

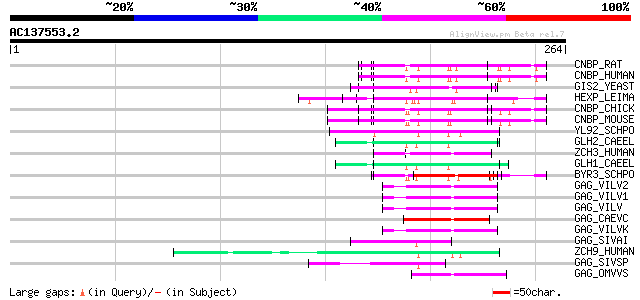
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137553.2 - phase: 0
(264 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP) (... 60 7e-09
CNBP_HUMAN (P62633) Cellular nucleic acid binding protein (CNBP)... 60 7e-09
GIS2_YEAST (P53849) Zinc-finger protein GIS2 59 9e-09
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 59 2e-08
CNBP_CHICK (O42395) Cellular nucleic acid binding protein (CNBP) 58 2e-08
CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)... 57 4e-08
YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I 57 6e-08
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 57 6e-08
ZCH3_HUMAN (Q9NUD5) Zinc finger CCHC domain containing protein 3 56 8e-08
GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-... 54 4e-07
BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog 51 3e-06
GAG_VILV2 (P23425) Gag polyprotein [Contains: Core protein p16; ... 50 6e-06
GAG_VILV1 (P23424) Gag polyprotein [Contains: Core protein p16; ... 50 6e-06
GAG_VILV (P03352) Gag polyprotein [Contains: Core protein p16; C... 50 6e-06
GAG_CAEVC (P33458) Gag polyprotein [Contains: Core protein p16; ... 49 1e-05
GAG_VILVK (P35955) Gag polyprotein [Contains: Core protein p16; ... 47 5e-05
GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17; ... 45 2e-04
ZCH9_HUMAN (Q8N567) Zinc finger CCHC domain containing protein 9 44 3e-04
GAG_SIVSP (P19504) Gag polyprotein [Contains: Core protein p17; ... 44 3e-04
GAG_OMVVS (P16900) Gag polyprotein [Contains: Core protein p16; ... 44 3e-04
>CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 59.7 bits (143), Expect = 7e-09
Identities = 29/93 (31%), Positives = 43/93 (46%), Gaps = 11/93 (11%)
Query: 173 VCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECK----RGDIVCFNCDEEGHLTSQCK 228
+CY+CGE GH + C+ +E C+ CG+ H +CK + C+NC + GHL C
Sbjct: 53 ICYRCGESGHLAKDCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCD 112
Query: 229 KPKK------GQASVYDSDYVLTKVMNKCGELG 255
+ G+ D K +CGE G
Sbjct: 113 HADEQKCYSCGEFGHIQKDCTKVKCY-RCGETG 144
Score = 56.6 bits (135), Expect = 6e-08
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query: 167 DDPAEIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAEC-KRGDIVCFNCDEEGHLTS 225
D E CY CGE GH C + KC+RCG+ H C K ++ C+ C E GHL
Sbjct: 112 DHADEQKCYSCGEFGHIQKDCT--KVKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 169
Query: 226 QC 227
+C
Sbjct: 170 EC 171
Score = 52.8 bits (125), Expect = 9e-07
Identities = 32/98 (32%), Positives = 43/98 (43%), Gaps = 10/98 (10%)
Query: 168 DPAEIVCYKCGEKGHKSNVC----NGEEKKCFRCGKKWHTIAECKRGD-IVCFNCDEEGH 222
D E CY CG GH + C E+ C+ CGK H +C D C++C E GH
Sbjct: 68 DLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFGH 127
Query: 223 LTSQCKKPK---KGQASVYDSDYVLTKVMN--KCGELG 255
+ C K K G+ + T +N +CGE G
Sbjct: 128 IQKDCTKVKCYRCGETGHVAINCSKTSEVNCYRCGESG 165
Score = 51.2 bits (121), Expect = 3e-06
Identities = 29/121 (23%), Positives = 43/121 (34%), Gaps = 39/121 (32%)
Query: 174 CYKCGEKGHKSNVC-------NGEEKK---------------------CFRCGKKWHTIA 205
C+KCG GH + C G + C+RCG+ H
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 206 ECKRGDIVCFNCDEEGHLTSQCKKPKKGQAS-----------VYDSDYVLTKVMNKCGEL 254
+C + C+NC GH+ CK+PK+ + D D+ + CGE
Sbjct: 66 DCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEF 125
Query: 255 G 255
G
Sbjct: 126 G 126
Score = 29.6 bits (65), Expect = 7.8
