

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137079.15 + phase: 0 /pseudo
(864 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
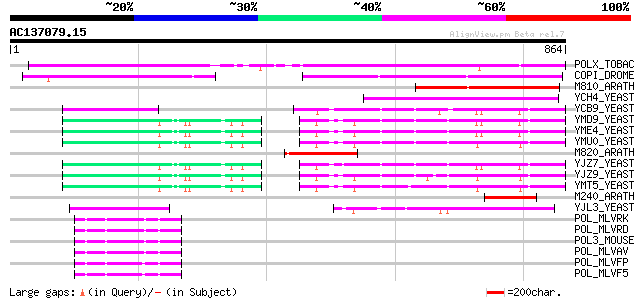
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 459 e-128
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 266 2e-70
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 196 2e-49
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 155 3e-37
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 107 2e-22
YMD9_YEAST (Q03434) Transposon Ty1 protein B 103 1e-21
YME4_YEAST (Q04711) Transposon Ty1 protein B 103 2e-21
YMU0_YEAST (Q04670) Transposon Ty1 protein B 103 3e-21
M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820... 101 7e-21
YJZ7_YEAST (P47098) Transposon Ty1 protein B 101 1e-20
YJZ9_YEAST (P47100) Transposon Ty1 protein B 100 1e-20
YMT5_YEAST (Q04214) Transposon Ty1 protein B 99 5e-20
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 99 6e-20
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 90 3e-17
POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.2... 54 1e-06
POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.2... 54 1e-06
POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein (Endonucl... 54 1e-06
POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.2... 51 2e-05
POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.2... 50 3e-05
POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.2... 50 3e-05
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 459 bits (1181), Expect = e-128
Identities = 304/864 (35%), Positives = 446/864 (51%), Gaps = 71/864 (8%)
Query: 30 DVWHMRLGHVSSSGLSVISKQFPFIPCIKNAP--PCDACHYAKQKRLPFPHSSIKSSAPF 87
D+WH R+GH+S GL +++K+ I K PCD C + KQ R+ F SS +
Sbjct: 423 DLWHKRMGHMSEKGLQILAKK-SLISYAKGTTVKPCDYCLFGKQHRVSFQTSSERKLNIL 481
Query: 88 DLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHLKHFISYVENQFH 147
DL+++D+ GP S G+KYF+T +DD SR WV LKTKD+ + + F + VE +
Sbjct: 482 DLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETG 541
Query: 148 TTLKCLRSDNGSEFIA--MTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAF 205
LK LRSDNG E+ + + S GI H KT TPQ NGV ER ++ I+ RS+
Sbjct: 542 RKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLR 601
Query: 206 HSHVPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHLKVFGCLAYASTLQ 265
+ +P + W VQ A ++INR PS L F+ P + + S HLKVFGC A+A +
Sbjct: 602 MAKLPKSFWGEAVQTACYLINRSPSVPLAFEIPERVWTNKEVSYSHLKVFGCRAFAHVPK 661
Query: 266 AHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIP 325
RTK + ++ IF+G+ + G L+D ++ SR+V+F E+
Sbjct: 662 EQRTKLDDKSIPCIFIGYGDEEFGYRLWDPVKKKVIRSRDVVFRESE------------- 708
Query: 326 TTSSHIDLGDPITD-LSPHPISAPEFQLTSTPPSQYVSAPAVQHAIPVTDSISEPTVRKS 384
+ D+ + + + + P+ ++ P TS P+ S TD +SE +
Sbjct: 709 -VRTAADMSEKVKNGIIPNFVTIPS---TSNNPTSAEST---------TDEVSEQGEQPG 755
Query: 385 TRISQ------------RPSYLADYHCNLPSKSCSNVSSGISSYPLSSF-LSYDNCSPTY 431
I Q P+ + H L V S YP + + L D+ P
Sbjct: 756 EVIEQGEQLDEGVEEVEHPTQGEEQHQPLRRSERPRVES--RRYPSTEYVLISDDREP-- 811
Query: 432 THFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVY 491
S+ E + + N+ +AM E+ +L KN T+ +V LP GK P+ CKWV+
Sbjct: 812 -------ESLKEVLSHPEKNQL---MKAMQEEMESLQKNGTYKLVELPKGKRPLKCKWVF 861
Query: 492 KVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQL 551
K+K + + RYKARLV +G+ Q +G+D+ + FSPV K+T+IR +LSLAA +EQL
Sbjct: 862 KLKKDGDCKLVRYKARLVVKGFEQKKGIDFDEIFSPVVKMTSIRTILSLAASLDLEVEQL 921
Query: 552 DVNNAFLHGDLHEEVYMALPPGYPTINSS-QVCKLNKSLYGLKQASRQWYSKLSTSLISF 610
DV AFLHGDL EE+YM P G+ VCKLNKSLYGLKQA RQWY K + + S
Sbjct: 922 DVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSFMKSQ 981
Query: 611 GYTQSLADYSLFVK-VSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQL 669
Y ++ +D ++ K S +F LL+YVDD+++ G I +K L F +KDLG
Sbjct: 982 TYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKDLGPA 1041
Query: 670 RYFLGFEIARSKSG--ILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTD 727
+ LG +I R ++ + L+Q KY +LE +KP +TP KL +
Sbjct: 1042 QQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCPTTVE 1101
Query: 728 ASS------YRRLIGRLLY-LTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSS 780
Y +G L+Y + TRPDI+++V +S+F+ P H++A + IL+YL+ +
Sbjct: 1102 EKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRYLRGT 1161
Query: 781 PAKGLFFSSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSST 840
L F S + L G+ D+D A D R+S TGY ISW+SK Q V+ S+T
Sbjct: 1162 TGDCLCFGGSDPI-LKGYTDADMAGDIDNRKSSTGYLFTFSGGAISWQSKLQKCVALSTT 1220
Query: 841 EAEYRALAHLTCELQWLNYLFHDL 864
EAEY A E+ WL +L
Sbjct: 1221 EAEYIAATETGKEMIWLKRFLQEL 1244
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 266 bits (679), Expect = 2e-70
Identities = 156/411 (37%), Positives = 235/411 (56%), Gaps = 8/411 (1%)
Query: 456 WREAMTTELNALAKNNTWSVVTLPPGKVPIGCKWVYKVKYHANGSIERYKARLVAQGYTQ 515
