

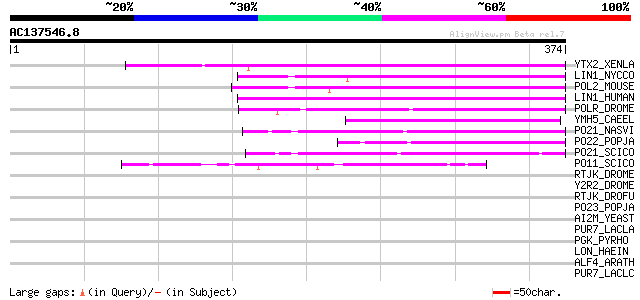
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137546.8 + phase: 0 /pseudo
(374 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 116 8e-26
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 100 1e-20
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 95 3e-19
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 86 2e-16
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 59 2e-08
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 59 3e-08
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 55 2e-07
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 52 2e-06
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 49 2e-05
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 45 2e-04
RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile elem... 42 0.003
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 40 0.007
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 37 0.062
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 34 0.53
AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein 34 0.69
PUR7_LACLA (O68830) Phosphoribosylaminoimidazole-succinocarboxam... 32 2.6
PGK_PYRHO (O58965) Phosphoglycerate kinase (EC 2.7.2.3) 31 4.5
LON_HAEIN (P43864) ATP-dependent protease La (EC 3.4.21.53) 31 4.5
ALF4_ARATH (Q84VX3) Aberrant root formation protein 4 31 4.5
PUR7_LACLC (Q9R7D5) Phosphoribosylaminoimidazole-succinocarboxam... 31 5.8
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 116 bits (291), Expect = 8e-26
Identities = 84/300 (28%), Positives = 143/300 (47%), Gaps = 5/300 (1%)
Query: 79 QSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVED-VRIEGVEPVRNAIFTHFAQHFM 137
+SR L + D S+FF++L + I + ED +E E +R+ + F Q+
Sbjct: 359 RSRMQLLCDMDRGSRFFYALEKKKGNRKQITCLFAEDGTPLEDPEAIRDRARS-FYQNLF 417
Query: 138 SHNVVRPSVSNLQFQALSVTEG---EGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFI 194
S + + P + L V E L P ++DE+ +A+ KS G DG+ F
Sbjct: 418 SPDPISPDACEELWDGLPVVSERRKERLETPITLDELSQALRLMPHNKSPGLDGLTIEFF 477
Query: 195 KEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKI 254
+ FW L D + ++E + G+L + L+ K + + + ++RP+SL+ + KI
Sbjct: 478 QFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSLLPKKGDLRLIKNWRPVSLLSTDYKI 537
Query: 255 IAKLLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEK 314
+AK ++ RL+ V+ V+ QS V R I D + + +++ A + L +D EK
Sbjct: 538 VAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVFLIRDLLHFARRTGLSLAFLSLDQEK 597
Query: 315 TYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
+D VD YL L+ SF + +++ +A V +N S T RG+RQG PL
Sbjct: 598 AFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYASAECLVKINWSLTAPLAFGRGVRQGCPL 657
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 99.8 bits (247), Expect = 1e-20
Identities = 72/227 (31%), Positives = 111/227 (48%), Gaps = 10/227 (4%)
Query: 154 LSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFVSEFH 213
LS E E L +P S E+ I + KS GPDG F EF+ K++++ +
Sbjct: 440 LSQKEVEMLNRPISSSEIASTIQNLPKKKSPGPDG----FTSEFYQTFKEELVPILLNLF 495
Query: 214 RNGKLAKGINTTF----IVLISKVD-NPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIG 268
+N + + TF I LI K +P R ++RPISL+ KI+ K+L NR++ I
Sbjct: 496 QNIEKEGILPNTFYEANITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIK 555
Query: 269 SVVSETQSAFVKKRQILDGILIANEVVDEAFKLK-KDLMLFKVDFEKTYDSVDWGYLGEV 327
++ Q F+ Q I + V+ KLK KD M+ +D EK +D++ ++
Sbjct: 556 KIIHHDQVGFIPGSQGWFNIRKSINVIQHINKLKNKDHMILSIDAEKAFDNIQHPFMIRT 615