Identities = 10/21 (47%), Positives = 14/21 (66%)
Query: 170 AEIVCYKCGEKGHKSNVCNGE 190
+E+ CY+CGE GH + C E
Sbjct: 154 SEVNCYRCGESGHLARECTIE 174
>CNBP_HUMAN (P62633) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 59.7 bits (143), Expect = 7e-09
Identities = 29/93 (31%), Positives = 43/93 (46%), Gaps = 11/93 (11%)
Query: 173 VCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECK----RGDIVCFNCDEEGHLTSQCK 228
+CY+CGE GH + C+ +E C+ CG+ H +CK + C+NC + GHL C
Sbjct: 53 ICYRCGESGHLAKDCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCD 112
Query: 229 KPKK------GQASVYDSDYVLTKVMNKCGELG 255
+ G+ D K +CGE G
Sbjct: 113 HADEQKCYSCGEFGHIQKDCTKVKCY-RCGETG 144
Score = 56.6 bits (135), Expect = 6e-08
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query: 167 DDPAEIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAEC-KRGDIVCFNCDEEGHLTS 225
D E CY CGE GH C + KC+RCG+ H C K ++ C+ C E GHL
Sbjct: 112 DHADEQKCYSCGEFGHIQKDCT--KVKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 169
Query: 226 QC 227
+C
Sbjct: 170 EC 171
Score = 52.8 bits (125), Expect = 9e-07
Identities = 32/98 (32%), Positives = 43/98 (43%), Gaps = 10/98 (10%)
Query: 168 DPAEIVCYKCGEKGHKSNVC----NGEEKKCFRCGKKWHTIAECKRGD-IVCFNCDEEGH 222
D E CY CG GH + C E+ C+ CGK H +C D C++C E GH
Sbjct: 68 DLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFGH 127
Query: 223 LTSQCKKPK---KGQASVYDSDYVLTKVMN--KCGELG 255
+ C K K G+ + T +N +CGE G
Sbjct: 128 IQKDCTKVKCYRCGETGHVAINCSKTSEVNCYRCGESG 165
Score = 51.2 bits (121), Expect = 3e-06
Identities = 29/121 (23%), Positives = 43/121 (34%), Gaps = 39/121 (32%)
Query: 174 CYKCGEKGHKSNVC-------NGEEKK---------------------CFRCGKKWHTIA 205
C+KCG GH + C G + C+RCG+ H
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 206 ECKRGDIVCFNCDEEGHLTSQCKKPKKGQAS-----------VYDSDYVLTKVMNKCGEL 254
+C + C+NC GH+ CK+PK+ + D D+ + CGE
Sbjct: 66 DCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEF 125
Query: 255 G 255
G
Sbjct: 126 G 126
Score = 29.6 bits (65), Expect = 7.8
Identities = 10/21 (47%), Positives = 14/21 (66%)
Query: 170 AEIVCYKCGEKGHKSNVCNGE 190
+E+ CY+CGE GH + C E
Sbjct: 154 SEVNCYRCGESGHLARECTIE 174
>GIS2_YEAST (P53849) Zinc-finger protein GIS2
Length = 153
Score = 59.3 bits (142), Expect = 9e-09
Identities = 25/71 (35%), Positives = 40/71 (56%), Gaps = 5/71 (7%)
Query: 163 PKRRDDPAEIVCYKCGEKGHKSNVCNGEEK----KCFRCGKKWHTIAECKRGDIVCFNCD 218
PK+ +++ CYKCG H + C E+ KC+ CG+ H +C+ D +C+NC+
Sbjct: 83 PKKTSRFSKVSCYKCGGPNHMAKDCMKEDGISGLKCYTCGQAGHMSRDCQN-DRLCYNCN 141
Query: 219 EEGHLTSQCKK 229
E GH++ C K
Sbjct: 142 ETGHISKDCPK 152
Score = 58.2 bits (139), Expect = 2e-08
Identities = 26/70 (37%), Positives = 39/70 (55%), Gaps = 6/70 (8%)
Query: 167 DDPAEIVCYKCGEKGHKSNVCNG----EEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGH 222
D +E +CY C + GH C E K+C+ CG+ H +EC CFNC++ GH
Sbjct: 18 DCDSERLCYNCNKPGHVQTDCTMPRTVEFKQCYNCGETGHVRSECTVQR--CFNCNQTGH 75
Query: 223 LTSQCKKPKK 232
++ +C +PKK
Sbjct: 76 ISRECPEPKK 85
Score = 44.3 bits (103), Expect = 3e-04
Identities = 18/69 (26%), Positives = 32/69 (46%), Gaps = 11/69 (15%)
Query: 174 CYKCGEKGHKSNVCNGEEK-------KCFRCGKKWHTIAECKRGD----IVCFNCDEEGH 222
C+ C + GH S C +K C++CG H +C + D + C+ C + GH
Sbjct: 67 CFNCNQTGHISRECPEPKKTSRFSKVSCYKCGGPNHMAKDCMKEDGISGLKCYTCGQAGH 126
Query: 223 LTSQCKKPK 231
++ C+ +
Sbjct: 127 MSRDCQNDR 135
Score = 40.8 bits (94), Expect = 0.003
