

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137602.5 - phase: 0
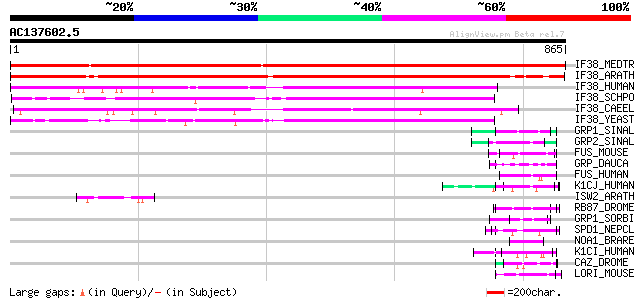
(865 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation factor 3 s... 1613 0.0
IF38_ARATH (O49160) Eukaryotic translation initiation factor 3 s... 1150 0.0
IF38_HUMAN (Q99613) Eukaryotic translation initiation factor 3 s... 494 e-139
IF38_SCHPO (O14164) Probable eukaryotic translation initiation f... 482 e-135
IF38_CAEEL (O02328) Probable eukaryotic translation initiation f... 455 e-127
IF38_YEAST (P32497) Eukaryotic translation initiation factor 3 9... 294 6e-79
GRP1_SINAL (P49310) Glycine-rich RNA-binding protein GRP1A 54 1e-06
GRP2_SINAL (P49311) Glycine-rich RNA-binding protein GRP2A 53 3e-06
FUS_MOUSE (P56959) RNA-binding protein FUS (Pigpen protein) 52 5e-06
GRP_DAUCA (Q03878) Glycine-rich RNA-binding protein 52 7e-06
FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncog... 52 7e-06
K1CJ_HUMAN (P13645) Keratin, type I cytoskeletal 10 (Cytokeratin... 51 2e-05
ISW2_ARATH (Q8RWY3) Putative chromatin remodelling complex ATPas... 51 2e-05
RB87_DROME (P48810) Heterogeneous nuclear ribonucleoprotein 87F ... 50 3e-05
GRP1_SORBI (Q99069) Glycine-rich RNA-binding protein 1 (Fragment) 50 3e-05
SPD1_NEPCL (P19837) Spidroin 1 (Dragline silk fibroin 1) (Fragment) 49 6e-05
NOA1_BRARE (Q7ZVE0) Nucleolar protein family A member 1 (snoRNP ... 49 6e-05
K1CI_HUMAN (P35527) Keratin, type I cytoskeletal 9 (Cytokeratin ... 49 7e-05
CAZ_DROME (Q27294) RNA-binding protein cabeza (Sarcoma-associate... 49 7e-05
LORI_MOUSE (P18165) Loricrin 48 1e-04
>IF38_MEDTR (Q9XHM1) Eukaryotic translation initiation factor 3
subunit 8 (eIF3 p110) (eIF3c)
Length = 935
Score = 1613 bits (4178), Expect = 0.0
Identities = 819/866 (94%), Positives = 840/866 (96%), Gaps = 3/866 (0%)
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
MASTVDQIKNAIKINDWVSLQE+FDKINKQL+KVMRVIESQKIPNLYIKALVMLEDFLAQ
Sbjct: 72 MASTVDQIKNAIKINDWVSLQESFDKINKQLEKVMRVIESQKIPNLYIKALVMLEDFLAQ 131
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQY 120
AS+NKDAKKKMSP+NAKAFNSMKQKLKKNNKQYEDLIIK RE+P+SE EK EDDEDSD+Y
Sbjct: 132 ASANKDAKKKMSPSNAKAFNSMKQKLKKNNKQYEDLIIKCRESPESEGEKDEDDEDSDEY 191
Query: 121 GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPS 180
SDD E+IEPDQL+KPEPVSDSETSELGNDRPGDD DAPWDQKL KKDRLLEKMFMKKPS
Sbjct: 192 ESDD-EMIEPDQLRKPEPVSDSETSELGNDRPGDDGDAPWDQKLSKKDRLLEKMFMKKPS 250
Query: 181 EITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKLQILFSVVSAQFDVN 240
EITWDTVN KFKEILEARGRKGTGRFEQVEQ TFLTKVAKTPAQKLQILFSVVSAQFDVN
Sbjct: 251 EITWDTVNKKFKEILEARGRKGTGRFEQVEQLTFLTKVAKTPAQKLQILFSVVSAQFDVN 310
Query: 241 PGLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKKGDDYDGPIHVWGN 300
PGLSGHMPI+VWKKCVQNMLVILDILVQHPNIKVDDSVELDENE+KKGDDY+GPI+VWGN
Sbjct: 311 PGLSGHMPISVWKKCVQNMLVILDILVQHPNIKVDDSVELDENETKKGDDYNGPINVWGN 370
Query: 301 LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESIGDFKASSKVA 360
LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEP FVVLAQNVQEYLESIGDFKASSKVA
Sbjct: 371 LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPQFVVLAQNVQEYLESIGDFKASSKVA 430
Query: 361 LKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVVTPELVARK 420
LKRVELIYYKPHEVYEATRKLAE+TV+GDNGE +SEE KGF DTRI APFVVT ELVARK
Sbjct: 431 LKRVELIYYKPHEVYEATRKLAEMTVEGDNGE-MSEEPKGFEDTRIPAPFVVTLELVARK 489
Query: 421 PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV 480
PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV
Sbjct: 490 PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV 549
Query: 481 HHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEK 540
HHMDISTQILFNRAMSQLGLCAFR GLVSEAHGCLSELYSGGRVKELLAQGVSQ+RYHEK
Sbjct: 550 HHMDISTQILFNRAMSQLGLCAFRAGLVSEAHGCLSELYSGGRVKELLAQGVSQSRYHEK 609
Query: 541 TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL 600
TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL
Sbjct: 610 TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL 669
Query: 601 EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDK 660
EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASL+VWKFVKNRD VLEMLKDK
Sbjct: 670 EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLDVWKFVKNRDAVLEMLKDK 729
Query: 661 IKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCI 720
IKEEALRTYLFTFSSSYDSLSV QLTNFFDLSLPR HSIVSRMM+NEELHASWDQPTGCI
Sbjct: 730 IKEEALRTYLFTFSSSYDSLSVVQLTNFFDLSLPRVHSIVSRMMVNEELHASWDQPTGCI 789
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGG 780
IFRNVE SRVQALAFQLTEKLSILAESNERA+EARLGGGGLDLPPRRRDGQDYAAAAAGG
Sbjct: 790 IFRNVEHSRVQALAFQLTEKLSILAESNERATEARLGGGGLDLPPRRRDGQDYAAAAAGG 849
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGY-GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSR 839
G GTSSGGRWQDLSYSQTRQGSGR GY GGRALSF+QAGGSGGYSRGRG GGGGYQNS R
Sbjct: 850 GSGTSSGGRWQDLSYSQTRQGSGRTGYGGGRALSFSQAGGSGGYSRGRGTGGGGYQNSGR 909
Query: 840 TQGGSALRGPHGDVSTRMVSLRGVRA 865
TQGGSALRGPHGD STRMVSLRGVRA
Sbjct: 910 TQGGSALRGPHGDTSTRMVSLRGVRA 935
>IF38_ARATH (O49160) Eukaryotic translation initiation factor 3
subunit 8 (eIF3 p110) (eIF3c) (p105)
Length = 900
Score = 1150 bits (2976), Expect = 0.0
Identities = 593/867 (68%), Positives = 692/867 (79%), Gaps = 33/867 (3%)
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
M TVDQ+KNA+KINDWVSLQE FDK+NKQL+KVMR+ E+ K P LYIK LVMLEDFL +
Sbjct: 63 MTYTVDQMKNAMKINDWVSLQENFDKVNKQLEKVMRITEAVKPPTLYIKTLVMLEDFLNE 122
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQY 120
A +NK+AKKKMS +N+KA NSMKQKLKKNNK YED I K+RE P+ EEEK +D+D D
Sbjct: 123 ALANKEAKKKMSTSNSKALNSMKQKLKKNNKLYEDDINKYREAPEVEEEKQPEDDDDDD- 181
Query: 121 GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPS 180
DDD+ +E D + D T + G+D D+ W++ L KKD+LLEK+ K P
Sbjct: 182 --DDDDEVEDDD----DSSIDGPTVDPGSDVDEPTDNLTWEKMLSKKDKLLEKLMNKDPK 235
Query: 181 EITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKLQILFSVVSAQFDVN 240
EITWD VN KFKEI+ ARG+KGT RFE V+Q T LTK+AKTPAQKL+ILFSV+SAQFDVN
Sbjct: 236 EITWDWVNKKFKEIVAARGKKGTARFELVDQLTHLTKIAKTPAQKLEILFSVISAQFDVN 295
Query: 241 PGLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKKGDDYDGPIHVWGN 300
PGLSGHMPINVWKKCV NML ILDILV++ NI VDD+VE DENE+ K DYDG I VWGN
Sbjct: 296 PGLSGHMPINVWKKCVLNMLTILDILVKYSNIVVDDTVEPDENETSKPTDYDGKIRVWGN 355
Query: 301 LVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESIGDFKASSKVA 360
LVAFLE++D EFFKSLQCIDPHTREYVERLRDEP+F+ LAQN+Q+Y E +GDFKA++KVA
Sbjct: 356 LVAFLERVDTEFFKSLQCIDPHTREYVERLRDEPMFLALAQNIQDYFERMGDFKAAAKVA 415
Query: 361 LKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVVTPELVARK 420
L+RVE IYYKP EVY+A RKLAEL + + EE EES F+V PE+V RK
Sbjct: 416 LRRVEAIYYKPQEVYDAMRKLAELVEEEEETEEAKEESGPPTS------FIVVPEVVPRK 469
Query: 421 PTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHLQENV 480
PTFPE+SR +MD+LVSLIY+ GDERTKARAMLCDI HHAL+D F ARDLLLMSHLQ+N+
Sbjct: 470 PTFPESSRAMMDILVSLIYRNGDERTKARAMLCDINHHALMDNFVTARDLLLMSHLQDNI 529
Query: 481 HHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNRYHEK 540
HMDISTQILFNR M+QLGLCAFR G+++E+H CLSELYSG RV+ELLAQGVSQ+RYHEK
Sbjct: 530 QHMDISTQILFNRTMAQLGLCAFRAGMITESHSCLSELYSGQRVRELLAQGVSQSRYHEK 589
Query: 541 TPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKIISKNFRRLL 600
TPEQER+ERRRQMPYHMH+NLELLE+VHL AMLLEVPNMAAN HDAKR++ISKNFRRLL
Sbjct: 590 TPEQERMERRRQMPYHMHLNLELLEAVHLICAMLLEVPNMAANSHDAKRRVISKNFRRLL 649
Query: 601 EVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTVLEMLKDK 660
E+SE+Q FT PPE VRDHVMAATR L GDFQKAF+++ SLEVW+ +KNRD++L+M+KD+
Sbjct: 650 EISERQAFTAPPENVRDHVMAATRALTKGDFQKAFEVLNSLEVWRLLKNRDSILDMVKDR 709
Query: 661 IKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCI 720
IKEEALRTYLFT+SSSY+SLS+DQL FD+S P+ HSIVS+MMINEELHASWDQPT CI
Sbjct: 710 IKEEALRTYLFTYSSSYESLSLDQLAKMFDVSEPQVHSIVSKMMINEELHASWDQPTRCI 769
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRD-GQDYAAAAAG 779
+F V+ SR+Q+LAFQLTEKLSILAESNERA E+R GGGGLDL RRRD QDYA AA+G
Sbjct: 770 VFHEVQHSRLQSLAFQLTEKLSILAESNERAMESRTGGGGLDLSSRRRDNNQDYAGAASG 829
Query: 780 GGGGTSSGGRWQD-LSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS 838
G GG WQD +Y Q RQG+ R+GYGG S Q G G +RG G G