Query: 328 LKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
LK + + K I TA++++NG FPL+ G RQG PL
Sbjct: 616 LKKIGIEGTFLKLIEAIYSKPTANIILNGVKLKSFPLRSGTRQGCPL 662
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 94.7 bits (234), Expect = 3e-19
Identities = 71/231 (30%), Positives = 113/231 (48%), Gaps = 10/231 (4%)
Query: 150 QFQALSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFV 209
Q L+ + + L P S E++ I + KS GPDG F EF+ K+D++ +
Sbjct: 463 QVPKLNQDQVDHLNSPISPKEIEAVINSLPTKKSPGPDG----FSAEFYQTFKEDLIPIL 518
Query: 210 SE-FHR---NGKLAKGINTTFIVLISKVD-NPQRLNDFRPISLVGSMNKIIAKLLANRLR 264
+ FH+ G L I LI K +P ++ +FRPISL+ KI+ K+LANR++
Sbjct: 519 HKLFHKIEVEGTLPNSFYEATITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQ 578
Query: 265 LVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLK-KDLMLFKVDFEKTYDSVDWGY 323
I +++ Q F+ Q I + V+ KLK K+ M+ +D EK +D + +
Sbjct: 579 EHIKAIIHPDQVGFIPGMQGWFNIRKSINVIHYINKLKDKNHMIISLDAEKAFDKIQHPF 638
Query: 324 LGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
+ +VL+ + I+ A++ VNG + PLK G RQG PL
Sbjct: 639 MIKVLERSGIQGPYLNMIKAIYSKPVANIKVNGEKLEAIPLKSGTRQGCPL 689
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 85.5 bits (210), Expect = 2e-16
Identities = 60/223 (26%), Positives = 105/223 (46%), Gaps = 2/223 (0%)
Query: 154 LSVTEGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFVSEFH 213
L+ E E L +P + E++ I + KS GP+G F + + L +++
Sbjct: 440 LNQEEVESLNRPITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIE 499
Query: 214 RNGKLAKGINTTFIVLISKVD-NPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVS 272
+ G L I+LI K + + +FRPISL+ KI+ K+LAN+++ I ++
Sbjct: 500 KEGILPNSFYEASIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIH 559
Query: 273 ETQSAFVKKRQILDGILIANEVVDEAFKLKK-DLMLFKVDFEKTYDSVDWGYLGEVLKCM 331
Q F+ Q I + ++ + K + M+ +D EK +D + ++ + L +
Sbjct: 560 HDQVGFIPAMQGWFNIRKSINIIQHINRTKDTNHMIISIDAEKAFDKIQQPFMLKPLNKL 619
Query: 332 SFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
+ K IR TA++++NG + PLK G RQG PL
Sbjct: 620 GIDGTYLKIIRAIYDKPTANIILNGQKLEAPPLKTGTRQGCPL 662
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 59.3 bits (142), Expect = 2e-08
Identities = 58/227 (25%), Positives = 97/227 (42%), Gaps = 12/227 (5%)
Query: 155 SVTEGEGLIKPFSVDEVKEAICDYD------SFKSS-GPDGVNFGFIKEFWIVLKDDIMQ 207
S GE + S++ V AI + D S SS GPDG+ +E V +++
Sbjct: 324 SSCSGEVIQMDHSLERVWSAITEQDLRASRVSLSSSPGPDGITPKSARE---VPSGIMLR 380
Query: 208 FVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVI 267
++ G L I V I K +R DFRPIS+ + + + +LA RL I
Sbjct: 381 IMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQDFRPISVPSVLVRQLNAILATRLNSSI 440
Query: 268 GSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEV 327
Q F+ D I + V+ + K + + +D K +DS+ + +
Sbjct: 441 NW--DPRQRGFLPTDGCADNATIVDLVLRHSHKHFRSCYIANLDVSKAFDSLSHASIYDT 498
Query: 328 LKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
L+ P + +++ S+ +G ++EF RG++QGDPL
Sbjct: 499 LRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSEEFVPARGVKQGDPL 545
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 58.5 bits (140), Expect = 3e-08
Identities = 38/146 (26%), Positives = 68/146 (46%), Gaps = 1/146 (0%)
Query: 227 IVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQILD 286
I+ I K NP +++RPISL +I+ +++ +R+R ++S Q F+ R
Sbjct: 670 IIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCPS 729
Query: 287 GILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVG 346