Identities = 20/65 (30%), Positives = 31/65 (46%), Gaps = 11/65 (16%)
Query: 191 EKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQASVYDSDYVLTKVMNK 250
+K C+ CGK H +C + +C+NC++ GH+ + C P+ V K
Sbjct: 3 QKACYVCGKIGHLAEDCD-SERLCYNCNKPGHVQTDCTMPRT----------VEFKQCYN 51
Query: 251 CGELG 255
CGE G
Sbjct: 52 CGETG 56
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 58.5 bits (140), Expect = 2e-08
Identities = 31/113 (27%), Positives = 44/113 (38%), Gaps = 23/113 (20%)
Query: 166 RDDPAEIVCYKCGEKGHKSNVCNGEEKK-------CFRCGKKWHTIAECKRG-------D 211
+ D C++CGE+GH S C E + CFRCG+ H +C
Sbjct: 37 KGDERSTTCFRCGEEGHMSRECPNEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKG 96
Query: 212 IVCFNCDEEGHLTSQCKKPKKGQASVY---------DSDYVLTKVMNKCGELG 255
C+ C +EGHL+ C + G Y Y + KCG+ G
Sbjct: 97 FECYKCGQEGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAG 149
Score = 55.8 bits (133), Expect = 1e-07
Identities = 25/71 (35%), Positives = 32/71 (44%), Gaps = 17/71 (23%)
Query: 174 CYKCGEKGHKSNVC------NGEEKKCFRCGKKWHTIAECKR-----------GDIVCFN 216
CYKCGE GH S C ++ C++CGK H EC GD C+
Sbjct: 198 CYKCGESGHMSRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGSYGGSRGGGDRTCYK 257
Query: 217 CDEEGHLTSQC 227
C E GH++ C
Sbjct: 258 CGEAGHISRDC 268
Score = 55.5 bits (132), Expect = 1e-07
Identities = 34/111 (30%), Positives = 49/111 (43%), Gaps = 24/111 (21%)
Query: 159 DERRPKRRDDPAEIVCYKCGEKGHKSNVC-----NGEEKK--CFRCGKKWHTIAECKR-- 209
D +RP+ + C CG++GH + C G+E+ CFRCG++ H EC
Sbjct: 6 DVKRPRTESSTS---CRNCGKEGHYARECPEADSKGDERSTTCFRCGEEGHMSRECPNEA 62
Query: 210 -----GDIVCFNCDEEGHLTSQCKKPKKGQASVYDSDYVLTKVMNKCGELG 255
G + CF C E GH++ C K A+ Y KCG+ G
Sbjct: 63 RSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECY-------KCGQEG 106
Score = 55.1 bits (131), Expect = 2e-07
Identities = 39/137 (28%), Positives = 57/137 (41%), Gaps = 27/137 (19%)
Query: 138 GQQG---RPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC-NGE--- 190
GQ+G R P S ++G R + + CYKCG+ GH S C NG+
Sbjct: 103 GQEGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQGGY 162
Query: 191 ----EKKCFRCGKKWHTIAECKRG--------DIVCFNCDEEGHLTSQCKKPKKGQASVY 238
++ C++CG H +C G D C+ C E GH++ +C P G
Sbjct: 163 SGAGDRTCYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSREC--PSAGSTGSG 220
Query: 239 DSDYVLTKVMNKCGELG 255
D + KCG+ G
Sbjct: 221 D------RACYKCGKPG 231
Score = 49.7 bits (117), Expect = 7e-06
Identities = 28/113 (24%), Positives = 42/113 (36%), Gaps = 37/113 (32%)
Query: 174 CYKCGEKGHKSNVCNGEE-----------------------KKCFRCGKKWHTIAECKRG 210
CYKCG++GH S C + + C++CG H +C G
Sbjct: 99 CYKCGQEGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNG 158
Query: 211 --------DIVCFNCDEEGHLTSQCKKPKKGQASVYDSDYVLTKVMNKCGELG 255
D C+ C + GH++ C + G + D + KCGE G
Sbjct: 159 QGGYSGAGDRTCYKCGDAGHISRDCPNGQGGYSGAGD------RKCYKCGESG 205
>CNBP_CHICK (O42395) Cellular nucleic acid binding protein (CNBP)
Length = 172
Score = 58.2 bits (139), Expect = 2e-08
Identities = 30/94 (31%), Positives = 44/94 (45%), Gaps = 12/94 (12%)
Query: 173 VCYKCGEKGHKSNVCN-GEEKKCFRCGKKWHTIAECK----RGDIVCFNCDEEGHLTSQC 227
+CY+CGE GH + C+ E+K C+ CG+ H +CK + C+NC + GHL C
Sbjct: 47 ICYRCGESGHLAKDCDLQEDKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 106
Query: 228 KKPKK------GQASVYDSDYVLTKVMNKCGELG 255
+ G+ D K +CGE G
Sbjct: 107 DHADEQKCYSCGEFGHIQKDCTKVKCY-RCGETG 139
Score = 57.4 bits (137), Expect = 4e-08
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 6/79 (7%)