Sbjct: 830 G------GGYWQDKANYGQGRQGN-RSGYGG-GRSSGQNGQWSGQNRGGGYAG------- 874
Query: 839 RTQGGSALRGPHGDVSTRMVSL-RGVR 864
+ GS RG D S+RMVSL RGVR
Sbjct: 875 --RVGSGNRGMQMDGSSRMVSLNRGVR 899
>IF38_HUMAN (Q99613) Eukaryotic translation initiation factor 3
subunit 8 (eIF3 p110) (eIF3c)
Length = 913
Score = 494 bits (1271), Expect = e-139
Identities = 308/853 (36%), Positives = 453/853 (52%), Gaps = 123/853 (14%)
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
+ + + I+NA+KI D E F+ + K K +++ + +P YI+ L LED+L +
Sbjct: 60 LTNLIRTIRNAMKIRDVTKCLEEFELLGKAYGKAKSIVDKEGVPRFYIRILADLEDYLNE 119
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQ--------------- 105
+K+ KKKM+ NAKA ++++QK++K N+ +E I +++ P+
Sbjct: 120 LWEDKEGKKKMNKNNAKALSTLRQKIRKYNRDFESHITSYKQNPEQSADEDAEKNEEDSE 179
Query: 106 --SEEEKFED---------------------------DEDSDQYGSDDDEIIEPDQLKKP 136
S+E++ ED DED D S+DDE +
Sbjct: 180 GSSDEDEDEDGVSAATFLKKKSEAPSGESRKFLKKMDDEDEDSEDSEDDEDWDTGSTSSD 239
Query: 137 EPVSDSE------TSELGNDRPGDDDDAPWDQKLR----------KKDRLLE-------- 172
+ E S P D+D +K R K RL E
Sbjct: 240 SDSEEEEGKQTALASRFLKKAPTTDEDKKAAEKKREDKAKKKHDRKSKRLDEEEEDNEGG 299
Query: 173 ----------------KMFMKKPSEITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLT 216
KMF K +EIT V K EIL+ARG+KGT R Q+E L
Sbjct: 300 EWERVRGGVPLVKEKPKMFAKG-TEITHAVVIKKLNEILQARGKKGTDRAAQIELLQLLV 358
Query: 217 KVAKT----PAQKLQILFSVVSAQFDVNPGLSGHMPINVWKKCVQNMLVILDILVQHPNI 272
++A ++I F+++++ +D NP L+ +M +W KC+ + ++DIL +PNI
Sbjct: 359 QIAAENNLGEGVIVKIKFNIIASLYDYNPNLATYMKPEMWGKCLDCINELMDILFANPNI 418
Query: 273 KVDDSVELDENESKKGDDYDGPIHVWGNLVAFLEKIDAEFFKSLQCIDPHTREYVERLRD 332
V +++ L+E+E+ D P+ V G ++ +E++D EF K +Q DPH++EYVE L+D
Sbjct: 419 FVGENI-LEESENLHNADQ--PLRVRGCILTLVERMDEEFTKIMQNTDPHSQEYVEHLKD 475
Query: 333 EPLFVVLAQNVQEYLESIGDFKASSKVALKRVELIYYKPHEVYEATRKLAELTVDGDNGE 392
E + + VQ YLE G + ++ L R+ YYK Y+A ++ E
Sbjct: 476 EAQVCAIIERVQRYLEEKGTTEEVCRIYLLRILHTYYKFD--YKAHQRQLTPPEGSSKSE 533
Query: 393 EVSEESKGFVDTRIIAPFVVTPELVARKPTFPENSRTLMDVLVSLIY-KYGDERTKARAM 451
+ E++G E+S LM+ L IY K +R + A+
Sbjct: 534 QDQAENEG------------------------EDSAVLMERLCKYIYAKDRTDRIRTCAI 569
Query: 452 LCDIYHHALLDEFAVARDLLLMSHLQENVHHMDISTQILFNRAMSQLGLCAFRVGLVSEA 511
LC IYHHAL + ARDL+LMSHLQ+N+ H D QIL+NR M QLG+CAFR GL +A
Sbjct: 570 LCHIYHHALHSRWYQARDLMLMSHLQDNIQHADPPVQILYNRTMVQLGICAFRQGLTKDA 629
Query: 512 HGCLSELYSGGRVKELLAQGVSQNRYHEKTPEQERLERRRQMPYHMHINLELLESVHLTS 571
H L ++ S GR KELL QG+ E+ EQE++ERRRQ+P+H+HINLELLE V+L S
Sbjct: 630 HNALLDIQSSGRAKELLGQGLLLRSLQERNQEQEKVERRRQVPFHLHINLELLECVYLVS 689
Query: 572 AMLLEVPNMAANVHDAKRKIISKNFRRLLEVSEKQTFTGPPETVRDHVMAATRVLINGDF 631
AMLLE+P MAA+ DA+R++ISK F L V E+Q GPPE++R+HV+AA++ + GD+
Sbjct: 690 AMLLEIPYMAAHESDARRRMISKQFHHQLRVGERQPLLGPPESMREHVVAASKAMKMGDW 749
Query: 632 QKAFDIIASL----EVWKFVKNRDTVLEMLKDKIKEEALRTYLFTFSSSYDSLSVDQLTN 687
+ I + +VW D V ML KI+EE+LRTYLFT+SS YDS+S++ L++
Sbjct: 750 KTCHSFIINEKMNGKVWDLFPEADKVRTMLVRKIQEESLRTYLFTYSSVYDSISMETLSD 809
Query: 688 FFDLSLPRAHSIVSRMMINEELHASWDQPTGCIIFRNVELSRVQALAFQLTEKLSILAES 747
F+L LP HSI+S+M+INEEL AS DQPT ++ E + Q LA QL EKL L E+
Sbjct: 810 MFELDLPTVHSIISKMIINEELMASLDQPTQTVVMHRTEPTAQQNLALQLAEKLGSLVEN 869
Query: 748 NERASEARLGGGG 760
NER + + G G
Sbjct: 870 NERVFDHKQGTYG 882
>IF38_SCHPO (O14164) Probable eukaryotic translation initiation
factor 3 93 kDa subunit (eIF3 p93)
Length = 918
Score = 482 bits (1241), Expect = e-135
Identities = 275/762 (36%), Positives = 442/762 (57%), Gaps = 71/762 (9%)
Query: 3 STVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQAS 62
S ++ IKNA+ N+W+ + FD +NK K E+ + P YI+ L L+ L S
Sbjct: 185 SCMETIKNAMSSNNWIVVSNEFDHLNKVSQKCK---EAGRNPPPYIEFLSALDQKLE--S 239
Query: 63 SNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQYGS 122
++K K + N +AFN++KQ+++KNN+Q++ D D+Y
Sbjct: 240 ADKAFIKSLDAANGRAFNALKQRVRKNNRQFQS--------------------DIDRYRK 279
Query: 123 DDDEIIEPDQLKK-PEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPSE 181
D + ++P +L + P+P + E+ D + + + + KP E
Sbjct: 280 DPEGFMKPAELNEIPKPAGKAGQDEVIVDG------------VATRGIVAPTEGLGKPEE 327
Query: 182 ITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKLQILFSVVSAQFDVNP 241
IT + + I EARG+K T R EQ+ L+ +A T Q+L++ ++++ +FD+N
Sbjct: 328 ITPADIFKYLRAIFEARGKKSTDRSEQIRLLEKLSTIAVTDYQRLRVKVALLAVRFDINT 387
Query: 242 GLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKK-----GDDYDGPIH 296
G +MPI+ W + + ILDI +P I + + VE DENE ++ ++ +G I
Sbjct: 388 GSGQYMPIDQWNAALTELHSILDIFDANPKIVIVEQVE-DENEEEEEAIAAAENNNGVIQ 446
Query: 297 VWGNLVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESIGDFKAS 356
V G++V+FLE++D EF +SLQ IDPHT EY++RL+DE L Q YLE IG + +
Sbjct: 447 VQGSVVSFLERLDDEFTRSLQMIDPHTPEYIDRLKDETSLYTLLVRSQGYLERIGVVENT 506
Query: 357 SKVALKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVVTPEL 416
+++ ++R++ +YYKP +V A ++A + F T +TP
Sbjct: 507 ARLIMRRLDRVYYKPEQVIRANEEVAW---------------RSFPPT---FDLTITPRA 548
Query: 417 VARKPTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLLLMSHL 476
P L+ L +Y G + RAMLC IYH AL + F ARD+LLMSHL
Sbjct: 549 TTTTPDI------LIHSLCVYLYNNGVSLLRTRAMLCHIYHEALQNRFYKARDMLLMSHL 602
Query: 477 QENVHHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGVSQNR 536
Q++VH DI+TQIL NR M Q+GLCAFR G+V E L ++ + GRVKELL QG+ +
Sbjct: 603 QDSVHAADIATQILHNRTMVQIGLCAFRNGMVQETQYALQDISTTGRVKELLGQGIQAPK 662
Query: 537 YHEKTPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAA---NVHDAKRKIIS 593
+ + TP+Q+RL+++ +P+HMHINLELLE V+LT +ML+E+P MAA D+++++IS
Sbjct: 663 FGQFTPDQDRLDKQLVLPFHMHINLELLECVYLTCSMLMEIPAMAAASSTASDSRKRVIS 722
Query: 594 KNFRRLLEVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFVKNRDTV 653
+ FRR+LE ++Q F GPPE R+++M A++ L +G++++ + I ++++W + + D +
Sbjct: 723 RPFRRMLEYIDRQLFVGPPENTREYIMQASKALADGEWRRCEEFIHAIKIWSLMPDADKI 782
Query: 654 LEMLKDKIKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASW 713
+ML +KI+EE LRTYL +++ YDS+S++ L FDL + R IVSR++ E+HA+
Sbjct: 783 KQMLSEKIREEGLRTYLLAYAAFYDSVSLEFLATTFDLPVQRVTVIVSRLLSKREIHAAL 842
Query: 714 DQPTGCIIFRNVELSRVQALAFQLTEKLSILAESNERASEAR 755
DQ G IIF VE++++++L L+EK + L E+NE+ E +
Sbjct: 843 DQVHGAIIFERVEINKLESLTVSLSEKTAQLNEANEKLYEQK 884
>IF38_CAEEL (O02328) Probable eukaryotic translation initiation
factor 3 subunit 8 (eIF3 p110) (eIF3c)
Length = 898
Score = 455 bits (1171), Expect = e-127
Identities = 299/868 (34%), Positives = 457/868 (52%), Gaps = 133/868 (15%)
Query: 6 DQIKNAIKIN-------DWVSLQETFDKINKQLDKVMRVIESQKI--PNLYIKALVMLED 56
D++K IK N D L FD + K DK V + Q + P YI++LV +ED
Sbjct: 60 DELKGIIKQNRDAKSNKDLNRLLTGFDSLAKAYDKSKTVFQRQNVANPRFYIRSLVEIED 119
Query: 57 FLAQASSNKDAKKKMSPTNAKAFNSMKQKLKK--NNKQYEDLIIKFRETPQSEEEKFEDD 114
++ + +KDAK +S NAKA ++QKLKK ++Q +DL+ +R P + + +D
Sbjct: 120 YVNKLWDDKDAKSALSKNNAKALPPLRQKLKKYIKDQQLQDLVTDYRVNPDEDGYETPED 179
Query: 115 EDSDQYGSDDDEIIE--PDQLKKPEPVSDSETSELGND----------RPGDDDDAPW-- 160
ED D +G + E P + + VSDS++ +D DD+D+
Sbjct: 180 EDDDDFGEVSESKAEKSPGKPSEKAAVSDSDSDSDDDDSSNWSSEPESNSSDDEDSVTKM 239
Query: 161 -----------------DQKLRKKDRLLE-KMFMKKPSEITWDTVNN------------- 189
D K KK R++ K +++ + W VN
Sbjct: 240 EQLRRYFLKKEFRVESKDDKKEKKKRVIRVKEAVEEDDDADWTPVNREKSVVHFDPKEEV 299
Query: 190 -------KFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKL----QILFSVVSAQFD 238
K E++ ARG++ T R + V L +V++ L +I F ++SA F+
Sbjct: 300 THDVMIKKLNEVMSARGKRTTDRNQHVANLRKLLEVSEEKELGLGINVKISFCIISALFE 359
Query: 239 VNPGLSGHMPINVWKKCVQNMLVILDILVQHPNIKVDDSVELDENESKKGDDYDGPIHVW 298
+N +S HM + +Q + +LD+L+ +K+ + +E+E+ K D + +
Sbjct: 360 LNAKISDHMEYETFMTTLQTVNSLLDLLIGTDRVKLSVTYA-EEDENLKDDTQE--YRIQ 416
Query: 299 GNLVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLE-----SIGDF 353
G+++ ++++D E K LQ D H+ +Y+E+L+ E L + ++Y+E I D
Sbjct: 417 GSILIAVQRLDGELAKILQNADCHSNDYIEKLKAEKDMCSLIEKAEKYVELRNDSGIFDK 476
Query: 354 KASSKVALKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFVVT 413
KV + R+E YYK + E
Sbjct: 477 HEVCKVYMMRIEHAYYKYQDQNE------------------------------------- 499
Query: 414 PELVARKPTFPENSRTLMDVLVSLIYKYGDE-RTKARAMLCDIYHHALLDEFAVARDLLL 472
E++ LMD L + IY DE R + RAMLC +Y+ A+ D++ ARDLLL
Sbjct: 500 -----------EDAGKLMDYLCNKIYTLDDEKRLRQRAMLCHVYYLAVHDKWHRARDLLL 548