W EA+ TELNA NNTW++ P K + +WV+ VKY+ G+ RYKARLVA+G+TQ
Sbjct: 906 WEEAINTELNAHKINNTWTITKRPENKNIVDSRWVFSVKYNELGNPIRYKARLVARGFTQ 965
Query: 516 TEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLHGDLHEEVYMALPPGYP 575
+DY +TF+PVA++++ R +LSL + Q+DV AFL+G L EE+YM LP G
Sbjct: 966 KYQIDYEETFAPVARISSFRFILSLVIQYNLKVHQMDVKTAFLNGTLKEEIYMRLPQGI- 1024
Query: 576 TINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYSLFVKVSG--ASFTAL 633
+ NS VCKLNK++YGLKQA+R W+ +L + S D +++ G +
Sbjct: 1025 SCNSDNVCKLNKAIYGLKQAARCWFEVFEQALKECEFVNSSVDRCIYILDKGNINENIYV 1084
Query: 634 LVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYTL 693
L+YVDD+V+A ++ + + K +L KF + DL ++++F+G I + I L+Q Y
Sbjct: 1085 LLYVDDVVIATGDMTRMNNFKRYLMEKFRMTDLNEIKHFIGIRIEMQEDKIYLSQSAYVK 1144
Query: 694 ELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTDASSYRRLIGRLLY-LTNTRPDISYSV 752
++L +TP PS + + R LIG L+Y + TRPD++ +V
Sbjct: 1145 KILSKFNMENCNAVSTPL-PSKINYELLNSDEDCNTPCRSLIGCLMYIMLCTRPDLTTAV 1203
Query: 753 QNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSS--SSELKLHGFADSDWACCPDTR 810
LS++ S+ +Q +R+L+YLK + L F + E K+ G+ DSDWA R
Sbjct: 1204 NILSRYSSKNNSELWQNLKRVLRYLKGTIDMKLIFKKNLAFENKIIGYVDSDWAGSEIDR 1263
Query: 811 RSVTGYCV-LLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQWLNYL 860
+S TGY + +LI W +K+Q++V+ SSTEAEY AL E WL +L
Sbjct: 1264 KSTTGYLFKMFDFNLICWNTKRQNSVAASSTEAEYMALFEAVREALWLKFL 1314
Score = 144 bits (363), Expect = 1e-33
Identities = 95/313 (30%), Positives = 158/313 (50%), Gaps = 15/313 (4%)
Query: 20 SCNSVFTDCFDVWHMRLGHVSSSGLSVISKQFPFIP--CIKN----APPCDACHYAKQKR 73
S N+ + F +WH R GH+S L I ++ F + N C+ C KQ R
Sbjct: 406 SINAKHKNNFRLWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQAR 465
Query: 74 LPFPHSSIKS--SAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDET 131
LPF K+ P ++H+D+ GP + + YF+ VD ++ + +K K +
Sbjct: 466 LPFKQLKDKTHIKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDV 525
Query: 132 QKHLKHFISYVENQFHTTLKCLRSDNGSEFIA--MTSFLLSKGIIHHKTCVETPQQNGVV 189
+ F++ E F+ + L DNG E+++ M F + KGI +H T TPQ NGV
Sbjct: 526 FSMFQDFVAKSEAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVS 585
Query: 190 ERKHQHILNVARSLAFHSHVPITMWNFTVQHAVHIINRIPSPLL--KFKSPFELLHKEPP 247
ER + I AR++ + + + W V A ++INRIPS L K+P+E+ H + P
Sbjct: 586 ERMIRTITEKARTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKP 645
Query: 248 SIIHLKVFGCLAYASTLQAHRTKFNPRARKTIFLGFKEGTKGSILYDLNSNELFVSRNVI 307
+ HL+VFG Y ++ + KF+ ++ K+IF+G++ G L+D + + V+R+V+
Sbjct: 646 YLKHLRVFGATVYVH-IKNKQGKFDDKSFKSIFVGYE--PNGFKLWDAVNEKFIVARDVV 702
Query: 308 FYENHFPFTLATK 320
E + + A K
Sbjct: 703 VDETNMVNSRAVK 715
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 196 bits (498), Expect = 2e-49
Identities = 101/225 (44%), Positives = 142/225 (62%), Gaps = 3/225 (1%)
Query: 633 LLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQLRYFLGFEIARSKSGILLNQRKYT 692
LL+YVDDI+L G+ + + + L + F +KDLG + YFLG +I SG+ L+Q KY
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYA 62
Query: 693 LELLEDAGTLGSKPAATPFDPSTKLGATTGTP-FTDASSYRRLIGRLLYLTNTRPDISYS 751
++L +AG L KP +TP KL ++ T + D S +R ++G L YLT TRPDISY+
Sbjct: 63 EQILNNAGMLDCKPMSTPLP--LKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYA 120
Query: 752 VQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKLHGFADSDWACCPDTRR 811
V + Q + P + + +R+L+Y+K + GL+ +S+L + F DSDWA C TRR
Sbjct: 121 VNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRR 180
Query: 812 SVTGYCVLLGSSLISWKSKKQSTVSRSSTEAEYRALAHLTCELQW 856
S TG+C LG ++ISW +K+Q TVSRSSTE EYRALA EL W
Sbjct: 181 STTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 155 bits (393), Expect = 3e-37
Identities = 98/308 (31%), Positives = 160/308 (51%), Gaps = 4/308 (1%)
Query: 551 LDVNNAFLHGDLHEEVYMALPPGYPTI-NSSQVCKLNKSLYGLKQASRQWYSKLSTSLIS 609
+DV+ AFL+ + E +Y+ PPG+ N V +L +YGLKQA W ++ +L
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 610 FGYTQSLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQL 669
G+ + ++ L+ + + + VYVDD+++A VK L + +KDLG++
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 670 RYFLGFEIARSKSG-ILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTPFTDA 728
FLG I +S +G I L+ + Y + ++ K TP S L TT D
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 729 SSYRRLIGRLLYLTNT-RPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFF 787
+ Y+ ++G+LL+ NT RPDISY V LS+F+ P H ++A+R+L+YL ++ + L +
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 788 SSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKK-QSTVSRSSTEAEYRA 846
S S++ L + D+ D S GY LL + ++W SKK + + STEAEY
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 847 LAHLTCEL 854
+ E+
Sbjct: 301 ASETVMEI 308
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 107 bits (266), Expect = 2e-22
Identities = 119/458 (25%), Positives = 201/458 (42%), Gaps = 54/458 (11%)
Query: 442 NEPKTFAQANKS-ECWREAMTTELNALAKNNTWSVVT------LPPGKVPIGCKWVYKVK 494