Query: 152 KGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC-NGEEKKCFRCGKKWHTIAECKRG 210
+G D + PKR E CY CG+ GH + C + +E+KC+ CG+ H +C +
Sbjct: 74 RGGHIAKDCKEPKRE---REQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK- 129
Query: 211 DIVCFNCDEEGHLTSQCKK 229
+ C+ C E GH+ C K
Sbjct: 130 -VKCYRCGETGHVAINCSK 147
Score = 56.6 bits (135), Expect = 6e-08
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query: 167 DDPAEIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAEC-KRGDIVCFNCDEEGHLTS 225
D E CY CGE GH C + KC+RCG+ H C K ++ C+ C E GHL
Sbjct: 107 DHADEQKCYSCGEFGHIQKDCT--KVKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 164
Query: 226 QC 227
+C
Sbjct: 165 EC 166
Score = 51.2 bits (121), Expect = 3e-06
Identities = 30/116 (25%), Positives = 44/116 (37%), Gaps = 34/116 (29%)
Query: 174 CYKCGEKGHKSNVC-------NGEEKK---------------CFRCGKKWHTIAECK-RG 210
C+KCG GH + C G + C+RCG+ H +C +
Sbjct: 6 CFKCGRTGHWARECPTGIGRGRGMRSRGRAGFQFMSSSLPDICYRCGESGHLAKDCDLQE 65
Query: 211 DIVCFNCDEEGHLTSQCKKPKKGQAS-----------VYDSDYVLTKVMNKCGELG 255
D C+NC GH+ CK+PK+ + D D+ + CGE G
Sbjct: 66 DKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFG 121
Score = 29.6 bits (65), Expect = 7.8
Identities = 10/21 (47%), Positives = 14/21 (66%)
Query: 170 AEIVCYKCGEKGHKSNVCNGE 190
+E+ CY+CGE GH + C E
Sbjct: 149 SEVNCYRCGESGHLARECTIE 169
>CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 178
Score = 57.4 bits (137), Expect = 4e-08
Identities = 27/79 (34%), Positives = 41/79 (51%), Gaps = 6/79 (7%)
Query: 152 KGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC-NGEEKKCFRCGKKWHTIAECKRG 210
+G D + PKR E CY CG+ GH + C + +E+KC+ CG+ H +C +
Sbjct: 80 RGGHIAKDCKEPKRE---REQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK- 135
Query: 211 DIVCFNCDEEGHLTSQCKK 229
+ C+ C E GH+ C K
Sbjct: 136 -VKCYRCGETGHVAINCSK 153
Score = 56.6 bits (135), Expect = 6e-08
Identities = 29/94 (30%), Positives = 44/94 (45%), Gaps = 12/94 (12%)
Query: 173 VCYKCGEKGHKSNVCN-GEEKKCFRCGKKWHTIAECK----RGDIVCFNCDEEGHLTSQC 227
+CY+CGE GH + C+ E++ C+ CG+ H +CK + C+NC + GHL C
Sbjct: 53 ICYRCGESGHLAKDCDLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 112
Query: 228 KKPKK------GQASVYDSDYVLTKVMNKCGELG 255
+ G+ D K +CGE G
Sbjct: 113 DHADEQKCYSCGEFGHIQKDCTKVKCY-RCGETG 145
Score = 56.6 bits (135), Expect = 6e-08
Identities = 25/62 (40%), Positives = 32/62 (51%), Gaps = 3/62 (4%)
Query: 167 DDPAEIVCYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAEC-KRGDIVCFNCDEEGHLTS 225
D E CY CGE GH C + KC+RCG+ H C K ++ C+ C E GHL
Sbjct: 113 DHADEQKCYSCGEFGHIQKDCT--KVKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 170
Query: 226 QC 227
+C
Sbjct: 171 EC 172
Score = 49.3 bits (116), Expect = 1e-05
Identities = 30/122 (24%), Positives = 44/122 (35%), Gaps = 40/122 (32%)
Query: 174 CYKCGEKGHKSNVC-------NGEEKK---------------------CFRCGKKWHTIA 205
C+KCG GH + C G + C+RCG+ H
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 206 ECK-RGDIVCFNCDEEGHLTSQCKKPKKGQAS-----------VYDSDYVLTKVMNKCGE 253
+C + D C+NC GH+ CK+PK+ + D D+ + CGE
Sbjct: 66 DCDLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGE 125
Query: 254 LG 255
G
Sbjct: 126 FG 127
Score = 29.6 bits (65), Expect = 7.8
Identities = 10/21 (47%), Positives = 14/21 (66%)
Query: 170 AEIVCYKCGEKGHKSNVCNGE 190
+E+ CY+CGE GH + C E
Sbjct: 155 SEVNCYRCGESGHLARECTIE 175
>YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I