Query: 473 MSHLQENVHHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQGV 532
MSH+Q V H D+ TQIL+NR + QLGLCAFR G + EAH LSE+ + R KELLAQ V
Sbjct: 549 MSHMQAIVDHSDVDTQILYNRTICQLGLCAFRHGFIREAHQGLSEIQNTQRAKELLAQAV 608
Query: 533 SQNRYHEKTPEQERLERRRQMPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRKII 592
R HEKT EQE+++R RQ+PYHMHIN+EL+E V+L +MLLE+P+MA+ + +R+++
Sbjct: 609 G-TRQHEKTAEQEKIDRSRQVPYHMHINVELMECVYLICSMLLEIPHMASCEFEMRRRML 667
Query: 593 SKNFRRLLEVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIA----SLEVWKFVK 648
S++F L+ SEK + TGPPE R+HV+AA++ ++NGD++K D I + +VW
Sbjct: 668 SRSFHYQLKQSEKASLTGPPENTREHVVAASKAMLNGDWKKCQDYIVNDKMNQKVWNLFH 727
Query: 649 NRDTVLEMLKDKIKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEE 708
N +TV M+ +I+EE+LRTYL T+S+ Y ++S+ +L + F+LS HSI+S+M+I EE
Sbjct: 728 NAETVKGMVVRRIQEESLRTYLLTYSTVYATVSLKKLADLFELSKKDVHSIISKMIIQEE 787
Query: 709 LHASWDQPTGCIIFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLP---- 764
L A+ D+PT C+I VE SR+Q LA L++KL LAE+NE+ E R G GG P
Sbjct: 788 LSATLDEPTDCLIMHRVEPSRLQMLALNLSDKLQTLAENNEQILEPRTGRGGYQGPGSWF 847
Query: 765 PRRRDGQDYAAAAAGGGGGTSSGGRWQD 792
P R + Q +GG G GG+ QD
Sbjct: 848 PGRNERQGDKQKGSGGYQGERRGGQGQD 875
>IF38_YEAST (P32497) Eukaryotic translation initiation factor 3 93
kDa subunit (eIF3 p93) (Nuclear transport protein NIP1)
Length = 812
Score = 294 bits (753), Expect = 6e-79
Identities = 216/769 (28%), Positives = 380/769 (49%), Gaps = 93/769 (12%)
Query: 1 MASTVDQIKNAIKINDWVSLQETFDKINKQLDKVMRVIESQKIPNLYIKALVMLEDFLAQ 60
M ++I A +DW+++ FD I++ L + + ++ PN++IK + +ED
Sbjct: 123 MQDVYNKISQAENSDDWLTISNEFDLISRLLVRAQQ--QNWGTPNIFIKVVAQVED---- 176
Query: 61 ASSNKDAKKKMSPTNAKAFNSMKQKLKKNNKQYEDLIIKFRETPQSEEEKFEDDEDSDQY 120
A +N + A+A+N+ KQ++KK +++ ED + KFR P+S +++ D D
Sbjct: 177 AVNNTQQADLKNKAVARAYNTTKQRVKKVSRENEDSMAKFRNDPESFDKEPTADLDISAN 236
Query: 121 GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDDDDAPWDQKLRKKDRLLEKMFMKKPS 180
G +S S+ GND+ +D
Sbjct: 237 GFT---------------ISSSQ----GNDQAVQED------------------------ 253
Query: 181 EITWDTVNNKFKEILEARGRKGTGRFEQVEQFTFLTKVAKTPAQKLQILFSVVSAQFDVN 240
+ + I+++RG+K + + L VA+ P + + +++ ++FD +
Sbjct: 254 ------FFTRLQTIIDSRGKKTVNQQSLISTLEELLTVAEKPYEFIMAYLTLIPSRFDAS 307
Query: 241 PGLSGHMPINVWKKCVQNMLVILDILVQHPNI----KVDDSVELDENESKKGDDYDGPIH 296
LS + PI+ WK ++ +L IL Q + + D ++ E+E K +D DG
Sbjct: 308 ANLS-YQPIDQWKSSFNDISKLLSILDQTIDTYQVNEFADPIDFIEDEPK--EDSDGVKR 364
Query: 297 VWGNLVAFLEKIDAEFFKSLQCIDPHTREYVERLRDEPLFVVLAQNVQEYLESI----GD 352
+ G++ +F+E++D EF KSL IDPH+ +Y+ RLRDE L Q Y E+ D
Sbjct: 365 ILGSIFSFVERLDDEFMKSLLNIDPHSSDYLIRLRDEQSIYNLILRTQLYFEATLKDEHD 424
Query: 353 F-KASSKVALKRVELIYYKPHEVYEATRKLAELTVDGDNGEEVSEESKGFVDTRIIAPFV 411
+A ++ +KR++ IYYK + + A + + + SK +D+ A +V
Sbjct: 425 LERALTRPFVKRLDHIYYKSENLIKIMETAAWNIIPAQFKSKFT--SKDQLDS---ADYV 479
Query: 412 VTPELVARKPTFPENSRTLMDVLVSLIYKYGDERTKARAMLCDIYHHALLDEFAVARDLL 471
L+D L +++ K + + RA+L +IY+ AL +F A+D+L
Sbjct: 480 ----------------DNLIDGLSTILSKQNNIAVQKRAILYNIYYTALNKDFQTAKDML 523
Query: 472 LMSHLQENVHHMDISTQILFNRAMSQLGLCAFRVGLVSEAHGCLSELYSGGRVKELLAQG 531
L S +Q N++ D S QILFNR + QLGL AF++ L+ E H L++L S ++E+L Q
Sbjct: 524 LTSQVQTNINQFDSSLQILFNRVVVQLGLSAFKLCLIEECHQILNDLLSSSHLREILGQQ 583
Query: 532 VSQNRYHEKTPEQERLERRRQ-MPYHMHINLELLESVHLTSAMLLEVPNMAANVHDAKRK 590
+ ER RQ +PYH HINL+L++ V LT ++L+E+P M A K K
Sbjct: 584 SLHRISLNSSNNASADERARQCLPYHQHINLDLIDVVFLTCSLLIEIPRMTAFYSGIKVK 643
Query: 591 II---SKNFRRLLEVSEKQTFTGPPETVRDHVMAATRVLINGDFQKAFDIIASLEVWKFV 647
I K+ RR LE +K +F GPPET+RD+V+ A + + G+++ + + ++ W +
Sbjct: 644 RIPYSPKSIRRSLEHYDKLSFQGPPETLRDYVLFAAKSMQKGNWRDSVKYLREIKSWALL 703
Query: 648 KNRDTVLEMLKDKIKEEALRTYLFTFSSSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINE 707
N +TVL L ++++ E+L+TY F+F Y S SV +L FDL + ++ ++
Sbjct: 704 PNMETVLNSLTERVQVESLKTYFFSFKRFYSSFSVAKLAELFDLPENKVVEVLQSVIAEL 763
Query: 708 ELHASW-DQPTGCIIFRNVELSRVQALAFQLTEKLSILAESNERASEAR 755
E+ A D+ T ++ + E+++++ +L ++ I E S R
Sbjct: 764 EIPAKLNDEKTIFVVEKGDEITKLEEAMVKLNKEYKIAKERLNPPSNRR 812
>GRP1_SINAL (P49310) Glycine-rich RNA-binding protein GRP1A
Length = 166
Score = 54.3 bits (129), Expect = 1e-06
Identities = 35/88 (39%), Positives = 41/88 (45%), Gaps = 4/88 (4%)
Query: 757 GGGGLDLPPRRRD-GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFN 815
G G DL R + + + GGGGG GG ++ S G G GYGG
Sbjct: 68 GMNGQDLDGRSITVNEAQSRGSGGGGGGRGGGGGYR--SGGGGGYGGGGGGYGGGGREGG 125
Query: 816 QAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
+GG GGYS RG GGGGY R GG
Sbjct: 126 YSGGGGGYS-SRGGGGGGYGGGGRRDGG 152
Score = 45.4 bits (106), Expect = 6e-04
Identities = 39/134 (29%), Positives = 51/134 (37%), Gaps = 11/134 (8%)
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGG 780
I + E R + F + + ++ E + L G + + + G GG
Sbjct: 39 IINDRETGRSRGFGFVTFKDEKSMKDAIEGMNGQDLDGRSITVNEAQSRGSGGGGGGRGG 98
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYS-RGRGMGG--GGYQNS 837
GGG SGG G G GYGG +GG GGYS RG G GG GG +
Sbjct: 99 GGGYRSGGGGG--------YGGGGGGYGGGGREGGYSGGGGGYSSRGGGGGGYGGGGRRD 150
Query: 838 SRTQGGSALRGPHG 851
GG G G
Sbjct: 151 GGEGGGYGGSGGGG 164
Score = 43.5 bits (101), Expect = 0.002
Identities = 35/94 (37%), Positives = 42/94 (44%), Gaps = 16/94 (17%)
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGG--GGGTSSGGRWQDLSYS-----QTRQ 800
NE S GGGG R G Y + GG GGG GG ++ YS + +
Sbjct: 82 NEAQSRGSGGGGG-----GRGGGGGYRSGGGGGYGGGGGGYGGGGREGGYSGGGGGYSSR 136
Query: 801 GSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGY 834
G G GYGG + GG GG G G GGGG+
Sbjct: 137 GGGGGGYGGGG---RRDGGEGGGYGGSG-GGGGW 166
>GRP2_SINAL (P49311) Glycine-rich RNA-binding protein GRP2A
Length = 169
Score = 53.1 bits (126), Expect = 3e-06
Identities = 40/132 (30%), Positives = 52/132 (39%), Gaps = 12/132 (9%)
Query: 721 IFRNVELSRVQALAFQLTEKLSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGG 780
I + E R + F + + ++ E + L G + + + G GG
Sbjct: 39 IINDRETGRSRGFGFVTFKDEKSMKDAIEGMNGQDLDGRSITVNEAQSRGSGAGGGGRGG 98
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGG-RALSFNQAGGSGGYSRGRGMGGGGYQNSSR 839
GGG GG + G G GYGG R +GG GGYS RG GGGGY R
Sbjct: 99 GGGYRGGGGY----------GGGGGGYGGGRREGGGYSGGGGGYS-SRGGGGGGYGGGGR 147
Query: 840 TQGGSALRGPHG 851
GG G G
Sbjct: 148 RDGGGYGGGEGG 159
Score = 48.1 bits (113), Expect = 1e-04
Identities = 32/88 (36%), Positives = 37/88 (41%), Gaps = 5/88 (5%)
Query: 746 ESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA 805
E+ R S A GGGG R G Y G GGG GG + + +G G
Sbjct: 83 EAQSRGSGA--GGGGRGGGGGYRGGGGYGGGGGGYGGGRREGGGYSGGGGGYSSRGGGGG 140
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGG 833
GYGG + GG G G G GGGG
Sbjct: 141 GYGGGG---RRDGGGYGGGEGGGYGGGG 165
Score = 41.2 bits (95), Expect = 0.012
Identities = 31/77 (40%), Positives = 34/77 (43%), Gaps = 18/77 (23%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG RR+G Y+ GGGG +S GG G G G GGR
Sbjct: 109 GGGGGGYGGGRREGGGYSG---GGGGYSSRGG------------GGGGYGGGGRRDGGGY 153
Query: 817 AGGSGGYSRGRGMGGGG 833
GG GG G G GGGG
Sbjct: 154 GGGEGG---GYGGGGGG 167
Score = 39.3 bits (90), Expect = 0.045
Identities = 31/80 (38%), Positives = 37/80 (45%), Gaps = 9/80 (11%)
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
A G G GR ++ +Q+R GSG AG GGR GG GGY G G GGGG
Sbjct: 65 AIEGMNGQDLDGRSITVNEAQSR-GSG-AGGGGR-------GGGGGYRGGGGYGGGGGGY 115
Query: 837 SSRTQGGSALRGPHGDVSTR 856
+ G G G S+R
Sbjct: 116 GGGRREGGGYSGGGGGYSSR 135
>FUS_MOUSE (P56959) RNA-binding protein FUS (Pigpen protein)
Length = 518
Score = 52.4 bits (124), Expect = 5e-06
Identities = 36/91 (39%), Positives = 44/91 (47%), Gaps = 7/91 (7%)
Query: 764 PPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY 823
PP+ Q+ +++GGGGG G QD S G G GYG + S GG GG
Sbjct: 153 PPQGYGQQNQYNSSSGGGGGGGGGNYGQD--QSSMSGGGGGGGYGNQDQSGGGGGGYGGG 210
Query: 824 SR---GRGMGGGGYQNSSRTQGGSALRGPHG 851
+ GRG GGGG N R+ GG RG G
Sbjct: 211 QQDRGGRGRGGGGGYN--RSSGGYEPRGRGG 239
Score = 45.1 bits (105), Expect = 8e-04
Identities = 40/113 (35%), Positives = 50/113 (43%), Gaps = 19/113 (16%)
Query: 746 ESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGG------TSSGGRWQDLSYSQTR 799
+ N+ S + GGGG GQD ++ + GGGGG S GG Q R
