

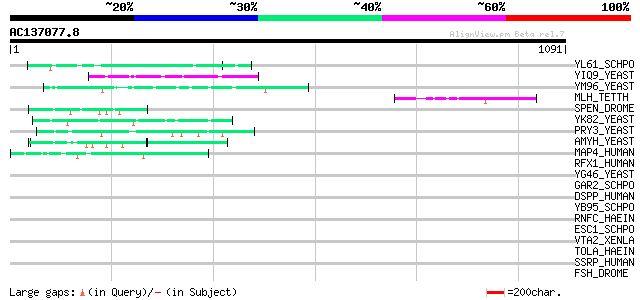
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC137077.8 - phase: 0
(1091 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein P... 58 2e-07
YIQ9_YEAST (P40442) Hypothetical 99.7 kDa protein in SDL1 5'regi... 53 5e-06
YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4 ... 50 3e-05
MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC ... 50 3e-05
SPEN_DROME (Q8SX83) Split ends protein 47 2e-04
YK82_YEAST (P36170) Hypothetical 122.2 kDa protein in SIR1 3'reg... 47 4e-04
PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3) 46 5e-04
AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3) (G... 46 5e-04
MAP4_HUMAN (P27816) Microtubule-associated protein 4 (MAP 4) 46 6e-04
RFX1_HUMAN (P22670) MHC class II regulatory factor RFX1 (RFX) (E... 45 0.001
YG46_YEAST (P53301) Hypothetical 52.8 kDa protein in BUB1-HIP1 i... 44 0.002
GAR2_SCHPO (P41891) Protein gar2 44 0.002
DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contai... 44 0.002
YB95_SCHPO (O42970) Hypothetical serine-rich protein C1E8.05 in ... 44 0.003
RNFC_HAEIN (P71397) Electron transport complex protein rnfC 43 0.005
ESC1_SCHPO (Q04635) Protein esc1 43 0.005
VTA2_XENLA (P18709) Vitellogenin A2 precursor (VTG A2) [Contains... 42 0.009
TOLA_HAEIN (P44678) TolA protein 42 0.009
SSRP_HUMAN (Q08945) Structure-specific recognition protein 1 (SS... 42 0.009
FSH_DROME (P13709) Female sterile homeotic protein (Fragile-chor... 42 0.009
>YL61_SCHPO (Q8TFG9) Hypothetical serine/threonine-rich protein
PB15E9.01c in chromosome I precursor
Length = 943
Score = 57.8 bits (138), Expect = 2e-07
Identities = 85/402 (21%), Positives = 148/402 (36%), Gaps = 37/402 (9%)
Query: 36 LCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPL---------------I 80
L S L + TS + S+ S+PL ++S T A+
Sbjct: 227 LASSSLNSTTSATATSSSISSTVSSSTPLTSSNSTTAATSASATSSSAQYNTSSLLPSST 286
Query: 81 PLSSPLWSLPTLSADSLQSSALAR-GSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQA 139
P S+PL S + +A S S+ L S S + TPL S + S++ +S
Sbjct: 287 PSSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSVSSANSTTATSTSSTPLSSV 346
Query: 140 PLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQS 199
S TP N+ + S SS L SV + S+S +T +S+ + S
Sbjct: 347 NSTTATSASSTPLTSVNSTTATSASS--TPLTSVNSTSATSASSTPLTSANSTTSTSVSS 404
Query: 200 T--FVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTS 257
T T S L ++V+ P ++ A LS + + + ++S ST+
Sbjct: 405 TAPSYNTSSVLPTSSVSSTPLSSANSTTATSASSTPLSSVNSTTATSASSTPLSSVNSTT 464
Query: 258 VSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASA 317
++A+ ++S + S +PL S S + A++
Sbjct: 465 ATSASSTPLTSVNSTTATSASSTPLT--------------SVNSTSATSASSTPLTSANS 510
Query: 318 LSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAA 377
++ +V+ + +N S S + S A++ + + A AS+
Sbjct: 511 TTSTSVSSTAPSYNTSSVLPTSSVSSTPLSSANSTTATSASSTPLTSVNSTTATSASSTP 570
Query: 378 FQAKLMADEALISSG-YENTSQGNNTFLPEGTSNLGQATPAS 418
F + A S+G + NT+ GN G+ +TP S
Sbjct: 571 FGNSTITSSASGSTGEFTNTNSGNGDV--SGSVTTPTSTPLS 610
Score = 51.2 bits (121), Expect = 1e-05
Identities = 82/442 (18%), Positives = 176/442 (39%), Gaps = 36/442 (8%)
Query: 36 LCSQDLAART-SDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSA 94
+ S LA+ + + S++ S ++S +++ ATPT + LSS T ++
Sbjct: 134 ITSSSLASSSITSSSLASSSTTSSSLASSSTNSTTSATPTSSATSSSLSS------TAAS 187
Query: 95 DSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPD 154
+S SS+LA S+ + A S + N S+ L + S T A
Sbjct: 188 NSATSSSLASSSLNSTTSATATSSSLSSTAASNSATSSS-------LASSSLNSTTSATA 240
Query: 155 NNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVT 214
++ +S++ SS T P +S T AS++ + + T S L ++ +
Sbjct: 241 TSSSISSTVSSST-----------PLTSSNSTTAATSASATSSSAQY-NTSSLLPSSTPS 288
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
P ++ A L+ + + + ++S +S + + AT + P+SSV
Sbjct: 289 STPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSVSSANSTTATSTSSTPLSSV-N 347
Query: 275 SVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S + S + + + S L V + ++ L++A S + +
Sbjct: 348 STTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTSTSVSSTAP 407
Query: 335 KHKNSGFM--SDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSG 392
+ S + S + + S+A + A +A ++ ++V S A A ++ S+
Sbjct: 408 SYNTSSVLPTSSVSSTPLSSANSTTATSA---SSTPLSSVNSTTATSASSTPLSSVNSTT 464
Query: 393 YENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATK 452
+ S T + T+ +TP + + + ++ + + +A V ++T
Sbjct: 465 ATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTSTSV----SSTA 520
Query: 453 RAENMDAILKAAELAAEAVSQA 474
+ N ++L + +++ +S A
Sbjct: 521 PSYNTSSVLPTSSVSSTPLSSA 542
Score = 43.1 bits (100), Expect = 0.004
Identities = 77/409 (18%), Positives = 152/409 (36%), Gaps = 48/409 (11%)
Query: 82 LSSPLWSLPTLSADSL---QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQ 138