++ + + K +K L + DF K +D V L + L L W +E +
Sbjct: 730 SLVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKEFLH 789
Query: 347 TATASVLVNG-SPTDEFPLKRGLRQG 371
T SV +N ++ +P+ G+ QG
Sbjct: 790 LRTFSVKINKFVSSNAYPISSGVPQG 815
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 55.5 bits (132), Expect = 2e-07
Identities = 52/217 (23%), Positives = 95/217 (42%), Gaps = 8/217 (3%)
Query: 158 EGEGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFVSEFHRNGK 217
E + L +P S DE+KE + ++GPDG+ W + + I + +G+
Sbjct: 311 EIKNLWRPISNDEIKEV--EACKRTAAGPDGMT----TTAWNSIDECIKSLFNMIMYHGQ 364
Query: 218 LAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSA 277
+ + VLI K FRP+S+ + ++LANR+ ++ Q A
Sbjct: 365 CPRRYLDSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRA 422
Query: 278 FVKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLW 337
F+ + + + + ++ EA K L + +D +K +DSV+ + + L+ P+
Sbjct: 423 FIVADGVAENTSLLSAMIKEARMKIKGLYIAILDVKKAFDSVEHRSILDALRRKKLPLEM 482
Query: 338 RKWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
R +I + + V + RG+RQGDPL
Sbjct: 483 RNYIMWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPL 519
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 52.0 bits (123), Expect = 2e-06
Identities = 34/154 (22%), Positives = 75/154 (48%), Gaps = 6/154 (3%)
Query: 222 INTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAFVKK 281
+ TT I ++NP +++RPI++ ++ +++ ++LA RL + + Q + +
Sbjct: 59 MRTTLIPKDGDLENP---SNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYARI 113
Query: 282 RQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWI 341
L L+ + + + +K + +D K +D+V + L+ + +I
Sbjct: 114 DGTLVNSLLLDTYISSRREQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTSNYI 173
Query: 342 RECVGTATASVLVN-GSPTDEFPLKRGLRQGDPL 374
+ +T ++ V GS T + ++RG++QGDPL
Sbjct: 174 TGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPL 207
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 49.3 bits (116), Expect = 2e-05
Identities = 52/216 (24%), Positives = 91/216 (42%), Gaps = 10/216 (4%)
Query: 160 EGLIKPFSVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFVSE-FHRNGKL 218
E L P S+ E+K A + K +GPDGV W L D + + F G++
Sbjct: 153 ETLWDPVSLIEIKSARASNE--KGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRV 206
Query: 219 AKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAF 278
I + V K++ FRP+S+ + + K+LA R V E Q+A+
Sbjct: 207 PSPIKGSRTVFTPKIEGGPDPGVFRPLSICSVILREFNKILAR--RFVSCYTYDERQTAY 264
Query: 279 VKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWR 338
+ + + + ++ EA +L+K+L + +D K ++SV L + + P
Sbjct: 265 LPIDGVCINVSMLTAIIAEAKRLRKELHIAILDLVKAFNSVYHSALIDAITEAGCPPGVV 324
Query: 339 KWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
+I + + G + + G+ QGDPL
Sbjct: 325 DYIADMYNNVITEMQFEGK-CELASILAGVYQGDPL 359
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 45.4 bits (106), Expect = 2e-04
Identities = 59/258 (22%), Positives = 107/258 (40%), Gaps = 34/258 (13%)
Query: 76 CWQQSRNHWLREGDMNSKFFHSLMSSRRRFNTICSVLVEDVRIEGVEPVRNAIFTHFAQH 135
CWQ+ E F L RR+ +CSV V+ V + E NA+
Sbjct: 326 CWQEFVASESNENPWGRVF--KLCRGRRKPVDVCSVKVDGVYTDTWEGSVNAMM------ 377
Query: 136 FMSHNVVRPSVSNLQFQALSVTEGEGLIKPF----SVDEVKEAICDYDSFKSSGPDGVNF 191
NV P+ + A + + + +P +DEV +++ KS GPDG+
Sbjct: 378 ----NVFFPASID---DASEIDRLKAIARPLPPDLEMDEVSDSVRRCKVRKSPGPDGIVG 430
Query: 192 GFIKEFWIVLKDDIM------QFVSEFHRNGKLAKGINTTFIVLISKVDNPQR-LNDFRP 244
++ W + + + S F + K+A + ++L+ +D + +RP