Sbjct: 159 QQNQYNSSSGGGGGG----GGGNYGQDQSSMSGGGGGGGYGNQDQSGGGGGGYGGGQQDR 214
Query: 800 QGSGRAGYGGRALSFNQAGGSGGYS---RGRGMGGGGYQNSSRTQGGSALRGP 849
G GR G GG +N++ SGGY RG G GG G S G + GP
Sbjct: 215 GGRGRGGGGG----YNRS--SGGYEPRGRGGGRGGRGGMGGSDRGGFNKFGGP 261
Score = 37.4 bits (85), Expect = 0.17
Identities = 26/75 (34%), Positives = 33/75 (43%), Gaps = 11/75 (14%)
Query: 780 GGGGTSSGGRWQDLSYSQTRQG-SGRAGYGGRALSFNQAGGSGGYSRGR----------G 828
GG G SG Q SY Q + + GYG + + +GG GG G G
Sbjct: 129 GGYGQQSGYGGQQQSYGQQQSSYNPPQGYGQQNQYNSSSGGGGGGGGGNYGQDQSSMSGG 188
Query: 829 MGGGGYQNSSRTQGG 843
GGGGY N ++ GG
Sbjct: 189 GGGGGYGNQDQSGGG 203
Score = 35.0 bits (79), Expect = 0.85
Identities = 23/57 (40%), Positives = 27/57 (47%), Gaps = 11/57 (19%)
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNS 837
GGG GGR + GR GYGG + GG GG+ G G GGGG Q +
Sbjct: 371 GGGNGRGGRGRG-------GPMGRGGYGGGG---SGGGGRGGFPSGGG-GGGGQQRA 416
Score = 33.5 bits (75), Expect = 2.5
Identities = 33/115 (28%), Positives = 43/115 (36%), Gaps = 26/115 (22%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRW-------QDLSYSQTRQ--------- 800
GGGG G+ + GGGGG G W +++++S +
Sbjct: 391 GGGGSG-----GGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNECNQCKAPKP 445
Query: 801 -GSGRAGYGGRALSFNQAG---GSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G G G GG + N G GGY RG G GG + R G RG G
Sbjct: 446 DGPG-GGPGGSHMGGNYGDDRRGRGGYDRGGYRGRGGDRGGFRGGRGGGDRGGFG 499
Score = 33.5 bits (75), Expect = 2.5
Identities = 17/42 (40%), Positives = 21/42 (49%), Gaps = 2/42 (4%)
Query: 804 RAGYGGRALSFNQAGGSGGYSRGRG--MGGGGYQNSSRTQGG 843
+ + R FN+ GG+G RGRG MG GGY GG
Sbjct: 358 KVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGG 399
>GRP_DAUCA (Q03878) Glycine-rich RNA-binding protein
Length = 157
Score = 52.0 bits (123), Expect = 7e-06
Identities = 41/104 (39%), Positives = 45/104 (42%), Gaps = 31/104 (29%)
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
NE S GGGG RR+G GGGG GG Y R+G G GY
Sbjct: 80 NEAQSRGSGGGGG------RREG---------GGGGYGGGG-----GYGGRREGGGGGGY 119
Query: 808 GGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GGR + GG GGY G GGGGY R +GG G G
Sbjct: 120 GGR-----REGGGGGY----GGGGGGY--GGRREGGDGGYGGGG 152
Score = 45.4 bits (106), Expect = 6e-04
Identities = 33/95 (34%), Positives = 38/95 (39%), Gaps = 13/95 (13%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G G +L R + + +GGGGG GG Y G GYGGR +
Sbjct: 66 GMNGQELDGRNITVNEAQSRGSGGGGGRREGGGG---GYG------GGGGYGGR----RE 112
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG GGY R GGGGY GG G G
Sbjct: 113 GGGGGGYGGRREGGGGGYGGGGGGYGGRREGGDGG 147
Score = 35.4 bits (80), Expect = 0.65
Identities = 29/80 (36%), Positives = 33/80 (41%), Gaps = 2/80 (2%)
Query: 777 AAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQN 836
A G G GR ++ +Q+R GSG G G R GG GGY R GGGG
Sbjct: 63 AIEGMNGQELDGRNITVNEAQSR-GSGGGG-GRREGGGGGYGGGGGYGGRREGGGGGGYG 120
Query: 837 SSRTQGGSALRGPHGDVSTR 856
R GG G G R
Sbjct: 121 GRREGGGGGYGGGGGGYGGR 140
>FUS_HUMAN (P35637) RNA-binding protein FUS (Oncogene FUS) (Oncogene
TLS) (Translocated in liposarcoma protein) (POMp75) (75
kDa DNA-pairing protein)
Length = 526
Score = 52.0 bits (123), Expect = 7e-06
Identities = 40/99 (40%), Positives = 50/99 (50%), Gaps = 14/99 (14%)
Query: 764 PPRRRDGQDYAAAAAGGGGGTSSGGRW-QDLSYSQTRQGSGRAGYGGRALSFNQAGGSGG 822
PP+ Q+ +++GGGGG GG + QD S + GSG GYG + S GGSGG
Sbjct: 151 PPQGYGQQNQYNSSSGGGGGGGGGGNYGQDQSSMSSGGGSG-GGYGNQDQS--GGGGSGG 207
Query: 823 Y------SRGR----GMGGGGYQNSSRTQGGSALRGPHG 851
Y RGR G GGGG +R+ GG RG G
Sbjct: 208 YGQQDRGGRGRGGSGGGGGGGGGGYNRSSGGYEPRGRGG 246
Score = 36.2 bits (82), Expect = 0.38
Identities = 34/116 (29%), Positives = 45/116 (38%), Gaps = 27/116 (23%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRW-------QDLSYSQTRQ--------- 800
GGGG G+ + GGGGG G W +++++S +
Sbjct: 398 GGGGSG-----GGGRGGFPSGGGGGGGQQRAGDWKCPNPTCENMNFSWRNECNQCKAPKP 452
Query: 801 -GSGRAGYGGRALSFN----QAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G G G GG + N + GG GGY RG G GG + R G RG G
Sbjct: 453 DGPG-GGPGGSHMGGNYGDDRRGGRGGYDRGGYRGRGGDRGGFRGGRGGGDRGGFG 507
Score = 35.0 bits (79), Expect = 0.85
Identities = 30/91 (32%), Positives = 39/91 (41%), Gaps = 9/91 (9%)
Query: 770 GQDYAAAAAGGGGGTSSG----GRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSG---- 821
GQ A ++ G G+SS G+ Q SYSQ G+ G+ S+N G G
Sbjct: 101 GQQPAPSSTSGSYGSSSQSSSYGQPQSGSYSQQPSYGGQQQSYGQQQSYNPPQGYGQQNQ 160
Query: 822 -GYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
S G G GGGG N + Q + G G
Sbjct: 161 YNSSSGGGGGGGGGGNYGQDQSSMSSGGGSG 191
Score = 35.0 bits (79), Expect = 0.85
Identities = 23/57 (40%), Positives = 27/57 (47%), Gaps = 11/57 (19%)
Query: 781 GGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNS 837
GGG GGR + GR GYGG + GG GG+ G G GGGG Q +
Sbjct: 378 GGGNGRGGRGRG-------GPMGRGGYGGGG---SGGGGRGGFPSGGG-GGGGQQRA 423
Score = 34.3 bits (77), Expect = 1.5
Identities = 23/67 (34%), Positives = 28/67 (41%), Gaps = 4/67 (5%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALS--- 813
GGGG ++ G + GGGGG G Y +G GR G GG S
Sbjct: 201 GGGGSGGYGQQDRGGRGRGGSGGGGGGGGGGYNRSSGGYEPRGRGGGRGGRGGMGGSDRG 260
Query: 814 -FNQAGG 819
FN+ GG
Sbjct: 261 GFNKFGG 267
Score = 33.5 bits (75), Expect = 2.5
Identities = 17/42 (40%), Positives = 21/42 (49%), Gaps = 2/42 (4%)
Query: 804 RAGYGGRALSFNQAGGSGGYSRGRG--MGGGGYQNSSRTQGG 843
+ + R FN+ GG+G RGRG MG GGY GG
Sbjct: 365 KVSFATRRADFNRGGGNGRGGRGRGGPMGRGGYGGGGSGGGG 406
>K1CJ_HUMAN (P13645) Keratin, type I cytoskeletal 10 (Cytokeratin
10) (K10) (CK 10)
Length = 593
Score = 50.8 bits (120), Expect = 2e-05
Identities = 60/207 (28%), Positives = 78/207 (36%), Gaps = 55/207 (26%)
Query: 675 SSYDSLSVDQLTNFFDLSLPRAHSIVSRMMINEELHASWDQPTGCIIFRNVELSRVQALA 734
SSY S + N L + + S++ + + L AS + G V+LS++QA
Sbjct: 359 SSYKSEITELRRNVQALEI----ELQSQLALKQSLEASLAETEGRYC---VQLSQIQAQI 411
Query: 735 FQLTEKLSILAESNERAS--------------------------EARLGGGGLDLPPRRR 768
L E+L + E + E GGGG R
Sbjct: 412 SALEEQLQQIRAETECQNTEYQQLLDIKIRLENEIQTYRSLLEGEGSSGGGG-------R 464
Query: 769 DGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRG 828
G + GG GG SSGG Y GS GYGG G SGG S G G
Sbjct: 465 GGGSFG----GGYGGGSSGGGSSGGGYGGGHGGSSGGGYGG--------GSSGGGSSGGG 512
Query: 829 MGGGGYQNSSRTQGGSALRGPHGDVST 855
GGG +SS GG + G HG S+
Sbjct: 513 YGGG---SSSGGHGGGSSSGGHGGSSS 536
Score = 47.8 bits (112), Expect = 1e-04
Identities = 33/96 (34%), Positives = 41/96 (42%), Gaps = 22/96 (22%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGG---------- 819
G + ++ GG GG SSGG GS GYGG + S GG
Sbjct: 488 GGGHGGSSGGGYGGGSSGG------------GSSGGGYGGGSSSGGHGGGSSSGGHGGSS 535
Query: 820 SGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVST 855
SGGY G GGGG + GGS+ G +G S+
Sbjct: 536 SGGYGGGSSGGGGGGYGGGSSGGGSSSGGGYGGGSS 571
Score = 46.2 bits (108), Expect = 4e-04
Identities = 33/95 (34%), Positives = 45/95 (46%), Gaps = 6/95 (6%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLS--YSQTRQGSGRAGYGGRALSFN 815
GGG G +++ G GGG+SSGG S Y G G GYGG + +
Sbjct: 500 GGGSSGGGSSGGGYGGGSSSGGHGGGSSSGGHGGSSSGGYGGGSSGGGGGGYGGGS---S 556
Query: 816 QAGGSGGYSRGRGMGGGGYQNSSR-TQGGSALRGP 849
G S G G G GG+++SS + G S+ +GP
Sbjct: 557 GGGSSSGGGYGGGSSSGGHKSSSSGSVGESSSKGP 591
Score = 45.8 bits (107), Expect = 5e-04
Identities = 36/106 (33%), Positives = 44/106 (40%), Gaps = 7/106 (6%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGG------GGTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
GGG G Y ++GGG GG SS G S S GS GYGG +
Sbjct: 484 GGGYGGGHGGSSGGGYGGGSSGGGSSGGGYGGGSSSGGHGGGSSSGGHGGSSSGGYGGGS 543
Query: 812 LSFNQAGGSGGYSRGRGMGGGGYQNSSRTQG-GSALRGPHGDVSTR 856
G GG S G GGGY S + G S+ G G+ S++
Sbjct: 544 SGGGGGGYGGGSSGGGSSSGGGYGGGSSSGGHKSSSSGSVGESSSK 589
Score = 40.4 bits (93), Expect = 0.020
Identities = 30/99 (30%), Positives = 43/99 (43%), Gaps = 16/99 (16%)
Query: 766 RRRDGQDYAAAAA---GGGGGTSSGGRWQDLSYSQTR----QGSGRAGYGGRALSFNQAG 818
R + Y+++ + GGGGG GG L S ++ G G+ G + S +G
Sbjct: 4 RYSSSKHYSSSRSGGGGGGGGCGGGGGVSSLRISSSKGSLGGGFSSGGFSGGSFSRGSSG 63
Query: 819 G------SGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G SGGY G GGG + S G S+ G +G