Length = 218
Score = 56.6 bits (135), Expect = 6e-08
Identities = 32/98 (32%), Positives = 48/98 (48%), Gaps = 17/98 (17%)
Query: 153 GEQRLNDERRPKRRDDPAEI----------VCYKCGEKGHKSNVCNGEEKK---CFRCGK 199
G + DER+ K+R + + C+ C ++GH C + CFRCG
Sbjct: 48 GSSKRYDERQKKKRSEYRRLRRINQRNRDKFCFACRQQGHIVQDCPEAKDNVSICFRCGS 107
Query: 200 KWHTIAEC-KRGDIV---CFNCDEEGHLTSQCKKPKKG 233
K H++ C K+G + CF C E GHL+ QC++ KG
Sbjct: 108 KEHSLNACSKKGPLKFAKCFICHENGHLSGQCEQNPKG 145
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 56.6 bits (135), Expect = 6e-08
Identities = 35/114 (30%), Positives = 44/114 (37%), Gaps = 39/114 (34%)
Query: 156 RLNDERRPKRRDDPAEIVCYKCGEKGHKSNVC---------------------------- 187
R ND PK+ +P VCY C + GH S C
Sbjct: 382 RSNDCPEPKKEREPR--VCYNCQQPGHNSRDCPEERKPREGRNGFTSGFGGGNDGGFGGG 439
Query: 188 ------NGEEK---KCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
N EE+ KCF C + H AEC CFNC E+GH +++C P K
Sbjct: 440 NAEGFGNNEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAK 493
Score = 44.7 bits (104), Expect = 2e-04
Identities = 24/102 (23%), Positives = 38/102 (36%), Gaps = 42/102 (41%)
Query: 174 CYKCGEKGHKSNVC-----NGEEKKCFRCGKKWHTIAEC--------------------- 207
C+ C + GH+SN C E + C+ C + H +C
Sbjct: 373 CFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPREGRNGFTSGFGGGN 432
Query: 208 ----------------KRGDIVCFNCDEEGHLTSQCKKPKKG 233
+RG + CFNC EGH +++C +P +G
Sbjct: 433 DGGFGGGNAEGFGNNEERGPMKCFNCKGEGHRSAECPEPPRG 474
Score = 38.9 bits (89), Expect = 0.013
Identities = 19/64 (29%), Positives = 31/64 (47%), Gaps = 18/64 (28%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQC---KKP 230
C+ C + GH+SN C +K+ R VC+NC + GH + C +KP
Sbjct: 259 CFNCQQPGHRSNDCPEPKKE---------------REPRVCYNCQQPGHNSRDCPEERKP 303
Query: 231 KKGQ 234
++G+
Sbjct: 304 REGR 307
Score = 33.1 bits (74), Expect = 0.71
Identities = 16/37 (43%), Positives = 19/37 (51%), Gaps = 2/37 (5%)
Query: 156 RLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEK 192
R ND PK+ +P VCY C + GH S C E K
Sbjct: 268 RSNDCPEPKKEREPR--VCYNCQQPGHNSRDCPEERK 302
Score = 32.0 bits (71), Expect = 1.6
Identities = 10/19 (52%), Positives = 14/19 (73%)
Query: 214 CFNCDEEGHLTSQCKKPKK 232
CFNC + GH ++ C +PKK
Sbjct: 373 CFNCQQPGHRSNDCPEPKK 391
>ZCH3_HUMAN (Q9NUD5) Zinc finger CCHC domain containing protein 3
Length = 404
Score = 56.2 bits (134), Expect = 8e-08
Identities = 24/56 (42%), Positives = 34/56 (59%), Gaps = 3/56 (5%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKK 229
C+KCG + H S C + +CFRCG++ H C++G IVC C + GH +QC K
Sbjct: 336 CFKCGSRTHMSGSCT--QDRCFRCGEEGHLSPYCRKG-IVCNLCGKRGHAFAQCPK 388
Score = 45.8 bits (107), Expect = 1e-04
Identities = 18/41 (43%), Positives = 24/41 (57%), Gaps = 2/41 (4%)
Query: 189 GEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKK 229
G+ K CF+CG + H C + CF C EEGHL+ C+K
Sbjct: 331 GQPKTCFKCGSRTHMSGSCTQDR--CFRCGEEGHLSPYCRK 369
>GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-)
(Germline helicase-1)
Length = 763
Score = 53.9 bits (128), Expect = 4e-07
Identities = 33/121 (27%), Positives = 44/121 (36%), Gaps = 41/121 (33%)
Query: 156 RLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEK----------------------- 192
R +D P++ +P VCY C + GH S C E K
Sbjct: 169 RSSDCPEPRKEREPR--VCYNCQQPGHTSRECTEERKPREGRTGGFGGGAGFGNNGGNDG 226
Query: 193 ----------------KCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQAS 236
KCF C + H AEC CFNC E+GH +++C P K +
