

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137079.8 + phase: 0
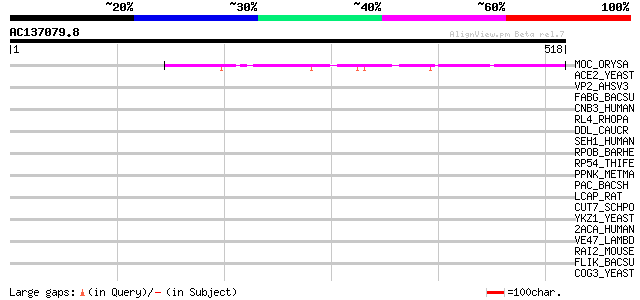
(518 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MOC_ORYSA (Q84MM9) Protein MONOCULM 1 144 5e-34
ACE2_YEAST (P21192) Metallothionein expression activator 38 0.072
VP2_AHSV3 (Q89508) Outer capsid protein VP2 35 0.36
FABG_BACSU (P51831) 3-oxoacyl-[acyl-carrier-protein] reductase (... 35 0.47
CNB3_HUMAN (Q9NQW8) Cyclic-nucleotide-gated cation channel beta ... 34 1.0
RL4_RHOPA (P61068) 50S ribosomal protein L4 33 1.8
DDL_CAUCR (Q9A5A9) D-alanine--D-alanine ligase (EC 6.3.2.4) (D-a... 33 2.3
SEH1_HUMAN (Q96EE3) Nucleoporin SEH1 (SEC13-like protein) 32 3.0
RPOB_BARHE (Q9KJG4) DNA-directed RNA polymerase beta chain (EC 2... 32 3.0
RP54_THIFE (P24695) RNA polymerase sigma-54 factor 32 3.0
PPNK_METMA (Q8PTD1) Probable inorganic polyphosphate/ATP-NAD kin... 32 4.0
PAC_BACSH (P12256) Penicillin acylase (EC 3.5.1.11) (Penicillin ... 32 5.2
LCAP_RAT (P97629) Leucyl-cystinyl aminopeptidase (EC 3.4.11.3) (... 32 5.2
CUT7_SCHPO (P24339) Kinesin-like protein cut7 32 5.2
YKZ1_YEAST (Q02209) Hypothetical 41.3 kDa protein in FOX2-YPT52 ... 31 6.8
2ACA_HUMAN (Q06190) Serine/threonine protein phosphatase 2A, 72/... 31 6.8
VE47_LAMBD (P03752) EA47 gene protein 31 8.8
RAI2_MOUSE (Q9QVY8) Retinoic acid-induced protein 2 (3f8) 31 8.8
FLIK_BACSU (P23451) Probable flagellar hook-length control protein 31 8.8
COG3_YEAST (P40094) Conserved oligomeric Golgi complex component... 31 8.8
>MOC_ORYSA (Q84MM9) Protein MONOCULM 1
Length = 441
Score = 144 bits (363), Expect = 5e-34
Identities = 116/411 (28%), Positives = 183/411 (44%), Gaps = 61/411 (14%)
Query: 145 KEILFTCAKAISENDMETAEWLMS-ELSKMVSVSGNPIQRLGAYMLEALVARI---ASSG 200
+++L CA + D+ A L+ S G+ RL + AL R+ A G
Sbjct: 51 RDLLLACADLLQRGDLPAARRAAEIVLAAAASPRGDAADRLAYHFARALALRVDAKAGHG 110
Query: 201 SIIYKSLKCKEPITATSKELLSHMHVLYEICPYLKFGYMSANGVIAEALKDESEIHIIDF 260
++ + A+S L+ +I P+L+F +++AN I EA+ +HI+D
Sbjct: 111 HVVVGGGAARP---ASSGAYLA----FNQIAPFLRFAHLTANQAILEAVDGARRVHILDL 163
Query: 261 QINQGIQWMSLIQALAGKPG---GPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESY 317
G+QW L+QA+A + GPP++R+TG R G RL A S
Sbjct: 164 DAVHGVQWPPLLQAIAERADPALGPPEVRVTGAGADRDTLLR------TGNRLRAFARSI 217
Query: 318 NVAFEF-------------HAIGVS-------PSEVRLEDLELRRGEAIAVNFAMMLHHV 357
++ F F H G S + LE E +AVN M LH++
Sbjct: 218 HLPFHFTPLLLSCATTAPHHVAGTSTGAAAAASTAAAATGLEFHPDETLAVNCVMFLHNL 277
Query: 358 PDEDVHGGKNHRDRLVRLAKCLSPKVVTLVEQES--------NTNELPFFARFVETMNYY 409
G + ++ K +SP VVT+ E+E+ + ++LP R M++Y