LS P + PT+ AD + + A SV + T + Y + S + S+S S
Sbjct: 25 LSDPA-NTPTILADDIVHGYTPATYLSSVPTLLKRATTSYNYNTSSASSSSLTSSSAASS 83
Query: 139 APLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQ 198
+ + S + N+ SASP+S ++ +S S SSS +SS L
Sbjct: 84 SLTSSSSLASSS----TNSTTSASPTSSSLTSSSATSSSLASSS---------TTSSSLA 130
Query: 199 STFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSV 258
S+ + T S L ++++T SS + + ++AS ++ S
Sbjct: 131 SSSI-TSSSLASSSITSSSLASSS----------------------TTSSSLASSSTNST 167
Query: 259 SAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASAL 318
++ATP + SS+ + S S + + + S A ++ +S+L
Sbjct: 168 TSATPTSSATSSSLSSTAASNSATSSSLASSSLNSTTSATATSSSLSSTAASNSATSSSL 227
Query: 319 SAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAF 378
+++++N + +S S + + + AA A A +++A +++
Sbjct: 228 ASSSLNSTTSA-----TATSSSISSTVSSSTPLTSSNSTTAATSASATSSSAQYNTSSLL 282
Query: 379 QAKLMADEALISSGYENTSQGNNTFLP---EGTSNLGQATPASILKGANGPNSPGSFIVA 435
+ + L S+ + ++T L T+ +TP S + AN + +
Sbjct: 283 PSSTPSSTPLSSANSTTATSASSTPLTSVNSTTTTSASSTPLSSVSSANSTTATSTSSTP 342
Query: 436 AKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQAGKIVTMGDPL 484
+AS+ + N A+ +V+ PL
Sbjct: 343 LSSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTPL 391
Score = 33.9 bits (76), Expect = 2.5
Identities = 37/150 (24%), Positives = 59/150 (38%), Gaps = 18/150 (12%)
Query: 895 SSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSS-NHTKNASQSES 953
SS S A+S T P L VN TS S+T ++ SS N T S S +
Sbjct: 284 SSTPSSTPLSSANSTTATSASSTP--LTSVNSTTTTSASSTPLSSVSSANSTTATSTSST 341
Query: 954 RVERAPYSTTDGATQVPI--VFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGG 1011
+ +T A+ P+ V S+ ATS ++ P ++ A+ P
Sbjct: 342 PLSSVNSTTATSASSTPLTSVNSTTATSASSTPLTSVNSTSATSASSTP----------- 390
Query: 1012 KALNDKPTTSTSEPDALEPRRSNRRIQPTS 1041
L +T+++ + P + + PTS
Sbjct: 391 --LTSANSTTSTSVSSTAPSYNTSSVLPTS 418
Score = 32.0 bits (71), Expect = 9.3
Identities = 28/117 (23%), Positives = 50/117 (41%), Gaps = 7/117 (5%)
Query: 930 TSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPI--VFSSQATSTNTLP--- 984
+SVS+T ++ S+N T S S + + +T A+ P+ V S+ ATS ++ P
Sbjct: 418 SSVSSTPLS--SANSTTATSASSTPLSSVNSTTATSASSTPLSSVNSTTATSASSTPLTS 475
Query: 985 TKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRSNRRIQPTS 1041
T + AS L + L +T+++ + P + + PTS
Sbjct: 476 VNSTTATSASSTPLTSVNSTSATSASSTPLTSANSTTSTSVSSTAPSYNTSSVLPTS 532
>YIQ9_YEAST (P40442) Hypothetical 99.7 kDa protein in SDL1 5'region
precursor
Length = 995
Score = 52.8 bits (125), Expect = 5e-06
Identities = 68/336 (20%), Positives = 156/336 (46%), Gaps = 23/336 (6%)
Query: 155 NNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVT 214
N+T +S++ SS ++ ++S GS+ SSSI + + +S++ + S+ + S + +
Sbjct: 28 NSTSISSNSSSTSV-VSSSSGSVSISSSIAETS----SSATDILSSITQSASSTSGVSSS 82
Query: 215 VPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
V P+ S + + +S D + QS + AS S+SVS + + SSV +
Sbjct: 83 VGPSSSSVVSSSVSQSSSSVS-DVSSSVSQS--SSSASDVSSSVSQSASSTSDVSSSVSQ 139
Query: 275 SVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
S S S ++ + + S V ++ A + S+ + + + + ++ + +
Sbjct: 140 SSSSASDVSSSVSQSSSSASDVSS-----SVSQSASSASDVSSSVSQSASSTSDVSSSVS 194
Query: 335 KHKNSGFMSDIEAKLA-SAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGY 393
+ +S SD+ + ++ S++ A +++V+++A++ ++V+S+ + A + + S
Sbjct: 195 QSSSSA--SDVSSSVSQSSSSASDVSSSVSQSASSTSDVSSSVSQSASSTSGVSSSGSQS 252
Query: 394 ENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKR 453
+++ G+++ P+ TS+ AS G+ NS S +A A + S++
Sbjct: 253 VSSASGSSSSFPQSTSS------ASTASGSATSNSLSSITSSASSASATASNSLSSSDGT 306
Query: 454 AENMDAILKA-AELAAEAVSQAGKIVTMGDPLPLIE 488
+ L + ++ G +V G L L++
Sbjct: 307 IYLPTTTISGDLTLTGKVIATEGVVVAAGAKLTLLD 342
Score = 33.5 bits (75), Expect = 3.2
Identities = 38/173 (21%), Positives = 72/173 (40%), Gaps = 3/173 (1%)
Query: 42 AARTSD--STVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQS 99
A+ TSD S+V QS +SS + ++SS A+ +++ + +S + + + S S
Sbjct: 127 ASSTSDVSSSVSQSSSSASDVSSSVSQSSSSASD-VSSSVSQSASSASDVSSSVSQSASS 185
Query: 100 SALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHL 159
++ SV S + + + S S + S+S A S + + + + +
Sbjct: 186 TSDVSSSVSQSSSSASDVSSSVSQSSSSASDVSSSVSQSASSTSDVSSSVSQSASSTSGV 245
Query: 160 SASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANN 212
S+S S + S P S+S G S+S T + + A+N
Sbjct: 246 SSSGSQSVSSASGSSSSFPQSTSSASTASGSATSNSLSSITSSASSASATASN 298
>YM96_YEAST (Q04893) Hypothetical 113.1 kDa protein in PRE5-FET4
intergenic region
Length = 1140
Score = 50.4 bits (119), Expect = 3e-05
Identities = 113/541 (20%), Positives = 209/541 (37%), Gaps = 70/541 (12%)
Query: 67 ASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALT-PLHPYQSPSP 125
+SS ++ TIA+ L S L S+ S+ S SS ++ SV D + + P + Y + S
Sbjct: 69 SSSLSSDTIASILS--SESLVSI--FSSLSYTSSDISSTSVNDVESSTSGPSNSYSALSS 124