Sbjct: 227 FGGDGGFGGGEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREG 286
Query: 237 V 237
V
Sbjct: 287 V 287
Score = 45.1 bits (105), Expect = 2e-04
Identities = 25/104 (24%), Positives = 39/104 (37%), Gaps = 44/104 (42%)
Query: 174 CYKCGEKGHKSNVC-----NGEEKKCFRCGKKWHTIAEC--------------------- 207
C+ C + GH+S+ C E + C+ C + HT EC
Sbjct: 160 CFNCQQPGHRSSDCPEPRKEREPRVCYNCQQPGHTSRECTEERKPREGRTGGFGGGAGFG 219
Query: 208 ------------------KRGDIVCFNCDEEGHLTSQCKKPKKG 233
+RG + CFNC EGH +++C +P +G
Sbjct: 220 NNGGNDGFGGDGGFGGGEERGPMKCFNCKGEGHRSAECPEPPRG 263
Score = 41.2 bits (95), Expect = 0.003
Identities = 21/65 (32%), Positives = 33/65 (50%), Gaps = 11/65 (16%)
Query: 178 GEKGHKSNVCNGEEKKCFRCGKKWHTIAEC-----KRGDIVCFNCDEEGHLTSQC---KK 229
GE GH N CF C + H ++C +R VC+NC + GH + +C +K
Sbjct: 147 GEGGHGGGERNNN---CFNCQQPGHRSSDCPEPRKEREPRVCYNCQQPGHTSRECTEERK 203
Query: 230 PKKGQ 234
P++G+
Sbjct: 204 PREGR 208
>BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog
Length = 179
Score = 51.2 bits (121), Expect = 3e-06
Identities = 20/63 (31%), Positives = 31/63 (48%), Gaps = 7/63 (11%)
Query: 173 VCYKCGEKGHKSNVCN--GEEKKCFRCGKKWHTIAEC-----KRGDIVCFNCDEEGHLTS 225
+CY C + GHK++ C +EK C+ CG H + +C R C+ C GH+
Sbjct: 37 ICYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDCPSSPNPRQGAECYKCGRVGHIAR 96
Query: 226 QCK 228
C+
Sbjct: 97 DCR 99
Score = 50.4 bits (119), Expect = 4e-06
Identities = 21/59 (35%), Positives = 31/59 (51%), Gaps = 3/59 (5%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDI--VCFNCDEEGHLTSQCKKP 230
CY CG GH++ C KC+ CGK H EC++ +C+ C++ GH+ C P
Sbjct: 118 CYACGSYGHQARDCT-MGVKCYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNCTSP 175
Score = 49.3 bits (116), Expect = 1e-05
Identities = 23/74 (31%), Positives = 36/74 (48%), Gaps = 14/74 (18%)
Query: 174 CYKCGEKGHKSNVC--NGEEK-----------KCFRCGKKWHTIAECKRGDIVCFNCDEE 220
CYKCG GH + C NG++ C+ CG H +C G + C++C +
Sbjct: 85 CYKCGRVGHIARDCRTNGQQSGGRFGGHRSNMNCYACGSYGHQARDCTMG-VKCYSCGKI 143
Query: 221 GHLTSQCKKPKKGQ 234
GH + +C++ GQ
Sbjct: 144 GHRSFECQQASDGQ 157
Score = 48.1 bits (113), Expect = 2e-05
Identities = 27/87 (31%), Positives = 39/87 (44%), Gaps = 18/87 (20%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAEC--KRGDIVCFNCDEEGHLTSQC---K 228
CY CGE GH++ C + C+ C + H +EC + + C+ C GHL C
Sbjct: 19 CYNCGENGHQARECT-KGSICYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDCPSSP 77
Query: 229 KPKKGQASVYDSDYVLTKVMNKCGELG 255
P++G A Y KCG +G
Sbjct: 78 NPRQG-AECY-----------KCGRVG 92
Score = 47.4 bits (111), Expect = 4e-05
Identities = 16/40 (40%), Positives = 27/40 (67%), Gaps = 1/40 (2%)
Query: 193 KCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
+C+ CG+ H EC +G I C+NC++ GH S+C +P++
Sbjct: 18 RCYNCGENGHQARECTKGSI-CYNCNQTGHKASECTEPQQ 56
Score = 38.9 bits (89), Expect = 0.013
Identities = 19/75 (25%), Positives = 29/75 (38%), Gaps = 18/75 (24%)
Query: 171 EIVCYKCGEKGHKSNVCNG-----EEKKCFRCGKKWHTIAECK-------------RGDI 212
E CY CG GH C + +C++CG+ H +C+ R ++
Sbjct: 57 EKTCYACGTAGHLVRDCPSSPNPRQGAECYKCGRVGHIARDCRTNGQQSGGRFGGHRSNM 116
Query: 213 VCFNCDEEGHLTSQC 227
C+ C GH C
Sbjct: 117 NCYACGSYGHQARDC 131
Score = 35.4 bits (80), Expect = 0.14
Identities = 14/44 (31%), Positives = 22/44 (49%), Gaps = 2/44 (4%)
Query: 166 RDDPAEIVCYKCGEKGHKSNVCN--GEEKKCFRCGKKWHTIAEC 207
RD + CY CG+ GH+S C + + C++C + H C
Sbjct: 129 RDCTMGVKCYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNC 172