+E T+ + NK + + EA E++ L K NTW + P KV I +++ K
Sbjct: 1250 DEAITYNKDNKEKDRYVEAYHKEISQLLKMNTWDTNKYYDRNDIDPKKV-INSMFIFNKK 1308
Query: 495 YHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLAAIKGWHLEQLDVN 554
+KAR VA+G Q D S + LS+A +++ QLD++
Sbjct: 1309 RDGT-----HKARFVARGDIQHPDTYDSDMQSNTVHHYALMTSLSIALDNDYYITQLDIS 1363
Query: 555 NAFLHGDLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQ 614
+A+L+ D+ EE+Y+ PP + ++ +L KSLYGLKQ+ WY + + LI+ Q
Sbjct: 1364 SAYLYADIKEELYIRPPPHLGL--NDKLLRLRKSLYGLKQSGANWYETIKSYLINCCDMQ 1421
Query: 615 SLADYSLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK--DLG----Q 668
+ +S K S + + ++VDD++L ++ K + T L ++ K +LG +
Sbjct: 1422 EVRGWSCVFKNSQVT---ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNE 1478
Query: 669 LRY-FLGFEIARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP--- 724
++Y LG EI +S KY +E + T P +P K G P
Sbjct: 1479 IQYDILGLEIKYQRS-------KYMKLGMEKSLTEKLPKLNVPLNPKGKKLRAPGQPGHY 1531
Query: 725 ------FTDASSYR-------RLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAA 770
D Y+ +LIG Y+ R D+ Y + L+Q + P
Sbjct: 1532 IDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMT 1591
Query: 771 QRILKYLKSSPAKGLFFSSSS----ELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLIS 826
+++++ + K L + + + KL +D+ + P +S G LL +I
Sbjct: 1592 YELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASYGNQP-YYKSQIGNIFLLNGKVIG 1650
Query: 827 WKSKKQSTVSRSSTEAEYRALAHLTCELQWLNYLFHDL 864
KS K S S+TEAE A++ L L++L +L
Sbjct: 1651 GKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQEL 1688
Score = 73.9 bits (180), Expect = 2e-12
Identities = 45/154 (29%), Positives = 76/154 (49%), Gaps = 4/154 (2%)
Query: 82 KSSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQ--KHLKHFI 139
+S PF LH D++GP YF++ D+ +RF WV L + E +
Sbjct: 656 ESYEPFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRREESILNVFTSIL 715
Query: 140 SYVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHIL 197
++++NQF+ + ++ D GSE+ T F ++GI T + +GV ER ++ +L
Sbjct: 716 AFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSRAHGVAERLNRTLL 775
Query: 198 NVARSLAFHSHVPITMWNFTVQHAVHIINRIPSP 231
N R+L S +P +W V+ + I N + SP
Sbjct: 776 NDCRTLLHCSGLPNHLWFSAVEFSTIIRNSLVSP 809
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 103 bits (258), Expect = 1e-21
Identities = 111/447 (24%), Positives = 197/447 (43%), Gaps = 53/447 (11%)
Query: 452 KSECWREAMTTELNALAKNNTWSV------VTLPPGKVPIGCKWVYKVKYHANGSIERYK 505
+ E + EA E+N L K TW + P +V I +++ K +K
Sbjct: 819 EKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRV-INSMFIFNKKRDGT-----HK 872
Query: 506 ARLVAQGYTQTEGVDYFDTFSPVAKLTTIR-----VLLSLAAIKGWHLEQLDVNNAFLHG 560
AR VA+G + + DT+ + T+ LSLA +++ QLD+++A+L+
Sbjct: 873 ARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYA 927
Query: 561 DLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYS 620
D+ EE+Y+ PP + ++ +L KSLYGLKQ+ WY + + LI + + +S
Sbjct: 928 DIKEELYIRPPPHLGM--NDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWS 985
Query: 621 LFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK--DLGQLRYFLGFEIA 678
K S + + ++VDD++L ++ K + T L ++ K +LG+ + ++I
Sbjct: 986 CVFKNSQVT---ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDIL 1042
Query: 679 RSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP---------FTDAS 729
+ I + KY +E++ T P +P + + G P D
Sbjct: 1043 GLE--IKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDED 1100
Query: 730 SYR-------RLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSP 781
Y+ +LIG Y+ R D+ Y + L+Q + P +++++ +
Sbjct: 1101 EYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTR 1160
Query: 782 AKGLFFSSSS----ELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSR 837
K L + + + KL +D+ + P +S G LL +I KS K S
Sbjct: 1161 DKQLIWHKNKPTEPDNKLVAISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCT 1219
Query: 838 SSTEAEYRALAHLTCELQWLNYLFHDL 864
S+TEAE A++ L L+YL +L
Sbjct: 1220 STTEAEIHAISESVPLLNNLSYLIQEL 1246
Score = 76.6 bits (187), Expect = 3e-13
Identities = 85/355 (23%), Positives = 141/355 (38%), Gaps = 54/355 (15%)
Query: 83 SSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFL--KTKDETQKHLKHFIS 140
S PF LH D++GP YF++ D+ ++F WV L + +D ++
Sbjct: 234 SYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILA 293
Query: 141 YVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHILN 198
+++NQF ++ ++ D GSE+ T FL GI T + +GV ER ++ +L+
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 199 VARSLAFHSHVPITMWNFTVQHAVHIINRIPSP-------------------LLKFKSPF 239
R+ S +P +W ++ + + N + SP LL F P
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQPV 413
Query: 240 ELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFN-----PRARKT------IFLGFKEGTK 288
+ P S IH + G YA L R + P +KT + L KE
Sbjct: 414 IVNDHNPNSKIHPR--GIPGYA--LHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL 469