Sbjct: 64 GGCFGGSSGGYGGLGGFGGGSFHGS---YGSSSFGGSYG 99
Score = 37.7 bits (86), Expect = 0.13
Identities = 34/121 (28%), Positives = 45/121 (37%), Gaps = 23/121 (19%)
Query: 741 LSILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSS----------GGRW 790
+S+ S++ S +R GGGG GGGGG SS GG +
Sbjct: 1 MSVRYSSSKHYSSSRSGGGG-------------GGGGCGGGGGVSSLRISSSKGSLGGGF 47
Query: 791 QDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPH 850
+S G +G G S GG GG+ G G G + + GGS G
Sbjct: 48 SSGGFSGGSFSRGSSGGGCFGGSSGGYGGLGGFGGGSFHGSYGSSSFGGSYGGSFGGGNF 107
Query: 851 G 851
G
Sbjct: 108 G 108
Score = 34.7 bits (78), Expect = 1.1
Identities = 32/93 (34%), Positives = 43/93 (45%), Gaps = 12/93 (12%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGG--GGTSSG----GRWQDLSYSQTRQGSGR 804
+S+ LGGG G ++ ++GGG GG+S G G + S+ + S
Sbjct: 38 SSKGSLGGG---FSSGGFSGGSFSRGSSGGGCFGGSSGGYGGLGGFGGGSFHGSYGSSSF 94
Query: 805 AG-YGGRALSFNQAGGS--GGYSRGRGMGGGGY 834
G YGG N GGS GG G G GGGG+
Sbjct: 95 GGSYGGSFGGGNFGGGSFGGGSFGGGGFGGGGF 127
>ISW2_ARATH (Q8RWY3) Putative chromatin remodelling complex ATPase
chain (EC 3.6.1.-) (ISW2-like) (Sucrose nonfermenting
protein 2 homolog)
Length = 1057
Score = 50.8 bits (120), Expect = 2e-05
Identities = 41/142 (28%), Positives = 65/142 (44%), Gaps = 35/142 (24%)
Query: 105 QSEEEKFEDDEDSDQY---------GSDDDEIIEPDQLKKPEPVSDSETSELGNDRPGDD 155
+ EEE+ +D+E+ D+ GSDDDE+ D+ PVSD E + + +D ++
Sbjct: 14 EEEEERVKDNEEEDEEELEAVARSSGSDDDEVAAADE----SPVSDGEAAPVEDDYEDEE 69
Query: 156 DDAPWDQKLRKKDRLLEKMFMKKPSEITWDTVNNKFKEILEAR--------GRKGTGR-- 205
D+ + R+K RL E +KK K +E+LE++ KG GR
Sbjct: 70 DEEKAEISKREKARLKEMQKLKK----------QKIQEMLESQNASIDADMNNKGKGRLK 119
Query: 206 --FEQVEQFTFLTKVAKTPAQK 225
+Q E F K + +QK
Sbjct: 120 YLLQQTELFAHFAKSDGSSSQK 141
>RB87_DROME (P48810) Heterogeneous nuclear ribonucleoprotein 87F
(HRP36.1 protein) (P11 protein)
Length = 386
Score = 49.7 bits (117), Expect = 3e-05
Identities = 34/95 (35%), Positives = 49/95 (50%), Gaps = 2/95 (2%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG R+ G ++ A GGG G +SGG + + + + G GG + NQ
Sbjct: 222 GGGGWGGQNRQNGGGNWGGAGGGGGFG-NSGGNFGGGQGGGSGGWNQQGGSGGGPWN-NQ 279
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GG+GG++ G G GG G NS+ + GG+ G G
Sbjct: 280 GGGNGGWNGGGGGGGYGGGNSNGSWGGNGGGGGGG 314
Score = 48.1 bits (113), Expect = 1e-04
Identities = 34/94 (36%), Positives = 39/94 (41%), Gaps = 11/94 (11%)
Query: 755 RLGGGGLDLPPRR--RDGQDYAAAAAGGGGGTS---SGGRWQDLSYSQTRQGSGRAGYGG 809
R GGGG PR R GQ GG GG + GG W G G G G
Sbjct: 198 RQGGGGGRGGPRAGGRGGQGDRGQGGGGWGGQNRQNGGGNWGGAG------GGGGFGNSG 251
Query: 810 RALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGG 843
Q GGSGG+++ G GGG + N GG
Sbjct: 252 GNFGGGQGGGSGGWNQQGGSGGGPWNNQGGGNGG 285
Score = 47.0 bits (110), Expect = 2e-04
Identities = 31/100 (31%), Positives = 44/100 (44%), Gaps = 3/100 (3%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG +G + GGGGG G +Q SY Q + G R ++Q
Sbjct: 289 GGGGGGYGGGNSNGS-WGGNGGGGGGGGGFGNEYQQ-SYGGGPQRNSNFG-NNRPAPYSQ 345
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTR 856
GG GG+++G GG G+ ++ GG G G + R
Sbjct: 346 GGGGGGFNKGNQGGGQGFAGNNYNTGGGGQGGNMGGGNRR 385
Score = 34.3 bits (77), Expect = 1.5
Identities = 23/53 (43%), Positives = 26/53 (48%), Gaps = 7/53 (13%)
Query: 799 RQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
RQG G G GG +AGG GG RG GGGG+ +R GG G G
Sbjct: 198 RQGGG-GGRGGP-----RAGGRGGQG-DRGQGGGGWGGQNRQNGGGNWGGAGG 243
Score = 33.1 bits (74), Expect = 3.2
Identities = 26/76 (34%), Positives = 29/76 (37%), Gaps = 15/76 (19%)
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGG----SGGYSRGRGMGGGGYQ 835
GGGG G R GR G G R GG +GG + G GGGG+
Sbjct: 200 GGGGGRGGPR-----------AGGRGGQGDRGQGGGGWGGQNRQNGGGNWGGAGGGGGFG 248
Query: 836 NSSRTQGGSALRGPHG 851
NS GG G G
Sbjct: 249 NSGGNFGGGQGGGSGG 264
>GRP1_SORBI (Q99069) Glycine-rich RNA-binding protein 1 (Fragment)
Length = 142
Score = 49.7 bits (117), Expect = 3e-05
Identities = 37/91 (40%), Positives = 38/91 (41%), Gaps = 16/91 (17%)
Query: 748 NERASEA-RLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAG 806
NE S R GGGG R G Y GGGG GG + G GR G
Sbjct: 61 NEAQSRGGRGGGGGGGYGGGRGGGGGYGRRDGGGGGYGGGGGGY----------GGGRGG 110
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGGYQNS 837
YGG GG GGY G GGGGY NS
Sbjct: 111 YGGGGYG----GGGGGYGGG-SRGGGGYGNS 136
Score = 48.9 bits (115), Expect = 6e-05
Identities = 31/66 (46%), Positives = 33/66 (49%), Gaps = 5/66 (7%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGY-SRGRGMGGGGYQNS 837
GGGGG GGR Y R+ G GYGG + GG GGY G G GGGGY
Sbjct: 71 GGGGGGYGGGRGGGGGYG--RRDGGGGGYGGGGGGY--GGGRGGYGGGGYGGGGGGYGGG 126
Query: 838 SRTQGG 843
SR GG
Sbjct: 127 SRGGGG 132
Score = 40.4 bits (93), Expect = 0.020
Identities = 29/79 (36%), Positives = 31/79 (38%), Gaps = 16/79 (20%)
Query: 775 AAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRAL----SFNQAGGSGGYSRGR-GM 829
A + GG GG GG G GR G GG GG GGY GR G
Sbjct: 63 AQSRGGRGGGGGGG-----------YGGGRGGGGGYGRRDGGGGGYGGGGGGYGGGRGGY 111
Query: 830 GGGGYQNSSRTQGGSALRG 848
GGGGY GG + G
Sbjct: 112 GGGGYGGGGGGYGGGSRGG 130
Score = 38.1 bits (87), Expect = 0.10
Identities = 30/79 (37%), Positives = 36/79 (44%), Gaps = 12/79 (15%)
Query: 776 AAAGGGGGTSSGGRWQDLSYSQTRQG---SGRAGYGGRALSFNQAGGSGGYSRGRGMGGG 832
+A G G GR ++ +Q+R G G GYGG GG GGY R G GGG
Sbjct: 43 SAIEGMNGKELDGRNITVNEAQSRGGRGGGGGGGYGG------GRGGGGGYGRRDG-GGG 95
Query: 833 GYQNSSRTQGGSALRGPHG 851
GY GG RG +G
Sbjct: 96 GYGGGGGGYGGG--RGGYG 112
Score = 37.4 bits (85), Expect = 0.17
Identities = 29/74 (39%), Positives = 30/74 (40%), Gaps = 16/74 (21%)
Query: 755 RLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSF 814
R GGGG RRDG GGGGG GGR Y G G GYGG
Sbjct: 81 RGGGGGYG----RRDG---GGGGYGGGGGGYGGGRG---GYGGGGYGGGGGGYGG----- 125
Query: 815 NQAGGSGGYSRGRG 828
+ G GGY G
Sbjct: 126 -GSRGGGGYGNSDG 138
>SPD1_NEPCL (P19837) Spidroin 1 (Dragline silk fibroin 1) (Fragment)
Length = 747
Score = 48.9 bits (115), Expect = 6e-05
Identities = 39/111 (35%), Positives = 49/111 (44%), Gaps = 10/111 (9%)
Query: 742 SILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQG 801
++ A + A + GG G R G AAAAAGG G GG QG
Sbjct: 373 AVAAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQRGYGGLGN--------QG 424
Query: 802 SGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSR-TQGGSALRGPHG 851
+GR G GG+ A +GG +G G GG G Q + R QG +A G G
Sbjct: 425 AGRGGLGGQGAGAAAAAAAGGAGQG-GYGGLGNQGAGRGGQGAAAAAGGAG 474
Score = 47.8 bits (112), Expect = 1e-04
Identities = 41/110 (37%), Positives = 51/110 (46%), Gaps = 23/110 (20%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR 810
A + LGG G G AAAAAGG G GG QG+GR G GG+
Sbjct: 324 AGQGGLGGQGAG------QGAGAAAAAAGGAGQGGYGGLGS--------QGAGRGGLGGQ 369
Query: 811 ---ALSFNQAGGS--GGY----SRGRGMGGGGYQNSSRTQGGSALRGPHG 851
A++ AGG+ GGY S+G G GG G ++ GG+ RG G
Sbjct: 370 GAGAVAAAAAGGAGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQRGYGG 419
Score = 47.8 bits (112), Expect = 1e-04
Identities = 37/96 (38%), Positives = 41/96 (42%), Gaps = 14/96 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GGL R GQ AAAAAGG G GG QG+GR G GG+
Sbjct: 122 GYGGLGNQGAGRGGQGAAAAAAGGAGQGGYGGLGS--------QGAGRGGLGGQG----- 168
Query: 817 AGGSGGYSRGRGMGG-GGYQNSSRTQGGSALRGPHG 851
AG + + G G GG GG QGG G G
Sbjct: 169 AGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGSQG 204
Score = 47.4 bits (111), Expect = 2e-04
Identities = 36/101 (35%), Positives = 43/101 (41%), Gaps = 9/101 (8%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR 810
A + GG G R G AAAAAGG G GG QG+GR G GG+
Sbjct: 54 AGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGS--------QGAGRGGLGGQ 105
Query: 811 ALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
A +GG +G G GG G Q + R G+A G
Sbjct: 106 GAGAAAAAAAGGAGQG-GYGGLGNQGAGRGGQGAAAAAAGG 145
Score = 46.6 bits (109), Expect = 3e-04
Identities = 36/111 (32%), Positives = 48/111 (42%), Gaps = 6/111 (5%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGG-----GTSSGGRWQDLSYSQTRQGSGRA 805
A + GG G R +G AAAAAGG G G G Q QG+GR
Sbjct: 250 AGQGGYGGLGSQGAGRGGEGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGSQGAGRG 309