Sbjct: 278 ------AGHDELAAFLKWVKAMSPAVVTIAEREAGGGGGGGDHIDDLP--RRVGVAMDHY 329
Query: 410 FAVFESIDVALPREHRERINVEQHCLAREVVNLVACEGAERVERHEVLKKWRSCFTMAGF 469
AVFE+++ +P RER+ VEQ L RE+ V G +++W AGF
Sbjct: 330 SAVFEALEATVPPGSRERLAVEQEVLGREIEAAVGPSGG---RWWRGIERWGGAARAAGF 386
Query: 470 TPYPLSSYINYSIQNLLE-NYQGH-YTLQEKDGALYLGWMNQPLITSSAWR 518
PLS++ + LL +Y Y +QE GA +LGW +PL++ SAW+
Sbjct: 387 AARPLSAFAVSQARLLLRLHYPSEGYLVQEARGACFLGWQTRPLLSVSAWQ 437
>ACE2_YEAST (P21192) Metallothionein expression activator
Length = 770
Score = 37.7 bits (86), Expect = 0.072
Identities = 49/180 (27%), Positives = 80/180 (44%), Gaps = 21/180 (11%)
Query: 8 RNLDSYCFLQNEN--LENYSSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSKSYTFTS 65
+N+D L +EN + +YS SH S A+ L + +S LQ +
Sbjct: 91 KNIDRTWNLGDENNKVSHYSKKSMSSHKRGLSGTAIFGFLGHNKTLSISSLQQSILNMSK 150
Query: 66 Q-NSLEIINDSLENESCLTHNQDDLWHKIREL--ENAMLGQDAADMLDIYNDTVIIQQES 122
+E+IN+ L N + + +N DD H IRE EN+ L Q ++ QQE
Sbjct: 151 DPQPMELINE-LGNHNTVKNNNDDFDH-IRENDGENSYLSQ-----------VLLKQQEE 197
Query: 123 DPLLLEAEK-WNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPI 181
+ LE +K N+ +E R + +I + + E E A+ L+S + S G+P+
Sbjct: 198 LRIALEKQKEVNEKLEKQLRDN--QIQQEKLRKVLEEQEEVAQKLVSGATNSNSKPGSPV 255
>VP2_AHSV3 (Q89508) Outer capsid protein VP2
Length = 1057
Score = 35.4 bits (80), Expect = 0.36
Identities = 22/73 (30%), Positives = 38/73 (51%), Gaps = 8/73 (10%)
Query: 70 EIINDSLENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEA 129
E I +NE CLTHN D ++ I+++ M+ + D Y ++Q+ +D +
Sbjct: 185 EQIEIERDNERCLTHNTDPIYQLIKKMRYGMMYPVHYMLNDRYK---VVQERAD---MGV 238
Query: 130 EKWNKMIEMISRG 142
EKW +++ I RG
Sbjct: 239 EKW--LLQKIGRG 249
>FABG_BACSU (P51831) 3-oxoacyl-[acyl-carrier-protein] reductase (EC
1.1.1.100) (3-ketoacyl-acyl carrier protein reductase)
Length = 246
Score = 35.0 bits (79), Expect = 0.47
Identities = 23/98 (23%), Positives = 49/98 (49%), Gaps = 11/98 (11%)
Query: 85 NQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMISRGDL 144
N +D+ + I+E + +DI + I +++ + ++ ++W+ +I + +G
Sbjct: 65 NPEDVQNMIKETLSVF------STIDILVNNAGITRDNLIMRMKEDEWDDVININLKG-- 116
Query: 145 KEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQ 182
+F C KA++ M+ + +S +V VSGNP Q
Sbjct: 117 ---VFNCTKAVTRQMMKQRSGRIINVSSIVGVSGNPGQ 151
>CNB3_HUMAN (Q9NQW8) Cyclic-nucleotide-gated cation channel beta 3
(CNG channel beta 3) (Cyclic nucleotide gated channel
beta 3) (Cone photoreceptor cGMP-gated channel beta
subunit) (Cyclic nucleotide-gated cation channel
modulatory subunit)
Length = 809