Query: 289 GSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHP---- 344
YD + + ++R Y++ + ++ S H D +D+ HP
Sbjct: 470 DQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDH----DFQSDIELHPEQPR 525
Query: 345 --ISAPEFQLTSTPPSQY------VSAPAVQHAIPVTDSISEPTVRKSTRISQRP 391
+S STPPS + VS ++ V +ISE + S + S P
Sbjct: 526 NVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNISESNILPSKKRSSTP 580
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 103 bits (257), Expect = 2e-21
Identities = 111/447 (24%), Positives = 198/447 (43%), Gaps = 53/447 (11%)
Query: 452 KSECWREAMTTELNALAKNNTWSV------VTLPPGKVPIGCKWVYKVKYHANGSIERYK 505
+ E + EA E+N L K NTW + P +V I +++ K +K
Sbjct: 819 EKEKYIEAYHKEVNQLLKMNTWDTDKYYDRKEIDPKRV-INSMFIFNRKRDGT-----HK 872
Query: 506 ARLVAQGYTQTEGVDYFDTFSPVAKLTTIR-----VLLSLAAIKGWHLEQLDVNNAFLHG 560
AR VA+G + + DT+ + T+ LSLA +++ QLD+++A+L+
Sbjct: 873 ARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYA 927
Query: 561 DLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYS 620
D+ EE+Y+ PP + ++ +L KSLYGLKQ+ WY + + LI + + +S
Sbjct: 928 DIKEELYIRPPPHLGM--NDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWS 985
Query: 621 LFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK--DLGQLRYFLGFEIA 678
K S + + ++VDD++L ++ K + T L ++ K +LG+ + ++I
Sbjct: 986 CVFKNSQVT---ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDIL 1042
Query: 679 RSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP---------FTDAS 729
+ I + KY +E++ T P +P + + G P D
Sbjct: 1043 GLE--IKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDED 1100
Query: 730 SYR-------RLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSP 781
Y+ +LIG Y+ R D+ Y + L+Q + P +++++ +
Sbjct: 1101 EYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTR 1160
Query: 782 AKGLFFSSSS----ELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSR 837
K L + + + KL +D+ + P +S G LL +I KS K S
Sbjct: 1161 DKQLIWHKNKPTEPDNKLVAISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCT 1219
Query: 838 SSTEAEYRALAHLTCELQWLNYLFHDL 864
S+TEAE A++ L L++L +L
Sbjct: 1220 STTEAEIHAISESVPLLNNLSHLVQEL 1246
Score = 76.6 bits (187), Expect = 3e-13
Identities = 85/355 (23%), Positives = 141/355 (38%), Gaps = 54/355 (15%)
Query: 83 SSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFL--KTKDETQKHLKHFIS 140
S PF LH D++GP YF++ D+ ++F WV L + +D ++
Sbjct: 234 SYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILA 293
Query: 141 YVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHILN 198
+++NQF ++ ++ D GSE+ T FL GI T + +GV ER ++ +L+
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 199 VARSLAFHSHVPITMWNFTVQHAVHIINRIPSP-------------------LLKFKSPF 239
R+ S +P +W ++ + + N + SP LL F P
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQPV 413
Query: 240 ELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFN-----PRARKT------IFLGFKEGTK 288
+ P S IH + G YA L R + P +KT + L KE
Sbjct: 414 IVNDHNPNSKIHPR--GIPGYA--LHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL 469
Query: 289 GSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHP---- 344
YD + + ++R Y++ + ++ S H D +D+ HP
Sbjct: 470 DQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDH----DFQSDIELHPEQPR 525
Query: 345 --ISAPEFQLTSTPPSQY------VSAPAVQHAIPVTDSISEPTVRKSTRISQRP 391
+S STPPS + VS ++ V +ISE + S + S P
Sbjct: 526 NVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNISESNILPSKKRSSTP 580
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 103 bits (256), Expect = 3e-21
Identities = 110/447 (24%), Positives = 199/447 (43%), Gaps = 53/447 (11%)
Query: 452 KSECWREAMTTELNALAKNNTWSV------VTLPPGKVPIGCKWVYKVKYHANGSIERYK 505
+ E + +A E+N L K TW + P +V I +++ K +K
Sbjct: 819 EKEKYIQAYHKEVNQLLKMKTWDTDRYYDRKEIDPKRV-INSMFIFNRKRDGT-----HK 872
Query: 506 ARLVAQGYTQTEGVDYFDTFSPVAKLTTIR-----VLLSLAAIKGWHLEQLDVNNAFLHG 560
AR VA+G + + DT+ P + T+ LSLA +++ QLD+++A+L+
Sbjct: 873 ARFVARG-----DIQHPDTYDPGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYA 927
Query: 561 DLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYS 620
D+ EE+Y+ PP + ++ +L KSLYGLKQ+ WY + + LI + + +S
Sbjct: 928 DIKEELYIRPPPHLGM--NDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWS 985
Query: 621 LFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK--DLGQLRYFLGFEIA 678
K S + + ++VDD++L ++ K + T L ++ K +LG+ + ++I
Sbjct: 986 CVFKNSQVT---ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDIL 1042
Query: 679 RSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP--FTD--------- 727
+ I + KY +E++ T P +P + + G P + D
Sbjct: 1043 GLE--IKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEED 1100
Query: 728 -----ASSYRRLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSP 781
++LIG Y+ R D+ Y + L+Q + P +++++ ++
Sbjct: 1101 DYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTR 1160