Query: 806 GYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTR 856
G GG+ AGG+G G G G G ++ G A +G +G + ++
Sbjct: 310 GLGGQGAGAAAAGGAGQGGLG-GQGAGQGAGAAAAAAGGAGQGGYGGLGSQ 359
Score = 43.5 bits (101), Expect = 0.002
Identities = 35/103 (33%), Positives = 38/103 (35%), Gaps = 15/103 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GGL R GQ AAAA G G R Q G+G+ GYGG +
Sbjct: 477 GYGGLGSQGAGRGGQGAGAAAAAAVGAGQEGIRGQ---------GAGQGGYGGLGSQGSG 527
Query: 817 AGGSGGYSRGR------GMGGGGYQNSSRTQGGSALRGPHGDV 853
GG GG G G G GG QG A G V
Sbjct: 528 RGGLGGQGAGAAAAAAGGAGQGGLGGQGAGQGAGAAAAAAGGV 570
Score = 43.5 bits (101), Expect = 0.002
Identities = 38/110 (34%), Positives = 47/110 (42%), Gaps = 11/110 (10%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG-G 809
A+ A GG G + GQ A AAA GG GG QG+GR G G G
Sbjct: 538 AAAAAAGGAGQGGLGGQGAGQG-AGAAAAAAGGVRQGGYG-----GLGSQGAGRGGQGAG 591
Query: 810 RALSFNQAGGSGGY----SRGRGMGGGGYQNSSRTQGGSALRGPHGDVST 855
A + G GGY +G G GG G Q + G A +G +G V +
Sbjct: 592 AAAAAAGGAGQGGYGGLGGQGVGRGGLGGQGAGAAAAGGAGQGGYGGVGS 641
Score = 43.5 bits (101), Expect = 0.002
Identities = 36/93 (38%), Positives = 40/93 (42%), Gaps = 10/93 (10%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GGL R GQ AAAAAGG G GG QG+GR G G A +
Sbjct: 450 GYGGLGNQGAGRGGQG-AAAAAGGAGQGGYGGLGS--------QGAGRGGQGAGAAAAAA 500
Query: 817 AGGSGGYSRGRGMGGGGYQN-SSRTQGGSALRG 848
G RG+G G GGY S+ G L G
Sbjct: 501 VGAGQEGIRGQGAGQGGYGGLGSQGSGRGGLGG 533
Score = 42.7 bits (99), Expect = 0.004
Identities = 35/107 (32%), Positives = 45/107 (41%), Gaps = 12/107 (11%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGG-----GTSSGGRWQDLSYSQTRQGSGRAGYGGRA 811
G GGL G AAAAAGG G G G Q QG+GR G GG+
Sbjct: 160 GRGGLG-----GQGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGSQGAGRGGLGGQG 214
Query: 812 LSFNQAGGSGGYSRG--RGMGGGGYQNSSRTQGGSALRGPHGDVSTR 856
A +GG +G G G G +S G A +G +G + ++
Sbjct: 215 AGAAAAAAAGGAGQGGLGGQGAGQGAGASAAAAGGAGQGGYGGLGSQ 261
Score = 42.0 bits (97), Expect = 0.007
Identities = 37/101 (36%), Positives = 44/101 (42%), Gaps = 19/101 (18%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GGL GQ AAAA GG GG QG+GR G G A +
Sbjct: 98 GRGGLG-------GQGAGAAAAAAAGGAGQGGYG-----GLGNQGAGRGGQGAAAAAAGG 145
Query: 817 AGGSGGY----SRGRGMGGGGYQ--NSSRTQGGSALRGPHG 851
A G GGY S+G G GG G Q ++ G A +G +G
Sbjct: 146 A-GQGGYGGLGSQGAGRGGLGGQGAGAAAAAAGGAGQGGYG 185
Score = 41.2 bits (95), Expect = 0.012
Identities = 33/95 (34%), Positives = 42/95 (43%), Gaps = 13/95 (13%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG-GRALSFNQAGGSGGY----S 824
GQ AAAA GG GG QG+GR G G G A + G GGY S
Sbjct: 40 GQGAGAAAAAAAGGAGQGGYG-----GLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGS 94
Query: 825 RGRGMGGGGYQN---SSRTQGGSALRGPHGDVSTR 856
+G G GG G Q ++ G A +G +G + +
Sbjct: 95 QGAGRGGLGGQGAGAAAAAAAGGAGQGGYGGLGNQ 129
Score = 40.4 bits (93), Expect = 0.020
Identities = 38/114 (33%), Positives = 47/114 (40%), Gaps = 25/114 (21%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAA-------GGGGGTSS--------GGRWQDLSYSQTRQG 801
G GGL R GQ AAAA GG GG S GG+ + + G
Sbjct: 58 GYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAAGG 117
Query: 802 SGRAGYGGRALSFNQAGGSGGY------SRGRGMGG-GGYQNSSRTQGGSALRG 848
+G+ GYGG NQ G GG + G G GG GG + +GG +G
Sbjct: 118 AGQGGYGGLG---NQGAGRGGQGAAAAAAGGAGQGGYGGLGSQGAGRGGLGGQG 168
Score = 40.4 bits (93), Expect = 0.020
Identities = 33/110 (30%), Positives = 38/110 (34%), Gaps = 18/110 (16%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSG---------------GRWQDLSYSQTRQG 801
G GGL R GQ AAAA GG G G+ + + G
Sbjct: 386 GYGGLGSQGAGRGGQGAGAAAAAAGGAGQRGYGGLGNQGAGRGGLGGQGAGAAAAAAAGG 445
Query: 802 SGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+G+ GYGG NQ G GG GG G A RG G
Sbjct: 446 AGQGGYGGLG---NQGAGRGGQGAAAAAGGAGQGGYGGLGSQGAGRGGQG 492
Score = 40.0 bits (92), Expect = 0.027
Identities = 34/104 (32%), Positives = 40/104 (37%), Gaps = 14/104 (13%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR 810
A + GG G R G A AAA GG GG QG+G+ GYGG
Sbjct: 146 AGQGGYGGLGSQGAGRGGLGGQGAGAAAAAAGGAGQGGYG-----GLGGQGAGQGGYGGL 200
Query: 811 ALSFNQAGGSGGY---------SRGRGMGGGGYQNSSRTQGGSA 845
GG GG + G G GG G Q + + G SA
Sbjct: 201 GSQGAGRGGLGGQGAGAAAAAAAGGAGQGGLGGQGAGQGAGASA 244
Score = 39.3 bits (90), Expect = 0.045
Identities = 33/98 (33%), Positives = 40/98 (40%), Gaps = 24/98 (24%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGG------------RALSFNQA 817
G AAAAAGG G GG QG+G+ GYGG A +
Sbjct: 2 GAGAAAAAAGGAGQGGYGG--------LGGQGAGQGGYGGLGGQGAGQGAGAAAAAAAGG 53
Query: 818 GGSGGY----SRGRGMGGGGYQNSSRTQGGSALRGPHG 851
G GGY S+G G GG G ++ GG+ G G
Sbjct: 54 AGQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGG 91
Score = 39.3 bits (90), Expect = 0.045
Identities = 33/104 (31%), Positives = 41/104 (38%), Gaps = 9/104 (8%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGG---GGTSSGGRWQDLSYSQTRQG-SGRAGYGGRAL 812
G GGL G AAAAA GG GG G Q S G +G+ GYGG
Sbjct: 206 GRGGLG-----GQGAGAAAAAAAGGAGQGGLGGQGAGQGAGASAAAAGGAGQGGYGGLGS 260
Query: 813 SFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTR 856
GG G + GG G G A +G +G + ++
Sbjct: 261 QGAGRGGEGAGAAAAAAGGAGQGGYGGLGGQGAGQGGYGGLGSQ 304
Score = 38.5 bits (88), Expect = 0.077
Identities = 30/87 (34%), Positives = 35/87 (39%), Gaps = 9/87 (10%)
Query: 774 AAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGG--------YSR 825
AAAAAGG G GG+ + +G AG GG +Q G GG +
Sbjct: 219 AAAAAGGAGQGGLGGQGAGQGAGASAAAAGGAGQGGYGGLGSQGAGRGGEGAGAAAAAAG 278
Query: 826 GRGMGG-GGYQNSSRTQGGSALRGPHG 851
G G GG GG QGG G G
Sbjct: 279 GAGQGGYGGLGGQGAGQGGYGGLGSQG 305
Score = 37.4 bits (85), Expect = 0.17
Identities = 35/105 (33%), Positives = 40/105 (37%), Gaps = 19/105 (18%)
Query: 751 ASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGR 810
A + LGG G G +AAAAGG G GG QG+GR G G
Sbjct: 226 AGQGGLGGQGAG------QGAGASAAAAGGAGQGGYGGLGS--------QGAGRGGEGAG 271
Query: 811 ALSFNQAG----GSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
A + G G GG G+G G GGY G G G
Sbjct: 272 AAAAAAGGAGQGGYGGLG-GQGAGQGGYGGLGSQGAGRGGLGGQG 315
Score = 37.0 bits (84), Expect = 0.22
Identities = 36/102 (35%), Positives = 38/102 (36%), Gaps = 16/102 (15%)
Query: 751 ASEARLGG-GGLDLPPRRRDGQDY--AAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGY 807
A R GG GGL R GQ AAAAAGG G GG QG GR G
Sbjct: 567 AGGVRQGGYGGLGSQGAGRGGQGAGAAAAAAGGAGQGGYGGLGG--------QGVGRGGL 618
Query: 808 GGRALSFNQAGGSG-----GYSRGRGMGGGGYQNSSRTQGGS 844
GG+ AGG+G G G S Q S
Sbjct: 619 GGQGAGAAAAGGAGQGGYGGVGSGASAASAAASRLSSPQASS 660
Score = 37.0 bits (84), Expect = 0.22
Identities = 35/110 (31%), Positives = 45/110 (40%), Gaps = 24/110 (21%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
G GGL GQ A AA GG GG QG+GR G G A +
Sbjct: 362 GRGGLG-------GQGAGAVAAAAAGGAGQGGYG-----GLGSQGAGRGGQGAGAAA-AA 408
Query: 817 AGGSG----------GYSRGRGMGGGGYQNSSRTQGGSALRGPHGDVSTR 856
AGG+G G RG G+GG G ++ G A +G +G + +
Sbjct: 409 AGGAGQRGYGGLGNQGAGRG-GLGGQGAGAAAAAAAGGAGQGGYGGLGNQ 457
Score = 36.6 bits (83), Expect = 0.29
Identities = 35/114 (30%), Positives = 41/114 (35%), Gaps = 20/114 (17%)
Query: 748 NERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGS----G 803
N+ A LGG G AAAAA GG G G + + QG+ G
Sbjct: 422 NQGAGRGGLGG----------QGAGAAAAAAAGGAGQGGYGGLGNQGAGRGGQGAAAAAG 471
Query: 804 RAGYGGRALSFNQAGGSGGYSRGR------GMGGGGYQNSSRTQGGSALRGPHG 851
AG GG +Q G GG G G G G + QGG G G
Sbjct: 472 GAGQGGYGGLGSQGAGRGGQGAGAAAAAAVGAGQEGIRGQGAGQGGYGGLGSQG 525
Score = 32.3 bits (72), Expect = 5.5
Identities = 19/57 (33%), Positives = 28/57 (48%), Gaps = 4/57 (7%)
Query: 801 GSGRAGYGGRALSFNQAGGSGGYSRGRGMGGG-GYQNSSRTQGGSALRGPHGDVSTR 856
G+G+ GYGG Q G GGY G G G G ++ G A +G +G + ++
Sbjct: 12 GAGQGGYGGLG---GQGAGQGGYGGLGGQGAGQGAGAAAAAAAGGAGQGGYGGLGSQ 65
Score = 32.3 bits (72), Expect = 5.5
Identities = 26/88 (29%), Positives = 33/88 (36%), Gaps = 10/88 (11%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGR-- 827
GQ GG GG +G + + + G+G+ GYGG +Q G GG G
Sbjct: 23 GQGAGQGGYGGLGGQGAG-QGAGAAAAAAAGGAGQGGYGGLG---SQGAGRGGQGAGAAA 78
Query: 828 ----GMGGGGYQNSSRTQGGSALRGPHG 851
G G GGY G G G