Sbjct: 431 EMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIA-----SLVILLKLLDRIRSDPGSYRP 485
Query: 245 ISLVGSMNKIIAKLLANRLRLVIGSV-VSETQSAFVKKRQILDGILIANEVVDEAFKLKK 303
I L+ ++ K++ ++ RL + V VS Q AF + D V E ++K
Sbjct: 486 ICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAFTYGKSTEDAWRCVQRHV-ECSEMKY 544
Query: 304 DLMLFKVDFEKTYDSVDW 321
+ L +DF+ +D++ W
Sbjct: 545 VIGL-NIDFQGAFDNLGW 561
>RTJK_DROME (P21328) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 41.6 bits (96), Expect = 0.003
Identities = 51/216 (23%), Positives = 96/216 (43%), Gaps = 14/216 (6%)
Query: 167 SVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFVSEFHRN----GKLAKGI 222
+++EVK I K+ G D ++ I+ +L D +QF++ + G K
Sbjct: 439 TLEEVKNLIAKLPLKKAPGEDLLDNRTIR----LLPDQALQFLALIFNSVLDVGYFPKAW 494
Query: 223 NTTFIVLISKVDN-PQRLNDFRPISLVGSMNKIIAKLLANRLRLV--IGSVVSETQSAFV 279
+ I++I K P ++ +RP SL+ S+ KI+ +L+ NRL + + + Q F
Sbjct: 495 KSASIIMIHKTGKTPTDVDSYRPTSLLPSLGKIMERLILNRLLTCKDVTKAIPKFQFGFR 554
Query: 280 KKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEVLKCMS-FPVLWR 338
+ + + EA + K+ + +D ++ +D V W + G + K FP
Sbjct: 555 LQHGTPEQLHRVVNFALEAMENKEYAVGAFLDIQQAFDRV-W-HPGLLYKAKRLFPPQLY 612
Query: 339 KWIRECVGTATASVLVNGSPTDEFPLKRGLRQGDPL 374
++ + T V V+G + P+ G+ QG L
Sbjct: 613 LVVKSFLEERTFHVSVDGYKSSIKPIAAGVPQGSVL 648
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 40.4 bits (93), Expect = 0.007
Identities = 52/209 (24%), Positives = 86/209 (40%), Gaps = 14/209 (6%)
Query: 168 VDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFI 227
V EV + S +S G DG+N K W + + + S R G +
Sbjct: 436 VFEVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKCPRV 495
Query: 228 VLISKVDNPQRL--NDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAFVKKRQIL 285
V + K + + + +R I L+ K++ ++ NR+R V+ Q F + R +
Sbjct: 496 VSLLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPEGC-RWQFGFRQGRCVE 554
Query: 286 DGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDW-GYLGEV--LKCMSFPVLWRKWIR 342
D V A + L F VDF+ +D+V+W L + L C LW+ +
Sbjct: 555 DAWRHVKSSVG-ASAAQYVLGTF-VDFKGAFDNVEWSAALSRLADLGCREMG-LWQSFF- 610
Query: 343 ECVGTATASVLVNGSPTDEFPLKRGLRQG 371
+ +V+ + S T E P+ RG QG
Sbjct: 611 ----SGRRAVIRSSSGTVEVPVTRGCPQG 635
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 37.4 bits (85), Expect = 0.062
Identities = 28/104 (26%), Positives = 53/104 (50%), Gaps = 9/104 (8%)
Query: 165 PFSVDEVKEAICDYDSFKSSGPDGVNFGFIKEFWIVLKDDIMQFV----SEFHRNGKLAK 220
P +++EVKE + K+ G D ++ I+ +L D + ++ + R G K
Sbjct: 437 PVTLEEVKELVSKLKPKKAPGEDLLDNRTIR----LLPDQALLYLVLIFNSILRVGYFPK 492
Query: 221 GINTTFIVLISKVDN-PQRLNDFRPISLVGSMNKIIAKLLANRL 263
T I++I K P ++ +RP SL+ S+ K++ +L+ NR+
Sbjct: 493 ARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRI 536
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 34.3 bits (77), Expect = 0.53
Identities = 16/72 (22%), Positives = 35/72 (48%)
Query: 303 KDLMLFKVDFEKTYDSVDWGYLGEVLKCMSFPVLWRKWIRECVGTATASVLVNGSPTDEF 362
K + +D K +D+V + ++ + +I + A +++V G T++
Sbjct: 21 KTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDFIMSTITDAYTNIVVGGRTTNKI 80
Query: 363 PLKRGLRQGDPL 374
++ G++QGDPL
Sbjct: 81 YIRNGVKQGDPL 92
>AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein
Length = 789
Score = 33.9 bits (76), Expect = 0.69
Identities = 27/114 (23%), Positives = 45/114 (38%), Gaps = 8/114 (7%)
Query: 217 KLAKGINTTFIVL--ISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSET 274
KL+K INT + +V+ P+ FRP+S+ KI+ + + L ++ + S