Query: 782 AKGLFFSSSSEL----KLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSR 837
K L + S + KL +D+ + P +S G LL +I KS K S
Sbjct: 1161 DKQLIWHKSKPVKPTNKLVVISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCT 1219
Query: 838 SSTEAEYRALAHLTCELQWLNYLFHDL 864
S+TEAE A++ L L++L +L
Sbjct: 1220 STTEAEIHAISESVPLLNNLSHLVQEL 1246
Score = 76.6 bits (187), Expect = 3e-13
Identities = 85/355 (23%), Positives = 141/355 (38%), Gaps = 54/355 (15%)
Query: 83 SSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFL--KTKDETQKHLKHFIS 140
S PF LH D++GP YF++ D+ ++F WV L + +D ++
Sbjct: 234 SYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILA 293
Query: 141 YVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHILN 198
+++NQF ++ ++ D GSE+ T FL GI T + +GV ER ++ +L+
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 199 VARSLAFHSHVPITMWNFTVQHAVHIINRIPSP-------------------LLKFKSPF 239
R+ S +P +W ++ + + N + SP LL F P
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQPV 413
Query: 240 ELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFN-----PRARKT------IFLGFKEGTK 288
+ P S IH + G YA L R + P +KT + L KE
Sbjct: 414 IVNDHNPNSKIHPR--GIPGYA--LHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL 469
Query: 289 GSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHP---- 344
YD + + ++R Y++ + ++ S H D +D+ HP
Sbjct: 470 DQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDH----DFQSDIELHPEQPR 525
Query: 345 --ISAPEFQLTSTPPSQY------VSAPAVQHAIPVTDSISEPTVRKSTRISQRP 391
+S STPPS + VS ++ V +ISE + S + S P
Sbjct: 526 NVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNISESNILPSKKRSSTP 580
>M820_ARATH (P92520) Hypothetical mitochondrial protein AtMg00820
(ORF170)
Length = 170
Score = 101 bits (252), Expect = 7e-21
Identities = 49/114 (42%), Positives = 76/114 (65%), Gaps = 1/114 (0%)
Query: 428 SPTYTHFCCTISSINEPKTFAQANKSECWREAMTTELNALAKNNTWSVVTLPPGKVPIGC 487
+P Y+ T + EPK+ A K W +AM EL+AL++N TW +V P + +GC
Sbjct: 13 NPKYS-LTITTTIKKEPKSVIFALKDPGWCQAMQEELDALSRNKTWILVPPPVNQNILGC 71
Query: 488 KWVYKVKYHANGSIERYKARLVAQGYTQTEGVDYFDTFSPVAKLTTIRVLLSLA 541
KWV+K K H++G+++R KARLVA+G+ Q EG+ + +T+SPV + TIR +L++A
Sbjct: 72 KWVFKTKLHSDGTLDRLKARLVAKGFHQEEGIYFVETYSPVVRTATIRTILNVA 125
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 101 bits (251), Expect = 1e-20
Identities = 112/444 (25%), Positives = 195/444 (43%), Gaps = 47/444 (10%)
Query: 452 KSECWREAMTTELNALAKNNTWSV------VTLPPGKVPIGCKWVYKVKYHANGSIERYK 505
+ E + EA E+N L K TW + P +V I +++ K +K
Sbjct: 1246 EKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRV-INSMFIFNKKRDGT-----HK 1299
Query: 506 ARLVAQGYTQTEGVDYFDTF--SPVAKLTTIRVLLSLAAIKGWHLEQLDVNNAFLHGDLH 563
AR VA+G Q D +DT S + LSLA +++ QLD+++A+L+ D+
Sbjct: 1300 ARFVARGDIQHP--DTYDTGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYADIK 1357
Query: 564 EEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYSLFV 623
EE+Y+ PP + ++ +L KS YGLKQ+ WY + + LI + + +S
Sbjct: 1358 EELYIRPPPHLGM--NDKLIRLKKSHYGLKQSGANWYETIKSYLIKQCGMEEVRGWSCVF 1415
Query: 624 KVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK--DLGQLRYFLGFEIARSK 681
K S + + ++VDD++L ++ K + T L ++ K +LG+ + ++I +
Sbjct: 1416 KNSQVT---ICLFVDDMILFSKDLNANKKIITTLKKQYDTKIINLGESDNEIQYDILGLE 1472
Query: 682 SGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP---------FTDASSYR 732
I + KY +E++ T P +P + + G P D Y+
Sbjct: 1473 --IKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQDELEIDEDEYK 1530
Query: 733 -------RLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKG 784
+LIG Y+ R D+ Y + L+Q + P +++++ + K
Sbjct: 1531 EKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSRQVLDMTYELIQFMWDTRDKQ 1590
Query: 785 LFFSSSS----ELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSST 840
L + + + KL +D+ + P +S G LL +I KS K S S+T
Sbjct: 1591 LIWHKNKPTEPDNKLVAISDASYGNQP-YYKSQIGNIFLLNGKVIGGKSTKASLTCTSTT 1649
Query: 841 EAEYRALAHLTCELQWLNYLFHDL 864
EAE A++ L L+YL +L
Sbjct: 1650 EAEIHAISESVPLLNNLSYLIQEL 1673
Score = 76.6 bits (187), Expect = 3e-13
Identities = 85/355 (23%), Positives = 141/355 (38%), Gaps = 54/355 (15%)
Query: 83 SSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFL--KTKDETQKHLKHFIS 140
S PF LH D++GP YF++ D+ ++F WV L + +D ++
Sbjct: 661 SYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILA 720
Query: 141 YVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHILN 198
+++NQF ++ ++ D GSE+ T FL GI T + +GV ER ++ +L+
Sbjct: 721 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 780
Query: 199 VARSLAFHSHVPITMWNFTVQHAVHIINRIPSP-------------------LLKFKSPF 239
R+ S +P +W ++ + + N + SP LL F P
Sbjct: 781 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQPV 840
Query: 240 ELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFN-----PRARKT------IFLGFKEGTK 288
+ P S IH + G YA L R + P +KT + L KE
Sbjct: 841 IVNDHNPNSKIHPR--GIPGYA--LHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL 896