Query: 126 RNF-LGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSS--- 181
N L ST+ SP + N S+ P + L + S ++
Sbjct: 125 TNAQLSSSTTETDSISSSAIQTSSPQTSSSNGGGSSSEPLGKSSVLETTASSSDTTAVTS 184
Query: 182 ----SIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSED 237
++ DV+ P SSSG T VGT S DA+ K+V S
Sbjct: 185 STFTTLTDVSSSPKISSSGSAVTSVGTTS--DAS-------------------KEVFSSS 223
Query: 238 HGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRIL 297
+ SL + +S AS+++S P + +S V S +P A +V
Sbjct: 224 TSD--VSSLLSSTSSPASSTISETLPFSSTILSITSSPVSSEAPSA----TSSSVSSEAS 277
Query: 298 SDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAVAIA 357
S S EA + + A + S+ + S S + ++++S
Sbjct: 278 SSTSSSVSSEAPLATSSVVSSEAPSSTSSV-----VSSEAPSSTSSSVSSEISST----T 328
Query: 358 AAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTSNLGQATPA 417
+++ ++A A ++V S+ A + + + ISS TS ++ P TS++ +
Sbjct: 329 SSSVSSEAPLATSSVVSSEAPSSTSSSVSSEISS---TTSSSVSSEAPLATSSVVSSEAP 385
Query: 418 SILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAILKAAELAAEAVSQAGKI 477
S + +P S + EA S+ + + + K++ +++E S +
Sbjct: 386 SSTSSSVSSEAPSSTSSSVSS------EAPSSTSSSVSSEISSTKSSVMSSEVSSATSSL 439
Query: 478 VTMGDPLPLIELIEAGPEGCWKAS-------RESSREVGLLKDMTRDLVNIDMVRDIPET 530
V+ P + L + S E+S L+ + L + ++ T
Sbjct: 440 VSSEAPSAISSLASSRLFSSKNTSVTSTLVATEASSVTSSLRPSSETLASNSIIESSLST 499
Query: 531 SHAQNRDILSSEISASI--MINEKNTR---GQQARTVSDLVKPVDMVLGSESETQDPSFT 585
+ +S S+++ ++ N+R + + T SDL K + S + T PS +
Sbjct: 500 GYNSTVSTTTSAASSTLGSKVSSSNSRMATSKTSSTSSDLSKSSVIFGNSSTVTTSPSAS 559
Query: 586 V 586
+
Sbjct: 560 I 560
Score = 39.7 bits (91), Expect = 0.045
Identities = 56/240 (23%), Positives = 88/240 (36%), Gaps = 44/240 (18%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S A+ +STV S L G S+P +++PT + +L T A S
Sbjct: 680 STSSASLAINSTVVSSSLAGYSFSTP------ESSPTTS-----------TLVTSEAPST 722
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
SS ++ S + P SPS +F+ STS IS PL + S A +N
Sbjct: 723 VSSMTTSAPFINNSTSARP-----SPSTASFITESTSSISSVPLASGDVTSSLAA--HNL 775
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPP 217
++PS+ + +L S S++ + P SG ST + L V
Sbjct: 776 TTFSAPSTSSAQLVS------KSTTSSSILVTPRIDRSGNSSTASRIATSLPNKTTFVSS 829
Query: 218 AQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVV 277
+S H + + S A A + T S P ++ K+V+
Sbjct: 830 LSSTSA--------------HARNIFNSTVLATAKQIETLTSTVNCSNPTPNYNITKTVI 875
>MLH_TETTH (P40631) Micronuclear linker histone polyprotein (MIC LH)
[Contains: Micronuclear linker histone-alpha;
Micronuclear linker histone-beta; Micronuclear linker
histone-delta; Micronuclear linker histone-gamma]
Length = 633
Score = 50.4 bits (119), Expect = 3e-05
Identities = 65/291 (22%), Positives = 123/291 (41%), Gaps = 39/291 (13%)
Query: 757 SSTHKGDTPHEKRPKLGSNAVEVKG-KDKMSKNIDAAESANPDEMRSLNLTENEIVFNIG 815
SS+ KG + + + S+A K +D+ K ++ SA DE
Sbjct: 343 SSSSKGRKSSKSQERKNSHADTSKQMEDEGQKRRQSSSSAKRDE---------------- 386
Query: 816 KSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDKNDS 875
S+ +S+++ ++ R+ +K +K K G K K SK + G S KN
Sbjct: 387 --SSKKSRRNSMKEART--KKANNKSASKASKSGSKSKGKSASK---SKGKSSSKGKNSK 439
Query: 876 VKIANFSMPQGSELRGWRNSSKN-DSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVS- 933
+ A S P+ + + N+ + DS E + ++ +T+ G+ VN N+ S
Sbjct: 440 SRSA--SKPKSNAAQNSNNTHQTADSSENASSTTQTRTR-GRQREQKDMVNEKSNSKSSS 496
Query: 934 ----NTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTF 989
N++ N S + +K+AS+S ++ TT+ Q S++ S + P K +
Sbjct: 497 KGKKNSKSNTRSKSKSKSASKSRKNASKSKKDTTNHGRQTRSKSRSESKSKSEAPNKPSN 556
Query: 990 TSRASKGKLAPASDKLR---KGGGGKALNDKPTTSTSEP---DALEPRRSN 1034
+ +SD+ R + K +DK + + S+ A +P+++N
Sbjct: 557 KMEVIEQPKEESSDRKRRESRSQSAKKTSDKKSKNRSDSKKMTAEDPKKNN 607
Score = 39.3 bits (90), Expect = 0.058
Identities = 44/197 (22%), Positives = 80/197 (40%), Gaps = 12/197 (6%)
Query: 850 KKRKFMEVSKHYVAHGSSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSK 909
K R+ K V+ G +K S + A+ S QG ++SS N K D K
Sbjct: 184 KSRRSSSKGKSSVSKGRTK---STSSKRRADSSASQGRS----QSSSSNRRKASSSKDQK 236
Query: 910 -PKTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQ 968
++ K GR N N S++ + SS+ K +S S+++ + +++
Sbjct: 237 GTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSSKNKKSSSSKNKKSSSSKGRKSSSSR 296
Query: 969 VPIVFSSQATSTNTLPTKRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDAL 1028
SS+ ++ ++ S +SKG+ + +S K K + K ++S +
Sbjct: 297 GRKASSSKNRKSSKSKDRK---SSSSKGRKSSSSSKSNKRKASSSRGRKSSSSKGRKSSK 353
Query: 1029 EPRRSNRRIQPTSRLLE 1045
R N TS+ +E
Sbjct: 354 SQERKNSHAD-TSKQME 369
Score = 33.5 bits (75), Expect = 3.2
Identities = 61/333 (18%), Positives = 129/333 (38%), Gaps = 34/333 (10%)
Query: 696 WIQESWREGVITEKNKKDETTLTVHIPASGETSVLRAWNLRPSLIWKDGQWLDFSKVGAN 755
W ++ + + +K+ T+ + + G++SV + S SK A+
Sbjct: 163 WNEKYGQAAQKKQTKRKNSTSKSRRSSSKGKSSVSKGRTKSTS-----------SKRRAD 211