Score = 33.9 bits (76), Expect = 1.0
Identities = 22/72 (30%), Positives = 32/72 (43%), Gaps = 3/72 (4%)
Query: 19 ENLENYSSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSKSYTFTSQNSLEIINDSLEN 78
EN EN SS ++PS Q+ + + N L++KS TS+ I D L
Sbjct: 16 ENNENEQSSRRNEEGSHPSNQSQQTTAQEENKGEEKSLKTKSTPVTSEEPHTNIQDKLSK 75
Query: 79 ESC---LTHNQD 87
++ LT N D
Sbjct: 76 KNSSGDLTTNPD 87
>RL4_RHOPA (P61068) 50S ribosomal protein L4
Length = 206
Score = 33.1 bits (74), Expect = 1.8
Identities = 21/81 (25%), Positives = 41/81 (49%), Gaps = 7/81 (8%)
Query: 282 PPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDLELR 341
P K+R+ + SA A+GGGL ++ + + A++ + F +G+ S + ++ E+
Sbjct: 101 PKKVRVLALRHALSAKAKGGGLIVLDKAELEAAKTKTLVGHFSGLGLE-SALIIDGAEVN 159
Query: 342 RGEAIAVNFAMMLHHVPDEDV 362
G FA ++P+ DV
Sbjct: 160 NG------FAAAARNIPNIDV 174
>DDL_CAUCR (Q9A5A9) D-alanine--D-alanine ligase (EC 6.3.2.4)
(D-alanylalanine synthetase) (D-Ala-D-Ala ligase)
Length = 324
Score = 32.7 bits (73), Expect = 2.3
Identities = 25/87 (28%), Positives = 41/87 (46%), Gaps = 16/87 (18%)
Query: 266 IQWMSLIQALAGKPGGPPKIRIT------GFDDSTSAYARGGGLGI--------VGERLS 311
IQ M L A+ G+ GP + +T GF D + Y+ GG + + V +R
Sbjct: 187 IQGMELAVAVLGESNGPRALAVTDIRASTGFYDYEAKYSEGGSIHVLPAPIPNAVRDRAM 246
Query: 312 KLAESYNVAFEFHAIGVSPSEVRLEDL 338
++AE + A GV+ S+ R +D+
Sbjct: 247 RMAELAHTALGCR--GVTRSDFRYDDI 271
>SEH1_HUMAN (Q96EE3) Nucleoporin SEH1 (SEC13-like protein)
Length = 421
Score = 32.3 bits (72), Expect = 3.0
Identities = 17/66 (25%), Positives = 35/66 (52%)
Query: 279 PGGPPKIRITGFDDSTSAYARGGGLGIVGERLSKLAESYNVAFEFHAIGVSPSEVRLEDL 338
P K++I ++++T YA+ L V + + +A + N+ FH + ++ +VR+ L
Sbjct: 191 PNAMAKVQIFEYNENTRKYAKAETLMTVTDPVHDIAFAPNLGRSFHILAIATKDVRIFTL 250
Query: 339 ELRRGE 344
+ R E
Sbjct: 251 KPVRKE 256
>RPOB_BARHE (Q9KJG4) DNA-directed RNA polymerase beta chain (EC
2.7.7.6) (RNAP beta subunit) (Transcriptase beta chain)
(RNA polymerase beta subunit)
Length = 1383
Score = 32.3 bits (72), Expect = 3.0
Identities = 25/97 (25%), Positives = 49/97 (49%), Gaps = 9/97 (9%)
Query: 101 LGQDAADMLDIYNDTVIIQQESD----PLLLEAEKWNKMIEMISRGDLKEILFTCAKAIS 156
LG DA+D+L + + V +++ D P ++ K K++ + D E++ K ++
Sbjct: 225 LGMDASDILSTFYNKVTYERDGDGWRIPYSVDRFKGMKLVSDLIDADSGEVVAEAGKKLT 284
Query: 157 ENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALV 193
+ A+ L + K V VS + + LG+Y+ E +V
Sbjct: 285 ---VRAAKALAEKGLKAVKVSEDDL--LGSYLAEDIV 316
>RP54_THIFE (P24695) RNA polymerase sigma-54 factor
Length = 475
Score = 32.3 bits (72), Expect = 3.0
Identities = 19/77 (24%), Positives = 35/77 (44%)