Query: 289 GSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHP---- 344
YD + + ++R Y++ + ++ S H D +D+ HP
Sbjct: 897 DQFNYDALTFDEDLNRLTASYQSFIASNEIQESNDLNIESDH----DFQSDIELHPEQPR 952
Query: 345 --ISAPEFQLTSTPPSQY------VSAPAVQHAIPVTDSISEPTVRKSTRISQRP 391
+S STPPS + VS ++ V +ISE + S + S P
Sbjct: 953 NVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNISESNILPSKKRSSTP 1007
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 100 bits (250), Expect = 1e-20
Identities = 113/449 (25%), Positives = 198/449 (43%), Gaps = 57/449 (12%)
Query: 452 KSECWREAMTTELNALAKNNTWSV------VTLPPGKVPIGCKWVYKVKYHANGSIERYK 505
+ E + EA E+N L K TW + P +V I +++ K +K
Sbjct: 1246 EKEKYIEAYHKEVNQLLKMKTWDTDEYYDRKEIDPKRV-INSMFIFNKKRDGT-----HK 1299
Query: 506 ARLVAQGYTQTEGVDYFDTFSPVAKLTTIR-----VLLSLAAIKGWHLEQLDVNNAFLHG 560
AR VA+G + + DT+ + T+ LSLA +++ QLD+++A+L+
Sbjct: 1300 ARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYA 1354
Query: 561 DLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYS 620
D+ EE+Y+ PP + ++ +L KSLYGLKQ+ WY + + LI + + +S
Sbjct: 1355 DIKEELYIRPPPHLGM--NDKLIRLKKSLYGLKQSGANWYETIKSYLIQQCGMEEVRGWS 1412
Query: 621 LFVKVSGASFTALLVYVDDIVLAGNCISE----IKSVKTFLDNKFCIKDLGQLRYFLGFE 676
K S + + ++VDD+VL ++ I+ +K D K I +LG+ + ++
Sbjct: 1413 CVFKNSQVT---ICLFVDDMVLFSKNLNSNKRIIEKLKMQYDTK--IINLGESDEEIQYD 1467
Query: 677 IARSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP--FTD------- 727
I + I + KY +E++ T P +P + + G P + D
Sbjct: 1468 ILGLE--IKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELE 1525
Query: 728 -------ASSYRRLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKS 779
++LIG Y+ R D+ Y + L+Q + P +++++ +
Sbjct: 1526 EDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWN 1585
Query: 780 SPAKGLFFSSSSEL----KLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTV 835
+ K L + S + KL +D+ + P +S G LL +I KS K S
Sbjct: 1586 TRDKQLIWHKSKPVKPTNKLVVISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLT 1644
Query: 836 SRSSTEAEYRALAHLTCELQWLNYLFHDL 864
S+TEAE A++ L L+YL +L
Sbjct: 1645 CTSTTEAEIHAISESVPLLNNLSYLIQEL 1673
Score = 76.6 bits (187), Expect = 3e-13
Identities = 85/355 (23%), Positives = 141/355 (38%), Gaps = 54/355 (15%)
Query: 83 SSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFL--KTKDETQKHLKHFIS 140
S PF LH D++GP YF++ D+ ++F WV L + +D ++
Sbjct: 661 SYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILA 720
Query: 141 YVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHILN 198
+++NQF ++ ++ D GSE+ T FL GI T + +GV ER ++ +L+
Sbjct: 721 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 780
Query: 199 VARSLAFHSHVPITMWNFTVQHAVHIINRIPSP-------------------LLKFKSPF 239
R+ S +P +W ++ + + N + SP LL F P
Sbjct: 781 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQPV 840
Query: 240 ELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFN-----PRARKT------IFLGFKEGTK 288
+ P S IH + G YA L R + P +KT + L KE
Sbjct: 841 IVNDHNPNSKIHPR--GIPGYA--LHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL 896
Query: 289 GSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHP---- 344
YD + + ++R Y++ + ++ S H D +D+ HP
Sbjct: 897 DQFNYDALTFDEDLNRLTASYQSFIASNEIQQSDDLNIESDH----DFQSDIELHPEQPR 952
Query: 345 --ISAPEFQLTSTPPSQY------VSAPAVQHAIPVTDSISEPTVRKSTRISQRP 391
+S STPPS + VS ++ V +ISE + S + S P
Sbjct: 953 NVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNISESNILPSKKRSSTP 1007
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 99.0 bits (245), Expect = 5e-20
Identities = 111/447 (24%), Positives = 196/447 (43%), Gaps = 53/447 (11%)
Query: 452 KSECWREAMTTELNALAKNNTWSV------VTLPPGKVPIGCKWVYKVKYHANGSIERYK 505
+ E + EA E+N L K TW + P +V I +++ K +K
Sbjct: 819 EKEKYIEAYHKEVNQLLKMKTWDTDKYYDRKEIDPKRV-INSMFIFNRKRDGT-----HK 872
Query: 506 ARLVAQGYTQTEGVDYFDTFSPVAKLTTIR-----VLLSLAAIKGWHLEQLDVNNAFLHG 560
AR VA+G + + DT+ + T+ LSLA +++ QLD+++A+L+
Sbjct: 873 ARFVARG-----DIQHPDTYDSGMQSNTVHHYALMTSLSLALDNNYYITQLDISSAYLYA 927
Query: 561 DLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADYS 620
D+ EE+Y+ PP + ++ +L KSLYGLKQ+ WY + + LI + + +S
Sbjct: 928 DIKEELYIRPPPHLGM--NDKLIRLKKSLYGLKQSGANWYETIKSYLIKQCGMEEVRGWS 985
Query: 621 LFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIK--DLGQLRYFLGFEIA 678
V S + ++VDD+VL ++ K + L ++ K +LG+ + ++I
Sbjct: 986 C---VFENSQVTICLFVDDMVLFSKNLNSNKRIIDKLKMQYDTKIINLGESDEEIQYDIL 1042
Query: 679 RSKSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGTP--FTD--------- 727
+ I + KY +E++ T P +P + + G P + D
Sbjct: 1043 GLE--IKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRKLSAPGQPGLYIDQQELELEED 1100
Query: 728 -----ASSYRRLIGRLLYL-TNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSP 781
++LIG Y+ R D+ Y + L+Q + P +++++ ++
Sbjct: 1101 DYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHILFPSKQVLDMTYELIQFIWNTR 1160