Query: 756 DSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSK---NIDAAESANPDEMRSLNLTENEIVF 812
S++ + S++ + KG S+ N +++ ++ S + ++
Sbjct: 212 SSASQGRSQSSSSNRRKASSSKDQKGTRSSSRKASNSKGRKNSTSNKRNSSSSSKRSSSS 271
Query: 813 NIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVNDK 872
KSS++++K+ + R G K + K K + S SS + K
Sbjct: 272 KNKKSSSSKNKKSSSSKGRKSSSSRGRKASSSKNRKSSKSKDRKSSSSKGRKSSS--SSK 329
Query: 873 NDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGAD-SKPKTKFGKPPGVLGRVNPPRNTS 931
++ K ++ + S +G R SSK+ ++ AD SK G+ +
Sbjct: 330 SNKRKASSSRGRKSSSSKG-RKSSKSQERKNSHADTSKQMEDEGQ-----------KRRQ 377
Query: 932 VSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFTS 991
S++ +SS ++ S E+R ++A + A++ S++ + +K +S
Sbjct: 378 SSSSAKRDESSKKSRRNSMKEARTKKANNKSASKASK----SGSKSKGKSASKSKGKSSS 433
Query: 992 RASKGKLAPASDKLRKGGGGKALNDKPTTSTSE 1024
+ K AS K + + N T +SE
Sbjct: 434 KGKNSKSRSAS-KPKSNAAQNSNNTHQTADSSE 465
>SPEN_DROME (Q8SX83) Split ends protein
Length = 5560
Score = 47.4 bits (111), Expect = 2e-04
Identities = 68/259 (26%), Positives = 101/259 (38%), Gaps = 33/259 (12%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
S D A +S + V S L P G S +P + N ++P+ + S
Sbjct: 1566 SSDFALGSSSNIVTGSSLVANVSRHPGGPCSGNTSPALPNLAATKATPI--IGNCSGGLG 1623
Query: 98 QSSALARGSVVDYSQALTPLH-------PYQSPSPRNFLGHSTSWISQAPLRGPWIGSPT 150
S+ ++ +L+P++ PY SPSP +G T+ + Q L P T
Sbjct: 1624 NSTGSKSAGLLQRLSSLSPMNSPQASMSPYNSPSPSPSVGGVTACLGQ--LTKP-AAPGT 1680
Query: 151 PAPDNNTHLSASPSSDTIKLASVKG------SLPPSSSIKDVTPG-----PPASSSGLQS 199
+ + +AS SS KG S PP + P PP G QS
Sbjct: 1681 ASAGLSGGTAASSSSPAANSGPTKGLQYPFPSHPPLPNTAAPPPAVQPAPPPLPEMGKQS 1740
Query: 200 TFVGTDSQLDANNVT----VPPAQQSSGPKAKKRKKDVL--SEDHGQKLLQSLTPAVAS- 252
G Q NN+T VP QSS + + +K + S + G SL AS
Sbjct: 1741 RLTG---QSSGNNLTKSLSVPDGPQSSPARVQLQKSASVPGSTNVGAPSSLSLDSTTASV 1797
Query: 253 RASTSVSAATPVGNVPMSS 271
S S+S++T GN ++S
Sbjct: 1798 ETSASISSSTSNGNSSLTS 1816
Score = 33.9 bits (76), Expect = 2.5
Identities = 37/132 (28%), Positives = 55/132 (41%), Gaps = 24/132 (18%)
Query: 176 SLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSS------------- 222
SLP +S P P + +Q+ D QLDA +++ PP + S
Sbjct: 3062 SLPNVASTSSAPPTPGKLTVNVQAASKHADLQLDAKHISSPPVCKPSPSLPCLIGDDDDD 3121
Query: 223 ---GPKAK-----KRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEK 274
PKAK R D L+ + L S P + A++S + +T P+SS
Sbjct: 3122 ALHTPKAKPTTPSSRGNDGLTPSREKPRLISPIPKTPTIANSS-TLSTQSAETPVSS--G 3178
Query: 275 SVVSVSPLADQP 286
+V+S S LA P
Sbjct: 3179 TVISSSALATTP 3190
>YK82_YEAST (P36170) Hypothetical 122.2 kDa protein in SIR1 3'region
precursor
Length = 1169
Score = 46.6 bits (109), Expect = 4e-04
Identities = 85/410 (20%), Positives = 163/410 (39%), Gaps = 33/410 (8%)
Query: 45 TSDSTVKQSVLQGKGIS---SPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
T S K S ++ K +S SP +S T + + + S S+ +S+ SL SS
Sbjct: 716 TITSEFKPSTMKTKVVSISSSPTNLITSYDTTSKDSTV---GSSTSSVSLISSISLPSSY 772
Query: 102 LARGSVVDYS-------QALTPLHPYQSPSPRNFLGHSTS--WISQAPLRGPWIGSPTPA 152
A + +S QALT + S + H TS S++ ++ + + +
Sbjct: 773 SASSEQIFHSSIVSSNGQALTSFSSTKVSSSESSESHRTSPTTSSESGIKSSGVEIESTS 832
Query: 153 PDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANN 212
+ + S +S +++++S + PSS I V P +S +ST ++ N+
Sbjct: 833 TSSFSFHETSTASTSVQISSQ--FVTPSSPISTVAPRSTGLNSQTESTNSSKETMSSENS 890
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQK-----LLQSLTPAVASRASTSVSAATPVGNV 267
+V P+ ++ PK K D S + + S TP+ + +T ++ ++ N
Sbjct: 891 ASVMPSSSATSPKTGKVTSDETSSGFSRDRTTVYRMTSETPS-TNEQTTLITVSSCESNS 949
Query: 268 PMSSVEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSL 327
++V +VVS + + LS L V +++ +EE + L S
Sbjct: 950 CSNTVSSAVVSTATTTINGITTEYTTWCPLSATELTTV--SKLESEEKTTLITVTSCES- 1006
Query: 328 ELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEA 387
+ + +S A + + + A A ++ N+A +A +++ A
Sbjct: 1007 ---GVCSETASPAIVSTATATVNDVVTVYSTWSPQATNKLAVSSDIENSASKASFVSEAA 1063
Query: 388 LISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVAAK 437
S S+ NN GT+++ T + N N S V++K
Sbjct: 1064 ETKS----ISRNNNFVPTSGTTSIETHTTTTSNASENSDNVSASEAVSSK 1109
>PRY3_YEAST (P47033) PRY3 protein (Pathogen related in Sc 3)
Length = 881
Score = 46.2 bits (108), Expect = 5e-04
Identities = 95/461 (20%), Positives = 177/461 (37%), Gaps = 52/461 (11%)
Query: 53 SVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQ 112
S + SS +S TI++ ++P + + T + S SS+ + + ++ ++
Sbjct: 163 STVSSSSSSSSSTSTTSDTVSTISSSIMPAVAQGY---TTTVSSAASSSSLKSTTINPAK 219
Query: 113 ALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTP--APDNNTHLSASPSSDTIKL 170
T + S + ST + + + S T A ++T +S+ +S T
Sbjct: 220 TAT-----LTASSSTVITSSTESVGSSTVSSASSSSVTTSYATSSSTVVSSDATSSTTTT 274
Query: 171 ASVKGSLP-----PSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPK 225