>GAG_VILV2 (P23425) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 50.1 bits (118), Expect = 6e-06
Identities = 20/55 (36%), Positives = 33/55 (59%), Gaps = 6/55 (10%)
Query: 178 GEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
G+ GHK G +KC+ CGK H +C++G I+C +C + GH+ C++ K+
Sbjct: 376 GKAGHK-----GVNQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 34.7 bits (78), Expect = 0.24
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKR 209
CY CG+ GH + C + C CGK+ H +C++
Sbjct: 387 CYNCGKPGHLARQCR-QGIICHHCGKRGHMQKDCRQ 421
>GAG_VILV1 (P23424) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 50.1 bits (118), Expect = 6e-06
Identities = 20/55 (36%), Positives = 33/55 (59%), Gaps = 6/55 (10%)
Query: 178 GEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
G+ GHK G +KC+ CGK H +C++G I+C +C + GH+ C++ K+
Sbjct: 376 GKAGHK-----GVNQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 34.7 bits (78), Expect = 0.24
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKR 209
CY CG+ GH + C + C CGK+ H +C++
Sbjct: 387 CYNCGKPGHLARQCR-QGIICHHCGKRGHMQKDCRQ 421
>GAG_VILV (P03352) Gag polyprotein [Contains: Core protein p16; Core
protein p25; Core protein p14]
Length = 442
Score = 50.1 bits (118), Expect = 6e-06
Identities = 20/55 (36%), Positives = 33/55 (59%), Gaps = 6/55 (10%)
Query: 178 GEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
G+ GHK G +KC+ CGK H +C++G I+C +C + GH+ C++ K+
Sbjct: 376 GKAGHK-----GVNQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 34.7 bits (78), Expect = 0.24
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKR 209
CY CG+ GH + C + C CGK+ H +C++
Sbjct: 387 CYNCGKPGHLARQCR-QGIICHHCGKRGHMQKDCRQ 421
>GAG_CAEVC (P33458) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 441
Score = 49.3 bits (116), Expect = 1e-05
Identities = 16/41 (39%), Positives = 28/41 (68%), Gaps = 1/41 (2%)
Query: 188 NGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCK 228
NG+ ++C+ CGK H +C++G I+C NC + GH+ +C+
Sbjct: 375 NGQPQRCYNCGKPGHQARQCRQG-IICHNCGKRGHMQKECR 414
Score = 36.6 bits (83), Expect = 0.064
Identities = 14/35 (40%), Positives = 20/35 (57%), Gaps = 1/35 (2%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECK 208
CY CG+ GH++ C + C CGK+ H EC+
Sbjct: 381 CYNCGKPGHQARQCR-QGIICHNCGKRGHMQKECR 414
>GAG_VILVK (P35955) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 47.0 bits (110), Expect = 5e-05
Identities = 19/55 (34%), Positives = 32/55 (57%), Gaps = 6/55 (10%)
Query: 178 GEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKK 232
G+ G K G +KC+ CGK H +C++G I+C +C + GH+ C++ K+
Sbjct: 376 GKAGQK-----GVNQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 34.7 bits (78), Expect = 0.24
Identities = 13/36 (36%), Positives = 20/36 (55%), Gaps = 1/36 (2%)
Query: 174 CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKR 209
CY CG+ GH + C + C CGK+ H +C++
Sbjct: 387 CYNCGKPGHLARQCR-QGIICHHCGKRGHMQKDCRQ 421
>GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 513
Score = 44.7 bits (104), Expect = 2e-04
Identities = 17/49 (34%), Positives = 26/49 (52%), Gaps = 1/49 (2%)
Query: 163 PKRRDDPAEIVCYKCGEKGHKSNVCNGEEK-KCFRCGKKWHTIAECKRG 210
P+++ + C+ CG+ GH C + KCF+CGK H +CK G
Sbjct: 381 PQKKGPRGPLKCFNCGKFGHMQRECKAPRQIKCFKCGKIGHMAKDCKNG 429
Score = 42.0 bits (97), Expect = 0.002
Identities = 21/45 (46%), Positives = 24/45 (52%), Gaps = 4/45 (8%)