Query: 782 AKGLFFSSSSEL----KLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSR 837
K L + S + KL +D+ + P +S G LL +I KS K S
Sbjct: 1161 DKQLIWHKSKPVKPTNKLVVISDASYGNQP-YYKSQIGNIYLLNGKVIGGKSTKASLTCT 1219
Query: 838 SSTEAEYRALAHLTCELQWLNYLFHDL 864
S+TEAE A++ L L+YL +L
Sbjct: 1220 STTEAEIHAISESVPLLNNLSYLIQEL 1246
Score = 75.9 bits (185), Expect = 4e-13
Identities = 85/355 (23%), Positives = 140/355 (38%), Gaps = 54/355 (15%)
Query: 83 SSAPFDLLHADLWGPYSTPSFLGHKYFLTLVDDYSRFTWVIFL--KTKDETQKHLKHFIS 140
S PF LH D++GP YF++ D+ ++F WV L + +D ++
Sbjct: 234 SYEPFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRREDSILDVFTTILA 293
Query: 141 YVENQFHTTLKCLRSDNGSEFIAMT--SFLLSKGIIHHKTCVETPQQNGVVERKHQHILN 198
+++NQF ++ ++ D GSE+ T FL GI T + +GV ER ++ +L+
Sbjct: 294 FIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSRAHGVAERLNRTLLD 353
Query: 199 VARSLAFHSHVPITMWNFTVQHAVHIINRIPSP-------------------LLKFKSPF 239
R+ S +P +W ++ + + N + SP LL F P
Sbjct: 354 DCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASPKSKKSARQHAGLAGLDISTLLPFGQPV 413
Query: 240 ELLHKEPPSIIHLKVFGCLAYASTLQAHRTKFN-----PRARKT------IFLGFKEGTK 288
+ P S IH + G YA L R + P +KT + L KE
Sbjct: 414 IVNDHNPNSKIHPR--GIPGYA--LHPSRNSYGYIIYLPSLKKTVDTTNYVILQGKESRL 469
Query: 289 GSILYDLNSNELFVSRNVIFYENHFPFTLATKQANIPTTSSHIDLGDPITDLSPHP---- 344
YD + + ++R Y + + ++ S H D +D+ HP
Sbjct: 470 DQFNYDALTFDEDLNRLTASYHSFIASNEIQQSNDLNIESDH----DFQSDIELHPEQLR 525
Query: 345 --ISAPEFQLTSTPPSQY------VSAPAVQHAIPVTDSISEPTVRKSTRISQRP 391
+S STPPS + VS ++ V +ISE + S + S P
Sbjct: 526 NVLSKAVSPTDSTPPSTHTEDSKRVSKTNIRAPREVDPNISESNILPSKKRSSTP 580
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 98.6 bits (244), Expect = 6e-20
Identities = 44/82 (53%), Positives = 61/82 (73%)
Query: 739 LYLTNTRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSPAKGLFFSSSSELKLHGF 798
+YLT TRPD++++V LSQF S QA ++L Y+K + +GLF+S++S+L+L F
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 799 ADSDWACCPDTRRSVTGYCVLL 820
ADSDWA CPDTRRSVTG+C L+
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLV 82
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 89.7 bits (221), Expect = 3e-17
Identities = 93/368 (25%), Positives = 164/368 (44%), Gaps = 37/368 (10%)
Query: 504 YKARLVAQGYTQTEGVDYFDTFSPVAKLTT----IRVLLSLAAIKGWHLEQLDVNNAFLH 559
YKAR+V +G TQ+ DT+S + + I++ L +A + ++ LD+N+AFL+
Sbjct: 1337 YKARIVCRGDTQSP-----DTYSVITTESLNHNHIKIFLMIANNRNMFMKTLDINHAFLY 1391
Query: 560 GDLHEEVYMALPPGYPTINSSQVCKLNKSLYGLKQASRQWYSKLSTSLISFGYTQSLADY 619
L EE+Y+ P + V KLNK+LYGLKQ+ ++W L L G + Y
Sbjct: 1392 AKLEEEIYIPHPH-----DRRCVVKLNKALYGLKQSPKEWNDHLRQYLNGIGLKDN--SY 1444
Query: 620 SLFVKVSGASFTALLVYVDDIVLAGNCISEIKSVKTFLDNKFCIKDLGQL------RYFL 673
+ + + + VYVDD V+A + + L + F +K G L L
Sbjct: 1445 TPGLYQTEDKNLMIAVYVDDCVIAASNEQRLDEFINKLKSNFELKITGTLIDDVLDTDIL 1504
Query: 674 GFEIARS----------KSGILLNQRKYTLELLEDAGTLGSKPAATPFDPSTKLGATTGT 723
G ++ + KS I +KY EL + + + DP + +
Sbjct: 1505 GMDLVYNKRLGTIDLTLKSFINRMDKKYNEELKKIRKSSIPHMSTYKIDPKKDVLQMSEE 1564
Query: 724 PFTD-ASSYRRLIGRLLYLTN-TRPDISYSVQNLSQFVSRPMVPHYQAAQRILKYLKSSP 781
F ++L+G L Y+ + R DI ++V+ +++ V+ P + +I++YL
Sbjct: 1565 EFRQGVLKLQQLLGELNYVRHKCRYDIEFAVKKVARLVNYPHERVFYMIYKIIQYLVRYK 1624
Query: 782 AKGLFF--SSSSELKLHGFADSDWACCPDTRRSVTGYCVLLGSSLISWKSKKQSTVSRSS 839
G+ + + + K+ D+ D + + G + G ++ + S K + SS
Sbjct: 1625 DIGIHYDRDCNKDKKVIAITDASVGSEYDAQSRI-GVILWYGMNIFNVYSNKSTNRCVSS 1683
Query: 840 TEAEYRAL 847
TEAE A+
Sbjct: 1684 TEAELHAI 1691
Score = 67.4 bits (163), Expect = 2e-10
Identities = 45/161 (27%), Positives = 80/161 (48%), Gaps = 5/161 (3%)
Query: 93 DLWGPYSTPSFLGHKYFLTLVDDYSRF--TWVIFLKTKDETQKHLKHFISYVENQFHTTL 150
D++GP S+ + +Y L +VD+ +R+ T F K + ++ I YVE QF +
Sbjct: 631 DIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQYVETQFDRKV 690
Query: 151 KCLRSDNGSEFI--AMTSFLLSKGIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSH 208
+ + SD G+EF + + +SKGI H T + NG ER + I+ A +L S+
Sbjct: 691 REINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITDATTLLRQSN 750
Query: 209 VPITMWNFTVQHAVHIINRIPSPLLKFKSPFELLHKEPPSI 249
+ + W + V A +I N + K P + + ++P ++
Sbjct: 751 LRVKFWEYAVTSATNIRNYLEHKSTG-KLPLKAISRQPVTV 790
>POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)] (Fragment)
Length = 581
Score = 54.3 bits (129), Expect = 1e-06
Identities = 49/172 (28%), Positives = 76/172 (43%), Gaps = 12/172 (6%)
Query: 101 PSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHL-KHFISYVENQFHTTLKCLRSDNGS 159
P G+KY L VD +S WV TK ET K + K + + +F + L +DNG
Sbjct: 307 PGLYGYKYLLVFVDTFS--GWVEAFPTKHETAKIVTKKLLEEIFPRFGMP-QVLGTDNGP 363
Query: 160 EFIAMTSFLLSK--GIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFT 217
F++ S ++K GI C PQ +G VER ++ I L + W
Sbjct: 364 AFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGT--RDWVLL 421
Query: 218 VQHAVHIINRIPSPLLKFKSPFELLHKEPPSII--HLKVFGCLAYASTLQAH 267