+SV S P+SS + PASSS S+ T++ +++ + S P
Sbjct: 275 SSVATSSSTTSSDPTSSTAAASSSDPASSSAAASSSASTENAASSSSAISSSSSMVSAPL 334
Query: 226 AKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVS-AATPVGNVPMSSVEKSVVSVSPLAD 284
+ S + +V S + SV A T V + +S + +S S AD
Sbjct: 335 SSTLTTSTAS-----------SRSVTSNSVNSVKFANTTVFSAQTTSSVSASLSSSVAAD 383
Query: 285 QPKNDQTVEKRILSDESLMKVKEARVHAEEASAL------SAAAVNHSLELWNQLDK--- 335
+ + E E V A A+ A+ L S+ AV+ S + L+
Sbjct: 384 DIQGSTSKEATSSVSEHTSIVTSATNAAQYATRLGSSSRSSSGAVSSSAVSQSVLNSVIA 443
Query: 336 ----HKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAA-----NVASNAAFQAKLMADE 386
+ S +A ++ A+ ++ A A+ N++++AA + + ++
Sbjct: 444 VNTDVSVTSVSSTAHTTKDTATTSVTASESITSETAQASSSTEKNISNSAATSSSIYSNS 503
Query: 387 ALISSGYENTSQGNNTFLPEGTSNLGQATPA----SILKGANGPNSPGSFIVAAKEAIRR 442
A + SG+ T E +S L + PA SI+K NS + I A ++
Sbjct: 504 ASV-SGHGVTYAAEYAITSEQSSALATSVPATNCSSIVKTTTLENSSTTTITAITKSTTT 562
Query: 443 RVEAASAATKRAE--NMDAILKAAELAAEAVSQAGKIVTMG 481
A+ +T+ A +D L + +A A T G
Sbjct: 563 LATTANNSTRAATAVTIDPTLDPTDNSASPTDNAKHTSTYG 603
Score = 34.7 bits (78), Expect = 1.4
Identities = 99/465 (21%), Positives = 164/465 (34%), Gaps = 66/465 (14%)
Query: 18 SAWKGSVVKNLILI-PQKHLCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIA 76
SA SV+ ++I + + S A T+ T SV + I+S +ASS I+
Sbjct: 431 SAVSQSVLNSVIAVNTDVSVTSVSSTAHTTKDTATTSVTASESITSETAQASSSTEKNIS 490
Query: 77 NPLIPLSSPLWSLPTLSA-----------DSLQSSALARGSVVDYSQALTPLHPYQSPSP 125
N SS + ++S S QSSALA ++ ++ S
Sbjct: 491 NSAATSSSIYSNSASVSGHGVTYAAEYAITSEQSSALATSVPATNCSSIVKTTTLENSST 550
Query: 126 RNFLGHSTSWISQA----------------PLRGPWIGSPTPAPDNNTHLSASPSSDT-I 168
+ S + A P P S +P DN H S SS T
Sbjct: 551 TTITAITKSTTTLATTANNSTRAATAVTIDPTLDPTDNSASPT-DNAKHTSTYGSSSTGA 609
Query: 169 KLASVK--GSLPPSSSIKDVTPGPPASS------SGLQSTFVGTDSQLDANNVTVPPAQQ 220
L S++ S+ SS+ + + S S ST T+S L N +T +
Sbjct: 610 SLDSLRTTTSISVSSNTTQLVSTCTSESDYSDSPSFAISTATTTESNLITNTITASCSTD 669
Query: 221 SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVS 280
S+ P + D + T +++ ST A+T + S ++ + +V
Sbjct: 670 SNFPTSAASSTDE----------TAFTRTISTSCSTLNGASTQTSELTTSPMKTN--TVV 717
Query: 281 PLADQP-------KNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQL 333
P + P +ND T I ++ + + E +S S A + +
Sbjct: 718 PASSFPSTTTTCLENDDTAFSSIYTEVNAATIINP---GETSSLASDFATSEKPNEPTSV 774
Query: 334 DKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEAL---IS 390
N G S + A A ++ + A SN A+ S
Sbjct: 775 KSTSNEGTSSTTTTYQQTVATLYAKPSST--SLGARTTTGSNGRSTTSQQDGSAMHQPTS 832
Query: 391 SGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNSPGSFIVA 435
S Y +G +T + ++ G ATP SI + + + +F+VA
Sbjct: 833 SIYTQLKEGTST-TAKLSAYEGAATPLSIFQCNSLAGTIAAFVVA 876
>AMYH_YEAST (P08640) Glucoamylase S1/S2 precursor (EC 3.2.1.3)
(Glucan 1,4-alpha-glucosidase) (1,4-alpha-D-glucan
glucohydrolase)
Length = 1367
Score = 46.2 bits (108), Expect = 5e-04
Identities = 75/404 (18%), Positives = 153/404 (37%), Gaps = 32/404 (7%)
Query: 42 AARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSA 101
++ T S+ + + S+P+ SS T + + P+ ++ S P S+ + SSA
Sbjct: 373 SSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSA 432
Query: 102 LARGSVVDYSQA-LTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGS---PTPAPDNNT 157
S + S A +T S +P ST+ S AP+ S P P P ++T
Sbjct: 433 PVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSST 492
Query: 158 HLSASPSSDTIKLASVKGSLPPSSSIKDVTPG------------PPASSSGLQSTFVGTD 205
S+S A V S SSS TP P+SS+ S+ T
Sbjct: 493 TESSS--------APVTSSTTESSSAPVPTPSSSTTESSSAPAPTPSSSTTESSSAPVTS 544
Query: 206 SQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVG 265
S ++++ VP S+ + +E S P +ST+ S++ PV
Sbjct: 545 STTESSSAPVPTPSSSTTESSSTPVTSSTTE-------SSSAPVPTPSSSTTESSSAPVP 597
Query: 266 NVPMSSVE-KSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVN 324
S+ E S + +P + ++ ++ S V E+S+ +
Sbjct: 598 TPSSSTTESSSAPAPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPS 657
Query: 325 HSLELWNQLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMA 384
S + S ++ + +++ +++A V + +++ + +
Sbjct: 658 SSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTES 717
Query: 385 DEALISSGYENTSQGNNTFLPEGTSNLGQATPASILKGANGPNS 428
A + + +T++ ++ +P +S+ +++ A + +S
Sbjct: 718 SSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSS 761
Score = 46.2 bits (108), Expect = 5e-04
Identities = 67/250 (26%), Positives = 101/250 (39%), Gaps = 32/250 (12%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
++ +A + ST + S SS +SS PT ++ SS +PT S+ +
Sbjct: 688 TESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSA--PVPTPSSSTT 745
Query: 98 QSS-ALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGS------PT 150
+SS A S + S A P +PS ST+ S AP+ P + P
Sbjct: 746 ESSSAPVTSSTTESSSA-----PVPTPSS------STTESSSAPVPTPSSSTTESSSAPV 794