Query: 193 KCFRCGKKWHTIAECKRG-DIVCFNCDEEGHLTSQCKKPKKGQAS 236
KCF CGK H ECK I CF C + GH+ C K GQA+
Sbjct: 391 KCFNCGKFGHMQRECKAPRQIKCFKCGKIGHMAKDC---KNGQAN 432
Score = 33.9 bits (76), Expect = 0.42
Identities = 11/24 (45%), Positives = 17/24 (70%)
Query: 209 RGDIVCFNCDEEGHLTSQCKKPKK 232
RG + CFNC + GH+ +CK P++
Sbjct: 387 RGPLKCFNCGKFGHMQRECKAPRQ 410
>ZCH9_HUMAN (Q8N567) Zinc finger CCHC domain containing protein 9
Length = 271
Score = 44.3 bits (103), Expect = 3e-04
Identities = 43/171 (25%), Positives = 66/171 (38%), Gaps = 34/171 (19%)
Query: 79 AETAEFSKCIKFENGLRADIKRAIGYQKIRNFSELVSSYMIYEEDTKAHYKIRNERRNKG 138
A A+ K K + L D+ + Y +R S++V + I D++ ++R E
Sbjct: 53 APQAKHKKNKKKKEYLNEDVNGFMEY--LRQNSQMVHNGQIIATDSE---EVREE----- 102
Query: 139 QQGRPKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEKK----- 193
A K + R R ++ +VC+ C + GH C +
Sbjct: 103 --------IAVALKKDSRREGRRLKRQAAKKNAMVCFHCRKPGHGIADCPAALENQDMGT 154
Query: 194 --CFRCGKKWHTIAECKR------GDIV---CFNCDEEGHLTSQCKKPKKG 233
C+RCG H I +CK G+ CF C E GHL+ C KG
Sbjct: 155 GICYRCGSTEHEITKCKAKVDPALGEFPFAKCFVCGEMGHLSRSCPDNPKG 205
>GAG_SIVSP (P19504) Gag polyprotein [Contains: Core protein p17;
Core protein p24]
Length = 507
Score = 44.3 bits (103), Expect = 3e-04
Identities = 19/66 (28%), Positives = 35/66 (52%), Gaps = 15/66 (22%)
Query: 143 PKPYSAPVNKGEQRLNDERRPKRRDDPAEIVCYKCGEKGHKSNVCNGEEKK-CFRCGKKW 201
P P++A KG++++ I C+ CG++GH + C ++ C++CGK
Sbjct: 377 PLPFAAVQQKGQRKI--------------IKCWNCGKEGHSARQCRAPRRQGCWKCGKAG 422
Query: 202 HTIAEC 207
H +A+C
Sbjct: 423 HVMAKC 428
Score = 35.0 bits (79), Expect = 0.19
Identities = 14/45 (31%), Positives = 27/45 (59%), Gaps = 7/45 (15%)
Query: 193 KCFRCGKKWHTIAEC----KRGDIVCFNCDEEGHLTSQCKKPKKG 233
KC+ CGK+ H+ +C ++G C+ C + GH+ ++C + + G
Sbjct: 393 KCWNCGKEGHSARQCRAPRRQG---CWKCGKAGHVMAKCPERQAG 434
Score = 31.6 bits (70), Expect = 2.1
Identities = 11/25 (44%), Positives = 17/25 (68%)
Query: 208 KRGDIVCFNCDEEGHLTSQCKKPKK 232
+R I C+NC +EGH QC+ P++
Sbjct: 388 QRKIIKCWNCGKEGHSARQCRAPRR 412
>GAG_OMVVS (P16900) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 446
Score = 44.3 bits (103), Expect = 3e-04
Identities = 16/45 (35%), Positives = 27/45 (59%), Gaps = 1/45 (2%)
Query: 192 KKCFRCGKKWHTIAECKRGDIVCFNCDEEGHLTSQCKKPKKGQAS 236
+KC+ CGK H +C++G I+C +C + GH+ C++ K S
Sbjct: 384 QKCYYCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKGNPTS 427
Score = 37.7 bits (86), Expect = 0.029
Identities = 23/80 (28%), Positives = 38/80 (46%), Gaps = 11/80 (13%)
Query: 155 QRLNDERRPKRRDDPAEIV-CYKCGEKGHKSNVCNGEEKKCFRCGKKWHTIAECKRGDIV 213
Q L RP R++ + CY CG+ GH + C + C CGK+ H +C++
Sbjct: 366 QLLAQALRPPRKEGKQGVQKCYYCGKPGHLARQCR-QGIICHHCGKRGHMQKDCRQ---- 420
Query: 214 CFNCDEEGHLTSQCKKPKKG 233
++G+ TSQ ++G
Sbjct: 421 -----KKGNPTSQQGNSRRG 435
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 32,744,971
Number of Sequences: 164201
Number of extensions: 1460273
Number of successful extensions: 4482
Number of sequences better than 10.0: 131
Number of HSP's better than 10.0 without gapping: 75
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 3889
Number of HSP's gapped (non-prelim): 377
length of query: 264
length of database: 59,974,054
effective HSP length: 108
effective length of query: 156
effective length of database: 42,240,346
effective search space: 6589493976
effective search space used: 6589493976
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC137553.2