Sbjct: 79 AAAGGAGQGGYGGLGSQGAGRGGLGGQG 106
>NOA1_BRARE (Q7ZVE0) Nucleolar protein family A member 1 (snoRNP
protein GAR1) (H/ACA ribonucleoprotein GAR1)
Length = 226
Score = 48.9 bits (115), Expect = 6e-05
Identities = 25/55 (45%), Positives = 29/55 (52%), Gaps = 1/55 (1%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSF-NQAGGSGGYSRGRGMGGG 832
G GGG GGR + G GR G+GGR F + GG GG+ GRG GGG
Sbjct: 165 GRGGGGGRGGRGGGFRGGRGANGGGRGGFGGRGGGFGGRGGGGGGFRGGRGGGGG 219
Score = 35.0 bits (79), Expect = 0.85
Identities = 22/56 (39%), Positives = 25/56 (44%), Gaps = 8/56 (14%)
Query: 801 GSGRAGYGG--RALSFNQAGGSGGYS------RGRGMGGGGYQNSSRTQGGSALRG 848
G GR G GG R GG GG+ GRG GGGG++ GG RG
Sbjct: 169 GGGRGGRGGGFRGGRGANGGGRGGFGGRGGGFGGRGGGGGGFRGGRGGGGGRGFRG 224
Score = 34.3 bits (77), Expect = 1.5
Identities = 21/50 (42%), Positives = 25/50 (50%), Gaps = 8/50 (16%)
Query: 802 SGRAGYGGRALSFNQAGG---SGGYSRGRGMG-----GGGYQNSSRTQGG 843
S R G GGR FN+ GG GG+ GRG G GGG+ +GG
Sbjct: 2 SFRGGGGGRGGGFNRGGGGGRGGGFGGGRGGGFGGGRGGGFGGGRGGRGG 51
Score = 32.3 bits (72), Expect = 5.5
Identities = 20/49 (40%), Positives = 21/49 (42%), Gaps = 6/49 (12%)
Query: 803 GRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
GR G GGR G GG+ GRG GGG GG RG G
Sbjct: 165 GRGGGGGRG------GRGGGFRGGRGANGGGRGGFGGRGGGFGGRGGGG 207
>K1CI_HUMAN (P35527) Keratin, type I cytoskeletal 9 (Cytokeratin 9)
(K9) (CK 9)
Length = 622
Score = 48.5 bits (114), Expect = 7e-05
Identities = 36/97 (37%), Positives = 44/97 (45%), Gaps = 18/97 (18%)
Query: 767 RRDGQDYAAAAAGGGGGTSSGGR----WQDLSYSQTRQGSGR----AGYGGRALSFNQAG 818
R+ Y + GGGGG SGG + S S R G GR +GYGG + G
Sbjct: 4 RQFSSSYLTSGGGGGGGLGSGGSIRSSYSRFSSSGGRGGGGRFSSSSGYGGGSSRVCGRG 63
Query: 819 GSG--GYSRGRGMG--------GGGYQNSSRTQGGSA 845
G G GYS G G G GGG+ SR GG++
Sbjct: 64 GGGSFGYSYGGGSGGGFSASSLGGGFGGGSRGFGGAS 100
Score = 46.6 bits (109), Expect = 3e-04
Identities = 36/108 (33%), Positives = 42/108 (38%), Gaps = 16/108 (14%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA---GYGGRALSF 814
GG R G Y +GGG G SG R GSG GYGG +
Sbjct: 481 GGSYGRGSRGGSGGSYGGGGSGGGYGGGSGSRGGSGGSYGGGSGSGGGSGGGYGGGSGGG 540
Query: 815 NQAGGSGGYSRGR-----------GMGGGGYQNSSRTQGGSALRGPHG 851
+ G GG+S G G GGGY S ++GGS G HG
Sbjct: 541 HSGGSGGGHSGGSGGNYGGGSGSGGGSGGGYGGGSGSRGGSG--GSHG 586
Score = 46.2 bits (108), Expect = 4e-04
Identities = 40/133 (30%), Positives = 55/133 (41%), Gaps = 11/133 (8%)
Query: 723 RNVELSRVQALAFQLTEKLSI---LAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAG 779
+N E S + ++ +L +++ L E + E+ G G + L R G Y + G
Sbjct: 432 QNQEYSLLLSIKMRLEKEIETYHNLLEGGQEDFESS-GAGKIGLGGRGGSGGSYGRGSRG 490
Query: 780 GGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSR 839
G GG+ GG S G G GG S+ GSGG S G G GGG S
Sbjct: 491 GSGGSYGGGG------SGGGYGGGSGSRGGSGGSYGGGSGSGGGSGG-GYGGGSGGGHSG 543
Query: 840 TQGGSALRGPHGD 852
GG G G+
Sbjct: 544 GSGGGHSGGSGGN 556
Score = 43.9 bits (102), Expect = 0.002
Identities = 36/97 (37%), Positives = 43/97 (44%), Gaps = 10/97 (10%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGG---TSSGGRWQDLSYSQTRQGSGRAGY------ 807
GGGGL R +++ G GGG +SS G S R G G GY
Sbjct: 17 GGGGLGSGGSIRSSYSRFSSSGGRGGGGRFSSSSGYGGGSSRVCGRGGGGSFGYSYGGGS 76
Query: 808 GGRALSFNQAGGSGGYSRG-RGMGGGGYQNSSRTQGG 843
GG + + GG GG SRG G GGGY +S GG
Sbjct: 77 GGGFSASSLGGGFGGGSRGFGGASGGGYSSSGGFGGG 113
Score = 38.1 bits (87), Expect = 0.10
Identities = 28/79 (35%), Positives = 32/79 (40%), Gaps = 2/79 (2%)
Query: 778 AGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGS-GGYSRGRGMGGGGYQN 836
+GG GG SGG + G GYGG + S +GGS GG S G GG Y
Sbjct: 542 SGGSGGGHSGGSGGNYGGGSGSGGGSGGGYGGGSGSRGGSGGSHGGGSGFGGESGGSYGG 601
Query: 837 SSRTQG-GSALRGPHGDVS 854
G G G G S
Sbjct: 602 GEEASGSGGGYGGGSGKSS 620
Score = 36.6 bits (83), Expect = 0.29
Identities = 28/87 (32%), Positives = 35/87 (40%), Gaps = 10/87 (11%)
Query: 755 RLGGGGLDLPPRRRDGQDYAAAAAGGG--------GGTSSGGRWQDLSYSQTRQGSGRAG 806
R GGG G ++A++ GGG GG S GG + G G
Sbjct: 62 RGGGGSFGYSYGGGSGGGFSASSLGGGFGGGSRGFGGASGGGYSSSGGFGGGFGGGSGGG 121
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGG 833
+GG S GG GG+ G G G GG
Sbjct: 122 FGGGYGS--GFGGLGGFGGGAGGGDGG 146
Score = 34.7 bits (78), Expect = 1.1
Identities = 35/118 (29%), Positives = 43/118 (35%), Gaps = 19/118 (16%)
Query: 742 SILAESNERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGG------GTSSGGRWQDLSY 795
SI + + +S GGGG G GGGG G SGG +
Sbjct: 26 SIRSSYSRFSSSGGRGGGGRFSSSSGYGGGSSRVCGRGGGGSFGYSYGGGSGG-----GF 80
Query: 796 SQTRQGSGRAGYGGRALSFNQAGG-----SGGYSRGRGMGGGGYQNSSRTQGGSALRG 848
S + G G +GG + F A G SGG+ G G G GG G L G
Sbjct: 81 SASSLGGG---FGGGSRGFGGASGGGYSSSGGFGGGFGGGSGGGFGGGYGSGFGGLGG 135
Score = 33.1 bits (74), Expect = 3.2
Identities = 20/57 (35%), Positives = 24/57 (42%), Gaps = 6/57 (10%)
Query: 776 AAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGMGGG 832
+ +GGG G GG S +R GSG + GG G GG G GGG
Sbjct: 561 SGSGGGSGGGYGGG------SGSRGGSGGSHGGGSGFGGESGGSYGGGEEASGSGGG 611
>CAZ_DROME (Q27294) RNA-binding protein cabeza (Sarcoma-associated
RNA-binding fly homolog) (P19)
Length = 399
Score = 48.5 bits (114), Expect = 7e-05
Identities = 43/127 (33%), Positives = 48/127 (36%), Gaps = 32/127 (25%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGR-------WQDLSYSQTR---------- 799
GGGG R G GGGGG GG W+ S + T
Sbjct: 239 GGGGGGRFDRGGGGGGGRYDRGGGGGGGGGGGNVQPRDGDWKCNSCNNTNFAWRNECNRC 298
Query: 800 -------QGSGRAGYGGRALSFNQAGGSGGYSRG--RGMGGGGYQNSSR---TQGGSALR 847
+GS G GG + GG GGY RG RG GGGGY N R +QGG
Sbjct: 299 KTPKGDDEGSSGGGGGG---GYGGGGGGGGYDRGNDRGSGGGGYHNRDRGGNSQGGGGGG 355
Query: 848 GPHGDVS 854
G G S
Sbjct: 356 GGGGGYS 362
Score = 45.1 bits (105), Expect = 8e-04
Identities = 32/82 (39%), Positives = 35/82 (42%), Gaps = 8/82 (9%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM 829
G D ++ GGGGG GG R GSG GY R N GG GG G
Sbjct: 303 GDDEGSSGGGGGGGYGGGGGGGGYDRGNDR-GSGGGGYHNRDRGGNSQGGGGG----GG- 356
Query: 830 GGGGYQNSSRTQGGSALRGPHG 851
GGGGY + GG RG G
Sbjct: 357 GGGGYSRFNDNNGGG--RGGRG 376
Score = 40.4 bits (93), Expect = 0.020
Identities = 31/82 (37%), Positives = 34/82 (40%), Gaps = 23/82 (28%)
Query: 779 GGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGSGG----YSRGRGMGGGGY 834
GGGGG GG G GR G+GGR GG GG + RG G GGG Y
Sbjct: 213 GGGGG---GG------------GGGRGGFGGRRGGGGGGGGGGGGGGRFDRGGGGGGGRY 257
Query: 835 QNSSRTQGGSALRGPHGDVSTR 856
GG G G+V R
Sbjct: 258 DRG----GGGGGGGGGGNVQPR 275
Score = 36.6 bits (83), Expect = 0.29
Identities = 28/79 (35%), Positives = 33/79 (41%), Gaps = 17/79 (21%)
Query: 758 GGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRAL-SFNQ 816
GGG D R + G G G GG W D +G G GG + S+N+
Sbjct: 45 GGGYDSGSGHR-------GSGGSGNGGGGGGSWND-------RGGNSYGNGGASKDSYNK 90
Query: 817 AGG--SGGYSRGRGMGGGG 833
G SGG G G GGGG
Sbjct: 91 GHGGYSGGGGGGGGGGGGG 109
Score = 35.8 bits (81), Expect = 0.50
Identities = 32/103 (31%), Positives = 38/103 (36%), Gaps = 19/103 (18%)
Query: 753 EARLGGGGLDLPPRRRDGQDYAAAAAGGG------GGTSSGGRWQDLSYSQTRQGSGRAG 806
E GGGG G Y GGG G+ GG ++ G G G
Sbjct: 306 EGSSGGGG---------GGGYGGGGGGGGYDRGNDRGSGGGGYHNRDRGGNSQGGGGGGG 356
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGGYQNSS--RTQGGSALR 847
GG FN +GG GRG GGG ++ R GG R
Sbjct: 357 GGGGYSRFND--NNGGGRGGRGGGGGNRRDGGPMRNDGGMRSR 397
Score = 32.7 bits (73), Expect = 4.2
Identities = 26/80 (32%), Positives = 33/80 (40%), Gaps = 20/80 (25%)
Query: 749 ERASEARLGGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYG 808
+R ++ GGGG +D + GGGGG GG YS+ +G G G
Sbjct: 327 DRGNDRGSGGGGY-------HNRDRGGNSQGGGGGGGGGG-----GYSRFNDNNG-GGRG 373
Query: 809 GRALSFNQAGGSGGYSRGRG 828
GR GG GG R G
Sbjct: 374 GR-------GGGGGNRRDGG 386
>LORI_MOUSE (P18165) Loricrin
Length = 486
Score = 48.1 bits (113), Expect = 1e-04
Identities = 35/94 (37%), Positives = 41/94 (43%), Gaps = 14/94 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G +GGGGG+S GG SYS G G G + ++
Sbjct: 107 GGGG---------GSSCGGGYSGGGGGSSCGGG----SYSGGGSSCGGGGGSGGGVKYSG 153
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPH 850
GG GG S G G GGG SS GGS G +
Sbjct: 154 GGGGGGSSCGGGSSGGGGGGSS-CGGGSGGGGSY 186
Score = 46.2 bits (108), Expect = 4e-04