Query: 151 PAPDNNTHLSA-----SPSSDTIKLASVKGSLPPSSSIKDVT-PGPPASSSGLQSTFVGT 204
P P ++T S+ +PSS + +S S P SSS + + P P SSS +S+
Sbjct: 795 PTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESSSAPV 854
Query: 205 DSQLDANNVTVPPAQQ------SSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSV 258
S ++V P SS P + S G + S + S+ TSV
Sbjct: 855 SSSTTESSVAPVPTPSSSSNITSSAPSSIPFSSTTESFSTGTTVTPSSSKYPGSQTETSV 914
Query: 259 SAATPVGNVP 268
S+ T VP
Sbjct: 915 SSTTETTIVP 924
Score = 46.2 bits (108), Expect = 5e-04
Identities = 58/239 (24%), Positives = 90/239 (37%), Gaps = 12/239 (5%)
Query: 38 SQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSL 97
++ +A + ST + S SS +SS PT ++ SS P+ S
Sbjct: 619 TESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTES 678
Query: 98 QSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNT 157
S+ + + S +T S +P ST+ S AP+ PTP+
Sbjct: 679 SSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPV-------PTPSSSTTE 731
Query: 158 HLSA---SPSSDTIKLAS--VKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANN 212
SA +PSS T + +S V S SSS TP + S + S ++++
Sbjct: 732 SSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSS 791
Query: 213 VTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSS 271
VP S+ + S + S TP +S S+SV TP + SS
Sbjct: 792 APVPTPSSSTTESSVAPVPTPSSSSNITSSAPSSTPFSSSTESSSVPVPTPSSSTTESS 850
Score = 40.4 bits (93), Expect = 0.026
Identities = 59/240 (24%), Positives = 89/240 (36%), Gaps = 21/240 (8%)
Query: 46 SDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARG 105
+ ST + S SS +SS PT ++ SS P+ S+ + SSA
Sbjct: 570 TSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPAPTPS-SSTTESSSAPVTS 628
Query: 106 SVVDYSQALTPLHPYQ----SPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
S + S A P S +P ST+ S AP+ PTP+ SA
Sbjct: 629 STTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPV-------PTPSSSTTESSSA 681
Query: 162 SPSSDTIKLAS--VKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQ 219
+S T + +S V S SSS TP + S + S ++++ VP
Sbjct: 682 PVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVPTPS 741
Query: 220 QSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSV 279
S+ + +E S P +ST+ S++ PV S+ E S V
Sbjct: 742 SSTTESSSAPVTSSTTE-------SSSAPVPTPSSSTTESSSAPVPTPSSSTTESSSAPV 794
Score = 40.0 bits (92), Expect = 0.034
Identities = 61/262 (23%), Positives = 104/262 (39%), Gaps = 14/262 (5%)
Query: 33 QKHLCSQDLAARTSDSTVKQSVLQGKGISSPLGRASSKATPTIANPLIPLSSPLWSLPT- 91
+K S+ +T+ S + S+P+ SS T + + P+ ++ S P
Sbjct: 298 KKTTTSKTCTKKTTTPVPTPSSSTTESSSAPVPTPSSSTTESSSAPVTSSTTESSSAPVP 357
Query: 92 --LSADSLQSSALARGSVVDYSQA-LTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWI-G 147
S+ + SSA S + S A +T S +P ST+ S AP+
Sbjct: 358 TPSSSTTESSSAPVTSSTTESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPVTSSTTES 417
Query: 148 SPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVT--PGPPASSSGLQSTFVG-T 204
S P + T S++P + + +S S P +SS + + P P SSS +S+ T
Sbjct: 418 SSAPVTSSTTESSSAPVTSSTTESS---SAPVTSSTTESSSAPVPTPSSSTTESSSAPVT 474
Query: 205 DSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPV 264
S ++++ VP S+ + V S TP+ ++ S+S A TP
Sbjct: 475 SSTTESSSAPVPTPSSST---TESSSAPVTSSTTESSSAPVPTPSSSTTESSSAPAPTPS 531
Query: 265 GNVPMSSVEKSVVSVSPLADQP 286
+ SS S + + P
Sbjct: 532 SSTTESSSAPVTSSTTESSSAP 553
>MAP4_HUMAN (P27816) Microtubule-associated protein 4 (MAP 4)
Length = 1152
Score = 45.8 bits (107), Expect = 6e-04
Identities = 89/407 (21%), Positives = 153/407 (36%), Gaps = 43/407 (10%)
Query: 1 MISAYGGTGTCGRMSGVSAWKGSVVKNLILIPQKHLCSQDLAARTSDSTVKQSVLQGKGI 60
MIS TGT G+ + A + SV++ L ++ C+ + +S+++ +G+ +
Sbjct: 622 MISPETITGT-GKKCSLPAEEDSVLEKL---GERKPCNSQPSELSSETSGIARPEEGRPV 677
Query: 61 SSPLGRASSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALT-PLHP 119
S G + TP N +P S + P + ++S S+A T P
Sbjct: 678 VS--GTGNDITTPP--NKELPPSPEKKTKPLATTQPAKTST---------SKAKTQPTSL 724
Query: 120 YQSPSPRNFLGHS-------TSWISQAPLRGPWIGSPTPAPDNNTHLSASPSSDTIKLAS 172
+ P+P G + + + AP + P + S P S PS D
Sbjct: 725 PKQPAPTTIGGLNKKPMSLASGLVPAAPPKRPAVASARP--------SILPSKDVKPKPI 776
Query: 173 VKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKD 232
P + PAS SG +ST + A + P+ +S K+
Sbjct: 777 ADAKAPEKRASPSKPASAPASRSGSKSTQTVAKTTTAAAVASTGPSSRSPSTLLPKKPTA 836
Query: 233 VLSED---HGQKLLQSLTPAVASR-ASTSVSA----ATPVGNVPMSSVEKSVVSVSPLAD 284
+ +E +K+ PA SR STS S+ T G P + V S V +P+
Sbjct: 837 IKTEGKPAEVKKMTAKSVPADLSRPKSTSTSSMKKTTTLSGTAPAAGVVPSRVKATPMPS 896
Query: 285 QPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSD 344
+P ++K+ S + A SA V + + KH+ G +
Sbjct: 897 RPSTTPFIDKKPTSAKPSSTTPRLSRLATNTSAPDLKNVRSKVGSTENI-KHQPGGGRAK 955
Query: 345 IEAKLASAAVAIAAAA-AVAKAAAAAANVASNAAFQAKLMADEALIS 390
+E K +AA + AV K A A+ A + ++++ + S
Sbjct: 956 VEKKTEAAATTRKPESNAVTKTAGPIASAQKQPAGKVQIVSKKVSYS 1002
>RFX1_HUMAN (P22670) MHC class II regulatory factor RFX1 (RFX)