Query: 25 SSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSKSYTFTSQNSLEIINDSLENESCLTH 84
S S NGS P F++ +S + + +T +N E+I D+++ L
Sbjct: 100 SGSGNGSDEDLPDFESRNSRTQSLQDYLRWQADMTHFTADERNMAELIIDAIDERGYLAD 159
Query: 85 NQDDLWHKIRELENAML 101
+ +DL + E+A+L
Sbjct: 160 SLEDLAATMNVQEDALL 176
>PPNK_METMA (Q8PTD1) Probable inorganic polyphosphate/ATP-NAD kinase
(EC 2.7.1.23) (Poly(P)/ATP NAD kinase)
Length = 289
Score = 32.0 bits (71), Expect = 4.0
Identities = 17/80 (21%), Positives = 41/80 (51%), Gaps = 1/80 (1%)
Query: 134 KMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGAYMLEALV 193
K I + SR D E+L + ++ + ++ + ++++ + G P++R+ +E LV
Sbjct: 18 KRIGIASRCDRPEVLDMVREILARFSSQVQIFVSTATAEVLGIEGTPVERMRDEGVE-LV 76
Query: 194 ARIASSGSIIYKSLKCKEPI 213
+ G+++ K K+P+
Sbjct: 77 ISVGGDGTVLRNIAKMKDPL 96
>PAC_BACSH (P12256) Penicillin acylase (EC 3.5.1.11) (Penicillin V
amidase)
Length = 338
Score = 31.6 bits (70), Expect = 5.2
Identities = 28/99 (28%), Positives = 45/99 (45%), Gaps = 6/99 (6%)
Query: 216 TSKELLSHMHVLYEICPYLKFGYMSANG--VIAEALKDESEIH--IIDFQINQ-GIQW-M 269
TS LL+ +++ P L + + A+G ++ E K IH I N G +W
Sbjct: 127 TSYTLLNEANIILGFAPPLHYTFTDASGESIVIEPDKTGITIHRKTIGVMTNSPGYEWHQ 186
Query: 270 SLIQALAGKPGGPPKIRITGFDDSTSAYARGGGLGIVGE 308
+ ++A G PP+ + G D T GGLG+ G+
Sbjct: 187 TNLRAYIGVTPNPPQDIMMGDLDLTPFGQGAGGLGLPGD 225
>LCAP_RAT (P97629) Leucyl-cystinyl aminopeptidase (EC 3.4.11.3)
(Cystinyl aminopeptidase) (Oxytocinase) (OTase)
(Insulin-regulated membrane aminopeptidase)
(Insulin-responsive aminopeptidase) (IRAP) (Placental
leucine aminopeptidase) (P-LAP) (Vesicle prot
Length = 1025
Score = 31.6 bits (70), Expect = 5.2
Identities = 38/142 (26%), Positives = 57/142 (39%), Gaps = 13/142 (9%)
Query: 2 FNIEPI-RNLDSY-CFLQNENLENYSSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSK 59
F++E I + L+SY FL S N SH S Q+ EQ E ++ K S
Sbjct: 496 FSVEKIFKELNSYEDFLDARFKTMRKDSLNSSHPISSSVQSSEQIEEMFDSLSYFKGASL 555
Query: 60 SYTFTSQNSLEIINDS----LENESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYN-- 113
S S ++ + L N S DDLW E+ L D M+ +
Sbjct: 556 LLMLKSYLSEDVFQHAIILYLHNHSYAAIQSDDLWDSFNEVTGKTL--DVKKMMKTWTLQ 613
Query: 114 ---DTVIIQQESDPLLLEAEKW 132
V +Q++ LLL+ E++
Sbjct: 614 KGFPLVTVQRKGTELLLQQERF 635
>CUT7_SCHPO (P24339) Kinesin-like protein cut7
Length = 1085
Score = 31.6 bits (70), Expect = 5.2
Identities = 33/156 (21%), Positives = 69/156 (44%), Gaps = 18/156 (11%)
Query: 38 FQALEQNLESSNNSPVSKLQSKSYTFTSQNSLEIINDSLENES-----CLTHNQDDLWHK 92
FQ+L++ L+S+ +S +SL++I L++ L H+ D+
Sbjct: 658 FQSLDEALQSARSSCA----------VPNSSLDLIVSELKDSKNSLLDALEHSLQDISMS 707