Identities = 39/106 (36%), Positives = 45/106 (41%), Gaps = 16/106 (15%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGG--GGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSF 814
GGGG Q Y + GGG GG+S GGR YS S GY G S
Sbjct: 267 GGGG--------SSQQYQCQSYGGGSSGGSSCGGR-----YSGGGGSSCGGGYSGGGGSS 313
Query: 815 NQAGGSGGYSRGRGMGGGGYQ-NSSRTQGGSALRGPHGDVSTRMVS 859
G SGG S G GGGGY + GG + G G S++ S
Sbjct: 314 CGGGSSGGGSSCGGSGGGGYSGGGGGSCGGGSSGGGGGYYSSQQTS 359
Score = 44.7 bits (104), Expect = 0.001
Identities = 37/109 (33%), Positives = 44/109 (39%), Gaps = 14/109 (12%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGG-----GGTSSGGRWQDLSYSQTRQGSG-RAGYGGR 810
GGGG G Y ++GGG GG S GG+ YS GS GY G
Sbjct: 170 GGGGSSCGGGSGGGGSYCGGSSGGGSSGGCGGGSGGGK-----YSGGGGGSSCGGGYSGG 224
Query: 811 ALSFNQAGGSGGYSRGRGM---GGGGYQNSSRTQGGSALRGPHGDVSTR 856
S + GGYS G G GGGGY + G G G S++
Sbjct: 225 GGSSGGSSCGGGYSGGGGSSCGGGGGYSGGGGSSCGGGSSGGGGGGSSQ 273
Score = 44.3 bits (103), Expect = 0.001
Identities = 32/99 (32%), Positives = 43/99 (43%), Gaps = 13/99 (13%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G ++ GGGGG+ G ++ + G G GG +
Sbjct: 55 GGGG---------GSYGGGSSCGGGGGSGGGVKYSGGGGGSSCGGGYSGGGGGSSCGGGY 105
Query: 817 AGGSGGYSRGRGMGGGGYQNS----SRTQGGSALRGPHG 851
+GG GG S G G GGG +S S + GGS+ G G
Sbjct: 106 SGGGGGSSCGGGYSGGGGGSSCGGGSYSGGGSSCGGGGG 144
Score = 44.3 bits (103), Expect = 0.001
Identities = 32/89 (35%), Positives = 39/89 (42%), Gaps = 13/89 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G Y+ + GGG SGG + YS G G + GG +
Sbjct: 121 GGGGSSC-----GGGSYSGGGSSCGGGGGSGG---GVKYSGGGGGGGSSCGGG-----SS 167
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSA 845
GG GG S G G GGGG + GGS+
Sbjct: 168 GGGGGGSSCGGGSGGGGSYCGGSSGGGSS 196
Score = 43.9 bits (102), Expect = 0.002
Identities = 35/91 (38%), Positives = 40/91 (43%), Gaps = 13/91 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSS--GGRWQDLSYSQTRQGSGRAG-YGGRALS 813
GGGG G ++GGGGG SS GG SY G G +G GG +
Sbjct: 155 GGGG---------GSSCGGGSSGGGGGGSSCGGGSGGGGSYCGGSSGGGSSGGCGGGSGG 205
Query: 814 FNQAGGSGGYSRGRGM-GGGGYQNSSRTQGG 843
+GG GG S G G GGGG S GG
Sbjct: 206 GKYSGGGGGSSCGGGYSGGGGSSGGSSCGGG 236
Score = 42.4 bits (98), Expect = 0.005
Identities = 38/101 (37%), Positives = 41/101 (39%), Gaps = 15/101 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGG---GGTSSGGRWQDLSYSQTRQGSGR--AGYGGRA 811
GGGG G Y+ GGG GG SSGG S G G G G
Sbjct: 141 GGGGSG------GGVKYSGGGGGGGSSCGGGSSGGGGGGSSCGGGSGGGGSYCGGSSGGG 194
Query: 812 LSFNQAGGSGG--YSRGRGMG--GGGYQNSSRTQGGSALRG 848
S GGSGG YS G G GGGY + GGS+ G
Sbjct: 195 SSGGCGGGSGGGKYSGGGGGSSCGGGYSGGGGSSGGSSCGG 235
Score = 41.2 bits (95), Expect = 0.012
Identities = 37/105 (35%), Positives = 42/105 (39%), Gaps = 22/105 (20%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQ--------DLSYSQTRQGS--GRAG 806
GGGG G +GGGGG+S GG + YS GS G
Sbjct: 81 GGGG---------GSSCGGGYSGGGGGSSCGGGYSGGGGGSSCGGGYSGGGGGSSCGGGS 131
Query: 807 YGGRALSFNQAGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
Y G S GGSGG + G GGGG S GGS+ G G
Sbjct: 132 YSGGGSSCGGGGGSGGGVKYSGGGGGG---GSSCGGGSSGGGGGG 173
Score = 40.8 bits (94), Expect = 0.016
Identities = 34/101 (33%), Positives = 39/101 (37%), Gaps = 15/101 (14%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G ++ GGGG SS Q Q G G +G GG +
Sbjct: 335 GGGG---------GSCGGGSSGGGGGYYSSQQTSQTSCAPQQSYGGGSSGGGGSCGGGSS 385
Query: 817 AGGSGG--YSRGRGMG----GGGYQNSSRTQGGSALRGPHG 851
GG GG YS G G GGGY GG + G G
Sbjct: 386 GGGGGGGCYSSGGGGSSGGCGGGYSGGGGGCGGGSSGGSGG 426
Score = 40.0 bits (92), Expect = 0.027
Identities = 32/86 (37%), Positives = 40/86 (46%), Gaps = 14/86 (16%)
Query: 778 AGGGGGTSSGGRWQDLSYSQ--TRQGSGRAGYGGRALSFNQAGGSGGYSRGRGM------ 829
+GGGGG+S GG + S + G G +G GG + GG GGYS G G
Sbjct: 209 SGGGGGSSCGGGYSGGGGSSGGSSCGGGYSGGGG-----SSCGGGGGYSGGGGSSCGGGS 263
Query: 830 -GGGGYQNSSRTQGGSALRGPHGDVS 854
GGGG +S + Q S G G S
Sbjct: 264 SGGGGGGSSQQYQCQSYGGGSSGGSS 289
Score = 39.7 bits (91), Expect = 0.035
Identities = 29/75 (38%), Positives = 33/75 (43%), Gaps = 6/75 (8%)
Query: 776 AAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQAGGS--GGYSRGRGMGGGG 833
+ GGGGG GG + YS G G GG + GGS GG S G G G GG
Sbjct: 20 SGGGGGGGGGGGGGY----YSGGGSGCGGGSSGGGSSCGGGGGGSYGGGSSCGGGGGSGG 75
Query: 834 YQNSSRTQGGSALRG 848
S GGS+ G
Sbjct: 76 GVKYSGGGGGSSCGG 90
Score = 39.7 bits (91), Expect = 0.035
Identities = 29/95 (30%), Positives = 34/95 (35%), Gaps = 4/95 (4%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G Y+ GG SSGG S Q G GG +
Sbjct: 239 GGGGSSCG----GGGGYSGGGGSSCGGGSSGGGGGGSSQQYQCQSYGGGSSGGSSCGGRY 294
Query: 817 AGGSGGYSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+GG G G GGGG + GG + G G
Sbjct: 295 SGGGGSSCGGGYSGGGGSSCGGGSSGGGSSCGGSG 329
Score = 39.3 bits (90), Expect = 0.045
Identities = 35/92 (38%), Positives = 40/92 (43%), Gaps = 16/92 (17%)
Query: 771 QDYAAAAAGGGG---GTSSGGRWQDLSYSQTRQGS----------GRAGYGGRALSFNQA 817
Q Y ++GGGG G SSGG YS GS G G GG + +
Sbjct: 367 QSYGGGSSGGGGSCGGGSSGGGGGGGCYSSGGGGSSGGCGGGYSGGGGGCGGGSSGGSGG 426
Query: 818 GGSGGYSRGRGMG-GGGYQNSSRTQGGSALRG 848
G GG S G G G GGGY S GGS+ G
Sbjct: 427 GCGGGSSGGSGGGCGGGY--SGGGGGGSSCGG 456
Score = 37.7 bits (86), Expect = 0.13
Identities = 30/87 (34%), Positives = 35/87 (39%), Gaps = 8/87 (9%)
Query: 770 GQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGS--GRAGYGGRALSFNQAGGSGGYSRGR 827
G Y+ G GG SSGG S QT Q S + YGG + G GG G
Sbjct: 330 GGGYSGGGGGSCGGGSSGGGGGYYSSQQTSQTSCAPQQSYGGGS------SGGGGSCGGG 383
Query: 828 GMGGGGYQNSSRTQGGSALRGPHGDVS 854
GGGG + GG + G G S
Sbjct: 384 SSGGGGGGGCYSSGGGGSSGGCGGGYS 410
Score = 36.2 bits (82), Expect = 0.38
Identities = 32/108 (29%), Positives = 41/108 (37%), Gaps = 13/108 (12%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRA-------GYGG 809
GGGG G ++ GGGG SGG + G G + YGG
Sbjct: 223 GGGGSSGGSSCGGGYSGGGGSSCGGGGGYSGGGGSSCGGGSSGGGGGGSSQQYQCQSYGG 282
Query: 810 RALSFNQAG----GSGGYSRGRGMGGGGYQN--SSRTQGGSALRGPHG 851
+ + G G GG S G G GGG + + GGS+ G G
Sbjct: 283 GSSGGSSCGGRYSGGGGSSCGGGYSGGGGSSCGGGSSGGGSSCGGSGG 330
Score = 35.8 bits (81), Expect = 0.50
Identities = 36/101 (35%), Positives = 41/101 (39%), Gaps = 21/101 (20%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGG----TSSGGRWQDLSYSQTRQGSGRAGYGGRAL 812
GGGG G Y+ +G GGG SS G SY GS G GG
Sbjct: 28 GGGG---------GGYYSGGGSGCGGGSSGGGSSCGGGGGGSYG---GGSSCGGGGGSGG 75
Query: 813 SFNQAGGSGGYSRGRGMGGGGYQNS-----SRTQGGSALRG 848
+GG GG S G G GGG +S S GGS+ G
Sbjct: 76 GVKYSGGGGGSSCGGGYSGGGGGSSCGGGYSGGGGGSSCGG 116
Score = 35.0 bits (79), Expect = 0.85
Identities = 34/96 (35%), Positives = 39/96 (40%), Gaps = 6/96 (6%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G ++GGGG SSGG S G G +G G
Sbjct: 375 GGGGSCGGGSSGGGGGGGCYSSGGGG--SSGGCGGGYSGGGGGCGGGSSGGSGGGCGGGS 432
Query: 817 AGGSGG-YSRGRGMGGGGYQNSSRTQGGSALRGPHG 851
+GGSGG G GGGG S GGS+ G G
Sbjct: 433 SGGSGGGCGGGYSGGGGG---GSSCGGGSSGGGSGG 465
Score = 32.3 bits (72), Expect = 5.5
Identities = 30/78 (38%), Positives = 33/78 (41%), Gaps = 25/78 (32%)
Query: 757 GGGGLDLPPRRRDGQDYAAAAAGGGGGTSSGGRWQDLSYSQTRQGSGRAGYGGRALSFNQ 816
GGGG G + + GG GG SSGG S G G +G GG
Sbjct: 412 GGGGC--------GGGSSGGSGGGCGGGSSGG-------SGGGCGGGYSGGGG------- 449
Query: 817 AGGS--GGYSRGRGMGGG 832
GGS GG S G G GGG
Sbjct: 450 -GGSSCGGGSSGGGSGGG 466
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.316 0.134 0.380
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 101,917,936
Number of Sequences: 164201
Number of extensions: 4647670
Number of successful extensions: 36928
Number of sequences better than 10.0: 666
Number of HSP's better than 10.0 without gapping: 199
Number of HSP's successfully gapped in prelim test: 494
Number of HSP's that attempted gapping in prelim test: 23529
Number of HSP's gapped (non-prelim): 4730
length of query: 865
length of database: 59,974,054
effective HSP length: 119
effective length of query: 746
effective length of database: 40,434,135
effective search space: 30163864710
effective search space used: 30163864710
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 70 (31.6 bits)
Medicago: description of AC137602.5