(Enhancer factor C) (EF-C)
Length = 979
Score = 44.7 bits (104), Expect = 0.001
Identities = 75/353 (21%), Positives = 137/353 (38%), Gaps = 77/353 (21%)
Query: 113 ALTPLHPY----QSPSPR-NFLGHSTSWISQ---APLRGPWIGSPTPAPDNNTHLSASPS 164
A TP Y QSP P+ G ++++ P G+PTP+P ++ + S
Sbjct: 47 AATPQPQYVTELQSPQPQAQPPGGQKQYVTELPAVPAPSQPTGAPTPSPAPQQYIVVTVS 106
Query: 165 SDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFV--------------------GT 204
+G++ S ++ + +PG AS +G+ + V G
Sbjct: 107 ---------EGAMRASETVSEASPGSTASQTGVPTQVVQQVQGTQQRLLVQTSVQAKPGH 157
Query: 205 DSQLDANNVTVP----PAQ----QSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRAST 256
S L N+ VP P Q QS+ P +K + + Q V + +S+
Sbjct: 158 VSPLQLTNIQVPQQALPTQRLVVQSAAPGSKGGQVSLTVHGTQQVHSPPEQSPVQANSSS 217
Query: 257 SVSAATPVGNVPMS------------SVEKSVVSVSPLADQPKNDQTVEKRILSD----- 299
S +A P G VP + E+SVV +P A +P Q + + L
Sbjct: 218 SKTAGAPTGTVPQQLQVHGVQQSVPVTQERSVVQATPQAPKPGPVQPLTVQGLQPVHVAQ 277
Query: 300 --ESLMKVKEARVHAEEA-------SALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLA 350
+ L +V V++ + ++ +A+A+ S + + + + S EA
Sbjct: 278 EVQQLQQVPVPHVYSSQVQYVEGGDASYTASAIRSSTYSYPETPLYTQTASTSYYEAAGT 337
Query: 351 SAAVAI-AAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNT 402
+ V+ A + AVA + + V+ ++++A A +G N+S G +
Sbjct: 338 ATQVSTPATSQAVASSGSMPMYVSG-----SQVVASSASTGAGASNSSGGGGS 385
>YG46_YEAST (P53301) Hypothetical 52.8 kDa protein in BUB1-HIP1
intergenic region
Length = 507
Score = 44.3 bits (103), Expect = 0.002
Identities = 50/204 (24%), Positives = 90/204 (43%), Gaps = 33/204 (16%)
Query: 161 ASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQ 220
+S +S T+ +SV ++ SSS+ + P+SS+ S + + S +++++ Q
Sbjct: 329 SSSASSTVS-SSVSSTVSSSSSVSSSSSTSPSSSTATSSKTLASSSVTTSSSISSFEKQS 387
Query: 221 SSGPKAKKRKKDVLSEDHGQKLLQSL-TPAVASRASTSVSAATPVGNVPMSSVEKSVVSV 279
SS KK V S + ++ S TPA S + S A T + + SV S
Sbjct: 388 SS-----SSKKTVASSSTSESIISSTKTPATVSSTTRSTVAPT--------TQQSSVSSD 434
Query: 280 SPLADQ-----PKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLD 334
SP+ D+ ND T +S + ++ + + EAS+ ++ +++ +L L
Sbjct: 435 SPVQDKGGVATSSNDVTSSTTQISSKYTSTIQSS---SSEASSTNSVQISNGADLAQSLP 491
Query: 335 KHKNSGFMSDIEAKLASAAVAIAA 358
+ E KL S VA+ A
Sbjct: 492 R----------EGKLFSVLVALLA 505
>GAR2_SCHPO (P41891) Protein gar2
Length = 500
Score = 43.9 bits (102), Expect = 0.002
Identities = 47/248 (18%), Positives = 99/248 (38%), Gaps = 33/248 (13%)
Query: 821 ESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSK--VNDKNDSVKI 878
+ K ++ V+ K+G+ + KP K +K + + +A SSK V+ K +
Sbjct: 4 KDKTSVKKSVKETASKKGA-----IEKPSKSKKITKEAAKEIAKQSSKTDVSPKKSKKEA 58
Query: 879 ANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSN---- 934
S P+ S+ + + K+ KE+ ++S+ ++ + + S S+
Sbjct: 59 KRASSPEPSK-KSVKKQKKSKKKEESSSESESESSSSESESSSSESESSSSESESSSSES 117
Query: 935 ------------TEMNKDSSNHTKNASQSES------RVERAPYSTTDGATQVPIVFSSQ 976
TE K+SS+ + ++S+SE ++E S++D +++ S
Sbjct: 118 SSSESEEEVIVKTEEKKESSSESSSSSESEEEEEAVVKIEEKKESSSDSSSESSSSESES 177
Query: 977 ATSTNTLPTKRTF---TSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSEPDALEPRRS 1033
+S++ + T +G +SD D ++S SE ++
Sbjct: 178 ESSSSESEEEEEVVEKTEEKKEGSSESSSDSESSSDSSSESGDSDSSSDSESESSSEDEK 237
Query: 1034 NRRIQPTS 1041
R+ +P S
Sbjct: 238 KRKAEPAS 245
>DSPP_HUMAN (Q9NZW4) Dentin sialophosphoprotein precursor [Contains:
Dentin phosphoprotein (Dentin phosphophoryn) (DPP);
Dentin sialoprotein (DSP)]
Length = 1253
Score = 43.9 bits (102), Expect = 0.002
Identities = 50/217 (23%), Positives = 88/217 (40%), Gaps = 17/217 (7%)
Query: 821 ESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKVND----KNDSV 876
E K+DP +V + + G+P+ ++ + K + H SK + K
Sbjct: 308 EGKEDPHNEVDGDKTSKSEENSAGIPEDNGSQRIEDTQK--LNHRESKRVENRITKESET 365
Query: 877 KIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTE 936
S +G E++G + ++N +KE +G ++ K G+ +LG+ N V N E
Sbjct: 366 HAVGKSQDKGIEIKGPSSGNRNITKE-VGKGNEGKEDKGQHGMILGKGNVKTQGEVVNIE 424
Query: 937 MNKDSS------NHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTFT 990
S H+ S S S + Y D + Q SS ++ N +
Sbjct: 425 GPGQKSEPGNKVGHSNTGSDSNSDGYDS-YDFDDKSMQGDDPNSSDESNGNDDANSESDN 483
Query: 991 SRASKGKLAPASDKLRKGGGG---KALNDKPTTSTSE 1024
+ +S+G + SD+ + G G K D + STS+
Sbjct: 484 NSSSRGDASYNSDESKDNGNGSDSKGAEDDDSDSTSD 520
Score = 33.5 bits (75), Expect = 3.2
Identities = 48/275 (17%), Positives = 103/275 (37%), Gaps = 27/275 (9%)
Query: 750 SKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANPDEMRSLNLTENE 809
SK +D S DT + G+N + DK ++S++ D S N +++
Sbjct: 507 SKGAEDDDSDSTSDTNNSDSNGNGNNGND--DNDKSDSGKGKSDSSDSDSSDSSNSSDSS 564
Query: 810 IVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSKV 869
+ S +N S S + + S + SS
Sbjct: 565 DSSDSDSSDSNSSSDSDSSDSDSSDSSDSDS--------SDSSNSSDSSDSSDSSDSSDS 616