Query: 93 IRELENAM---LGQDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMISRGDLKEILF 149
++L N + L + DM + Y V + L E+ K + R D+ ++
Sbjct: 708 SQKLGNGISSELIELQKDMKESYRQLVQELRSLYNLQHTHEESQKELMYGVRNDIDALVK 767
Query: 150 TCAKAISENDMETAEWLMSELSKMVSVSGNPIQRLG 185
TC ++++ D+ ++++ + SK S + I +G
Sbjct: 768 TCTTSLNDADIILSDYISDQKSKFESKQQDLIANIG 803
>YKZ1_YEAST (Q02209) Hypothetical 41.3 kDa protein in FOX2-YPT52
intergenic region
Length = 353
Score = 31.2 bits (69), Expect = 6.8
Identities = 27/95 (28%), Positives = 46/95 (48%), Gaps = 13/95 (13%)
Query: 2 FNIEPI---RNLDSYCFLQNENLENY----SSSDNGSHSTYPSFQALEQNLES------S 48
FN+E +N+ S ++ + L N ++ N +S + EQNL+ S
Sbjct: 177 FNMEVFDNFKNIVSIAIIKTKVLSNKRLLTTTLKNYHNSMKKKYNIQEQNLKENSLASCS 236
Query: 49 NNSPVSKLQSKSYTFTSQNSLEIINDSLENESCLT 83
NN P + L+S+S F+ NSL + S ++E+ T
Sbjct: 237 NNEPSASLESESRHFSPVNSLSPSSLSTDDEAVST 271
>2ACA_HUMAN (Q06190) Serine/threonine protein phosphatase 2A, 72/130
kDa regulatory subunit B (PP2A, subunit B,
B''-PR72/PR130) (PP2A, subunit B, B72/B130 isoforms)
(PP2A, subunit B, PR72/PR130 isoforms) (PP2A, subunit B,
R3 isoform)
Length = 1150
Score = 31.2 bits (69), Expect = 6.8
Identities = 33/174 (18%), Positives = 73/174 (40%), Gaps = 14/174 (8%)
Query: 25 SSSDNGSHSTYPSFQALEQNLESSNNSPVSKLQSKSYT-----FTSQNSLEI-----IND 74
SSS ++ Q +E+S P++K ++ ++ T Q +++ ++
Sbjct: 503 SSSQEEIDKLLMDLESFSQKMETSLREPLAKGKNSNFLNSHSQLTGQTLVDLEPKSKVSS 562
Query: 75 SLE--NESCLTHNQDDLWHKIRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEKW 132
+E + SCLT + HKI E + A+L + + D + V + L E E
Sbjct: 563 PIEKVSPSCLTRIIETNGHKIEEEDRALLLRILESIEDFAQELVECKSSRGSLSQEKEMM 622
Query: 133 NKMIEMISRGDLKEILFTCAKAISENDME-TAEWLMSELSKMVSVSGNPIQRLG 185
+ E ++ + L C + + + T+ L+ + +++ + P ++ G
Sbjct: 623 QILQETLTTSS-QANLSVCRSPVGDKAKDTTSAVLIQQTPEVIKIQNKPEKKPG 675
>VE47_LAMBD (P03752) EA47 gene protein
Length = 410
Score = 30.8 bits (68), Expect = 8.8
Identities = 32/112 (28%), Positives = 63/112 (55%), Gaps = 21/112 (18%)
Query: 93 IRELENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAEKWNKMIEMISRGDLKEILFTCA 152
I++ +N ++G D YN+ VI + ++DPL+ A+ IE +GDL+ +++ A
Sbjct: 48 IQKNKNQIIGYDIW-----YNNNVIEKWKNDPLMAWAKNSRNTIE--KQGDLE--MYSEA 98
Query: 153 KA------ISENDME--TAEWLMS-ELSKMVSVSGNPIQRLGAYMLEALVAR 195
KA I END+E T E +++ + K+V ++ ++L +Y+ E+ + +
Sbjct: 99 KATLISSYIEENDIEFITNESMLNIGIKKLVRLAQ---KKLPSYLTESSIIK 147
>RAI2_MOUSE (Q9QVY8) Retinoic acid-induced protein 2 (3f8)
Length = 529
Score = 30.8 bits (68), Expect = 8.8