Sbjct: 263 KLSKDINTNMFKFSPVRRVEIPKTSGGFRPLSVGNPREKIVQESMRMMLEIIYNNSFSYY 322
Query: 275 QSAFVKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYDSVDWGYLGEVL 328
F L I+ + KVD K +D++ L VL
Sbjct: 323 SHGFRPNLSCLTAIIQCKNYMQYC------NWFIKVDLNKCFDTIPHNMLINVL 370
>PUR7_LACLA (O68830) Phosphoribosylaminoimidazole-succinocarboxamide
synthase (EC 6.3.2.6) (SAICAR synthetase)
Length = 236
Score = 32.0 bits (71), Expect = 2.6
Identities = 24/77 (31%), Positives = 35/77 (45%), Gaps = 2/77 (2%)
Query: 219 AKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAF 278
A+G+ T FI +SK + R P+ +V + ++A A R L G V+ E F
Sbjct: 63 AEGLETHFIEKLSKTEQLNRKVSIIPLEVV--LRNVVAGSFAKRFGLEEGIVLQEPIVEF 120
Query: 279 VKKRQILDGILIANEVV 295
K LD I +E V
Sbjct: 121 YYKDDALDDPFINDEHV 137
>PGK_PYRHO (O58965) Phosphoglycerate kinase (EC 2.7.2.3)
Length = 410
Score = 31.2 bits (69), Expect = 4.5
Identities = 18/63 (28%), Positives = 32/63 (50%)
Query: 258 LLANRLRLVIGSVVSETQSAFVKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEKTYD 317
L+AN L G + F+KK+ +LD + A E++DE + + + F VD++
Sbjct: 225 LVANVFTLAKGFDLGRKNVEFMKKKGLLDYVKHAEEILDEFYPYIRTPVDFAVDYKGERV 284
Query: 318 SVD 320
+D
Sbjct: 285 EID 287
>LON_HAEIN (P43864) ATP-dependent protease La (EC 3.4.21.53)
Length = 803
Score = 31.2 bits (69), Expect = 4.5
Identities = 27/101 (26%), Positives = 45/101 (43%), Gaps = 8/101 (7%)
Query: 186 PDGVNFGFIKEFWIVLKDDIMQFVSEFHRNGKLAKGINTTFIVLISKVDNPQRLNDFR-- 243
P +G KE +V K ++ SEF L K + T + + ++D+ RL D
Sbjct: 114 PIETTYGDEKEL-VVAKSAVL---SEFENYLTLNKKVPTDILNALQRIDDVDRLADTMAA 169
Query: 244 --PISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAFVKKR 282
P+S+ N + + RL ++G + SE V+KR
Sbjct: 170 HLPVSIRHKQNALELANVQERLEYLLGMMESEADILQVEKR 210
>ALF4_ARATH (Q84VX3) Aberrant root formation protein 4
Length = 626
Score = 31.2 bits (69), Expect = 4.5
Identities = 28/100 (28%), Positives = 46/100 (46%), Gaps = 3/100 (3%)
Query: 218 LAKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKL---LANRLRLVIGSVVSET 274
L G +T ++ D+ + + F ISL S+ I AK+ +A V+GSVV E
Sbjct: 300 LITGNDTEKLMSTVAGDDDEFITSFPDISLGASLLFICAKISHEVAEAANAVLGSVVDEL 359
Query: 275 QSAFVKKRQILDGILIANEVVDEAFKLKKDLMLFKVDFEK 314
Q+ VK+ Q + D ++ K+ + F +D K
Sbjct: 360 QNNPVKRWQAYGMLKYILSSGDLLWEFKRHAIEFLLDITK 399
>PUR7_LACLC (Q9R7D5) Phosphoribosylaminoimidazole-succinocarboxamide
synthase (EC 6.3.2.6) (SAICAR synthetase)
Length = 236
Score = 30.8 bits (68), Expect = 5.8
Identities = 23/77 (29%), Positives = 35/77 (44%), Gaps = 2/77 (2%)
Query: 219 AKGINTTFIVLISKVDNPQRLNDFRPISLVGSMNKIIAKLLANRLRLVIGSVVSETQSAF 278
A+G+ T FI +SK + + P+ +V + ++A A R L G V+ E F
Sbjct: 63 AEGLETHFIKKLSKTEQLNKKVSIIPLEVV--LRNVVAGSFAKRFGLEEGIVLEEPIVEF 120
Query: 279 VKKRQILDGILIANEVV 295
K LD I +E V
Sbjct: 121 YYKDDALDDPFINDEHV 137
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.333 0.145 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,435,334
Number of Sequences: 164201
Number of extensions: 1596554
Number of successful extensions: 5851
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 5830
Number of HSP's gapped (non-prelim): 22
length of query: 374
length of database: 59,974,054
effective HSP length: 112
effective length of query: 262
effective length of database: 41,583,542
effective search space: 10894888004
effective search space used: 10894888004
T: 11
A: 40
X1: 15 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.5 bits)
S2: 66 (30.0 bits)
Medicago: description of AC137546.8