+ A++ P P +P+E+L+ PP ++ H + +LQAH
Sbjct: 422 LPLALYRARNTPGP--HGLTPYEILYGAPPPLVNFHDPEMSKFTNSPSLQAH 471
>POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 54.3 bits (129), Expect = 1e-06
Identities = 49/172 (28%), Positives = 76/172 (43%), Gaps = 12/172 (6%)
Query: 101 PSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHL-KHFISYVENQFHTTLKCLRSDNGS 159
P G+KY L VD +S WV TK ET K + K + + +F + L +DNG
Sbjct: 922 PGLYGYKYLLVFVDTFS--GWVEAFPTKHETAKIVTKKLLEEIFPRFGMP-QVLGTDNGP 978
Query: 160 EFIAMTSFLLSK--GIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFT 217
F++ S ++K GI C PQ +G VER ++ I L + W
Sbjct: 979 AFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGT--RDWVLL 1036
Query: 218 VQHAVHIINRIPSPLLKFKSPFELLHKEPPSII--HLKVFGCLAYASTLQAH 267
+ A++ P P +P+E+L+ PP ++ H + +LQAH
Sbjct: 1037 LPLALYRARNTPGP--HGLTPYEILYGAPPPLVNFHDPEMSKFTNSPSLQAH 1086
>POL3_MOUSE (P11367) Retrovirus-related Pol polyprotein
(Endonuclease) (Fragment)
Length = 390
Score = 54.3 bits (129), Expect = 1e-06
Identities = 49/172 (28%), Positives = 76/172 (43%), Gaps = 12/172 (6%)
Query: 101 PSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHL-KHFISYVENQFHTTLKCLRSDNGS 159
P G+KY L VD +S WV TK ET K + K + + +F + L +DNG
Sbjct: 131 PGLYGYKYLLVFVDTFS--GWVEAFPTKHETAKIVTKKLLEEIFPRFGMP-QVLGTDNGP 187
Query: 160 EFIAMTSFLLSK--GIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFT 217
F++ S ++K GI C PQ +G VER ++ I L + W
Sbjct: 188 AFVSQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLATGT--RDWVLL 245
Query: 218 VQHAVHIINRIPSPLLKFKSPFELLHKEPPSII--HLKVFGCLAYASTLQAH 267
+ A++ P P +P+E+L+ PP ++ H + +LQAH
Sbjct: 246 LPLALYRARNTPGP--HGLTPYEILYGAPPPLVNFHDPEMSKFTNSPSLQAH 295
>POL_MLVAV (P03356) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 50.8 bits (120), Expect = 2e-05
Identities = 49/172 (28%), Positives = 75/172 (43%), Gaps = 12/172 (6%)
Query: 101 PSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHL-KHFISYVENQFHTTLKCLRSDNGS 159
P G+KY L VD +S WV TK ET + + K + + +F + L SDNG
Sbjct: 922 PGLYGYKYLLVFVDTFS--GWVEAFPTKRETARVVSKKLLEEIFPRFGMP-QVLGSDNGP 978
Query: 160 EFIAMTSFLLSK--GIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFT 217
F + S ++ GI C PQ +G VER ++ I L + W
Sbjct: 979 AFTSQVSQSVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLAAGT--RDWVLL 1036
Query: 218 VQHAVHIINRIPSPLLKFKSPFELLHKEPPSII--HLKVFGCLAYASTLQAH 267
+ A++ P P +P+E+L+ PP ++ H L + +LQAH
Sbjct: 1037 LPLALYRARNTPGP--HGLTPYEILYGAPPPLVNFHDPDMSELTNSPSLQAH 1086
>POL_MLVFP (P26808) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 50.1 bits (118), Expect = 3e-05
Identities = 46/172 (26%), Positives = 76/172 (43%), Gaps = 12/172 (6%)
Query: 101 PSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHL-KHFISYVENQFHTTLKCLRSDNGS 159
P G+KY L VD +S WV TK ET K + K + + +F + L +DNG
Sbjct: 927 PGLYGYKYLLVFVDTFS--GWVEAFPTKKETAKVVTKKLLEEIFPRFGMP-QVLGTDNGP 983
Query: 160 EFIAMTSFLLSK--GIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFT 217
F++ S ++ G+ C PQ +G VER ++ I L + W
Sbjct: 984 AFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLAT--GSRDWVLL 1041
Query: 218 VQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHL--KVFGCLAYASTLQAH 267
+ A++ P P +P+E+L+ PP +++ + + +LQAH
Sbjct: 1042 LPLALYRARNTPGP--HGLTPYEILYGAPPPLVNFPDPDMAKVTHNPSLQAH 1091
>POL_MLVF5 (P26810) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1204
Score = 50.1 bits (118), Expect = 3e-05
Identities = 46/172 (26%), Positives = 76/172 (43%), Gaps = 12/172 (6%)
Query: 101 PSFLGHKYFLTLVDDYSRFTWVIFLKTKDETQKHL-KHFISYVENQFHTTLKCLRSDNGS 159
P G+KY L VD +S WV TK ET K + K + + +F + L +DNG
Sbjct: 927 PGLYGYKYLLVFVDTFS--GWVEAFPTKKETAKVVTKKLLEEIFPRFGMP-QVLGTDNGP 983
Query: 160 EFIAMTSFLLSK--GIIHHKTCVETPQQNGVVERKHQHILNVARSLAFHSHVPITMWNFT 217
F++ S ++ G+ C PQ +G VER ++ I L + W
Sbjct: 984 AFVSKVSQTVADLLGVDWKLHCAYRPQSSGQVERMNRTIKETLTKLTLAT--GSRDWVLL 1041
Query: 218 VQHAVHIINRIPSPLLKFKSPFELLHKEPPSIIHL--KVFGCLAYASTLQAH 267
+ A++ P P +P+E+L+ PP +++ + + +LQAH
Sbjct: 1042 LPLALYRARNTPGP--HGLTPYEILYGAPPPLVNFPDPDMAKVTHNPSLQAH 1091
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.134 0.410
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 102,139,913
Number of Sequences: 164201
Number of extensions: 4311337
Number of successful extensions: 10275
Number of sequences better than 10.0: 62
Number of HSP's better than 10.0 without gapping: 18
Number of HSP's successfully gapped in prelim test: 44
Number of HSP's that attempted gapping in prelim test: 10145
Number of HSP's gapped (non-prelim): 106
length of query: 864
length of database: 59,974,054
effective HSP length: 119
effective length of query: 745
effective length of database: 40,434,135
effective search space: 30123430575
effective search space used: 30123430575
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 70 (31.6 bits)
Medicago: description of AC137079.15