Identities = 40/156 (25%), Positives = 65/156 (41%), Gaps = 13/156 (8%)
Query: 37 SFQALEQNLESSNNSPVSKLQSKSYTFTSQNSLEIINDSLENESCLTHNQDDLWHKIREL 96
SF+ + LE P+ LQSK Y S+++ +I ENE+ + W K E
Sbjct: 275 SFKGTQTTLEKDELKPLDILQSKEYFQLSRHT--VIKMGSENEALDLSMKSVPWLKAGEA 332
Query: 97 ENAMLGQDAADMLDIYNDTVIIQQESDPLLLEAE------KWNKMIEMISRGDLKEILFT 150
+ +DAA L + + E+ P L + + ++E +S G E+ F
Sbjct: 333 SPPVFQEDAALDLSL---AAHRKAEAPPETLYNNSGSADIQGHTILEKLSSG--MEMPFA 387
Query: 151 CAKAISENDMETAEWLMSELSKMVSVSGNPIQRLGA 186
AK + M + S ++MVS +P L A
Sbjct: 388 PAKCSEASAMMESHSSNSNGTEMVSQPSHPGSELKA 423
>FLIK_BACSU (P23451) Probable flagellar hook-length control protein
Length = 429
Score = 30.8 bits (68), Expect = 8.8
Identities = 47/182 (25%), Positives = 77/182 (41%), Gaps = 21/182 (11%)
Query: 17 QNENLENYSSSDNGSHSTYPSFQALEQNLESSNNSPVS----------KLQSKSYTFTSQ 66
QN +L S G + Q L+ L++ + VS K+ S++ T S
Sbjct: 20 QNADLLQTLSKLTGKQLDANALQMLQNLLQAVESKNVSGTDQLLTETEKIFSEAKTALSA 79
Query: 67 N-SLEIINDSLENESCLTHNQDDLWHKIREL-------ENAMLGQDAADMLDIYNDTVII 118
N S IN +L+++ ++ ++++ +EL + G D L N+ I
Sbjct: 80 NDSASDINGALKSDKEQSNQENEVSEPAKELIYIQMFISQLVEGNKLTD-LGNGNEAHAI 138
Query: 119 QQESDPLLLEAEKWNKMIEMISRGDLKEILFTCAKAISENDMETAEWLMSELSKMVSVSG 178
Q D L EK ++I DLK+ +FT A++ S+ TA L S S M +S
Sbjct: 139 YQNGDQFLSALEKKGVSQQLIQ--DLKQQIFTKAESSSKLYSMTASELKSFQSLMDQMSM 196
Query: 179 NP 180
P
Sbjct: 197 LP 198
>COG3_YEAST (P40094) Conserved oligomeric Golgi complex component 3
(SEC34 protein)
Length = 801
Score = 30.8 bits (68), Expect = 8.8
Identities = 27/101 (26%), Positives = 50/101 (48%), Gaps = 9/101 (8%)
Query: 44 NLESSNNSPVSKLQSKSYTFTSQNSLEIINDSLE---NESCLTHNQDDLWHKIRELENAM 100
N+ + +S+L++ FT SLE+I+D+L+ +E L+ N K+RE A
Sbjct: 662 NMVDARTELISELRNVIKDFTESTSLELIDDTLDINSDEDLLSKNV-----KLRENIKAR 716
Query: 101 LGQDAADMLDIYNDTVIIQQESDPLL-LEAEKWNKMIEMIS 140
L + +L+ +D I+ D + L + ++K E I+
Sbjct: 717 LPRIYEQILNYIDDQEIVTNLLDAVQELITQSYSKYYETIT 757
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 60,614,068
Number of Sequences: 164201
Number of extensions: 2551914
Number of successful extensions: 7333
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 18
Number of HSP's that attempted gapping in prelim test: 7320
Number of HSP's gapped (non-prelim): 25
length of query: 518
length of database: 59,974,054
effective HSP length: 115
effective length of query: 403
effective length of database: 41,090,939
effective search space: 16559648417
effective search space used: 16559648417
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 68 (30.8 bits)
Medicago: description of AC137079.8


