNRIAASKCRKRKLERIARLEEKVK 288
Query: 315 SLNAESASLRSEINRLAENSERLRME 340
+L A+++ L S N L E +L+ +
Sbjct: 289 TLKAQNSELASTANMLREQVAQLKQK 314
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.305 0.123 0.346
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,269,675
Number of Sequences: 164201
Number of extensions: 2506341
Number of successful extensions: 11069
Number of sequences better than 10.0: 818
Number of HSP's better than 10.0 without gapping: 170
Number of HSP's successfully gapped in prelim test: 666
Number of HSP's that attempted gapping in prelim test: 9421
Number of HSP's gapped (non-prelim): 1896
length of query: 425
length of database: 59,974,054
effective HSP length: 113
effective length of query: 312
effective length of database: 41,419,341
effective search space: 12922834392
effective search space used: 12922834392
T: 11
A: 40
X1: 16 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 43 (22.0 bits)
S2: 67 (30.4 bits)
Medicago: description of AC137602.6