Query: 870 NDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRN 929
+D +DS ++ S S+ + SK+DS + +DS + + +
Sbjct: 617 SDSSDSKSDSSKSESDSSD-----SDSKSDSSDSNSSDSSDNS------------DSSDS 659
Query: 930 TSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRTF 989
++ SN+ + DSS+ + ++S S+S +++D + SS+++ ++ +
Sbjct: 660 SNSSNSSDSSDSSDSSDSSSSSDSSSSSDSSNSSDSSDSSDSSNSSESSDSSDSSDSDSS 719
Query: 990 TSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE 1024
S S + SD + + ++++S+
Sbjct: 720 DSSDSSNSNSSDSDSSNSSDSSDSSDSSDSSNSSD 754
Score = 33.5 bits (75), Expect = 3.2
Identities = 49/279 (17%), Positives = 100/279 (35%), Gaps = 33/279 (11%)
Query: 748 DFSKVGANDS-STHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANP-DEMRSLNL 805
D S ++DS S++ D+ SN+ + S + D+++S++ D S N
Sbjct: 723 DSSNSNSSDSDSSNSSDSSDSSDSSDSSNSSDSSDSSDSSNSSDSSDSSDSSDSSDSSNS 782
Query: 806 TENEIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHG 865
+++ N SS + + D S + S + S +
Sbjct: 783 SDSNDSSNSSDSSDSSNSSDSSNSSDSSDSSDSSD--------SDSSNSSDSSNSSDSSD 834
Query: 866 SSKVNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVN 925
SS +D +DS ++ S S R+ S N S +DS
Sbjct: 835 SSNSSDSSDSSDSSDSSDSDSSN----RSDSSNSSDSSDSSDSS---------------- 874
Query: 926 PPRNTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPT 985
N+S S+ + SN + N+S S + ++D + SS ++ ++
Sbjct: 875 ---NSSDSSDSSDSSDSNESSNSSDSSDSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSN 931
Query: 986 KRTFTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE 1024
++ + + +SD + N ++++S+
Sbjct: 932 SSDNSNSSDSSNSSDSSDSSDSSNSSDSSNSGDSSNSSD 970
Score = 32.3 bits (72), Expect = 7.1
Identities = 44/276 (15%), Positives = 96/276 (33%), Gaps = 6/276 (2%)
Query: 750 SKVGANDSSTHKGDTPHEKRPKLGSNAVEVKGKDKMSKNIDAAESANP-DEMRSLNLTEN 808
S +N S + D+ + SN+ + S N ++++S+N D S + + +
Sbjct: 897 SSDSSNSSDSDSSDSSNSSDSSDSSNSSDSSESSNSSDNSNSSDSSNSSDSSDSSDSSNS 956
Query: 809 EIVFNIGKSSTNESKQDPQRQVRSGLQKEGSKVIFGVPKPGKKRKFMEVSKHYVAHGSSK 868
N G SS + D S + S + SS
Sbjct: 957 SDSSNSGDSSNSSDSSDSNSSDSSDSSNSSDS-----SDSSDSSDSSDSSDSSNSSDSSD 1011
Query: 869 VNDKNDSVKIANFSMPQGSELRGWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPR 928
+D +DS ++ S S + S + S +DS + G +
Sbjct: 1012 SSDSSDSSNSSDSSNSSDSSDSSDSSDSSDSSDSSNSSDSSDSSDSSDSSDSSGSSDSSD 1071
Query: 929 NTSVSNTEMNKDSSNHTKNASQSESRVERAPYSTTDGATQVPIVFSSQATSTNTLPTKRT 988
++ S++ + DSS+ + ++ SES ++D + SS ++ ++
Sbjct: 1072 SSDSSDSSDSSDSSDSSDSSDSSESSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSN 1131
Query: 989 FTSRASKGKLAPASDKLRKGGGGKALNDKPTTSTSE 1024
+ + + +SD + + ++ +S+
Sbjct: 1132 SSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSDSSD 1167
>YB95_SCHPO (O42970) Hypothetical serine-rich protein C1E8.05 in
chromosome II precursor
Length = 317
Score = 43.5 bits (101), Expect = 0.003
Identities = 62/239 (25%), Positives = 96/239 (39%), Gaps = 16/239 (6%)
Query: 48 STVKQSVLQGKGISSPLGRASSKA-TP--TIANPLIPLSSPLWSLPTL---SADSLQSSA 101
+TV S L ++ L R + A TP TIA+ + + LW++ T + D L +
Sbjct: 36 NTVSWSNLTSSTVTLTLYRGGNSALTPIETIASDIDNTGTYLWNIATYYEAADDYLLGLS 95
Query: 102 LARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRGPWIGSPTPAPDNNTHLSA 161
G YSQ T + L +S + IS + IG+ T + T S+
Sbjct: 96 FDGGET--YSQYFTLQACSTCTISTSSLSYSGT-ISSTSIAPSMIGTRTSSSYFITSSSS 152
Query: 162 SPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQS 221
+PSS + S S P SSS K + +SSS S + S ++ + + +S
Sbjct: 153 TPSSSS----SSSSSSPSSSSSKSSSSSKSSSSSSSSSKSSSSSSSSSKSSSSSSSSSKS 208
Query: 222 SGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSSVEKSVVSVS 280
S + + S + S TPA +S + VS A SS + SVS
Sbjct: 209 SASPSSSKSSSKFSSS---SFITSTTPASSSSSGAIVSNAKTASTDDSSSASSATSSVS 264
Score = 39.7 bits (91), Expect = 0.045
Identities = 45/195 (23%), Positives = 80/195 (40%), Gaps = 3/195 (1%)
Query: 84 SPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRNFLGHSTSWISQAPLRG 143
S ++L S ++ +S+L+ + S ++ P S F+ S+S S +
Sbjct: 103 SQYFTLQACSTCTISTSSLSYSGTIS-STSIAPSMIGTRTSSSYFITSSSSTPSSSSSSS 161
Query: 144 PWIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVG 203
S + + +++ S+S SS + K +S S SSS + AS S +S+
Sbjct: 162 SSSPSSSSSKSSSSSKSSSSSSSSSKSSSSSSSSSKSSSSSSSSSKSSASPSSSKSSSKF 221
Query: 204 TDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATP 263
+ S + T P + SSG K + S +V S AS+++SA+
Sbjct: 222 SSSSFITS--TTPASSSSSGAIVSNAKTASTDDSSSASSATSSVSSVVSSASSALSASAS 279
Query: 264 VGNVPMSSVEKSVVS 278
+ +SS S S
Sbjct: 280 SASASVSSSASSDAS 294
>RNFC_HAEIN (P71397) Electron transport complex protein rnfC
Length = 819
Score = 42.7 bits (99), Expect = 0.005
Identities = 58/252 (23%), Positives = 108/252 (42%), Gaps = 21/252 (8%)
Query: 244 QSLTPAVASRASTSVSAATPVGN--VPMSSVEKSVVSVSPLADQPKNDQTVEKRILSDES 301
Q P A +A+ + + A +S +++ + +Q + ++ ++ + +
Sbjct: 559 QQTQPTNAKKAAVAAALARAKAKKLAQANSTSEAISNSQTAENQVEKTKSAVEKTQENST 618
Query: 302 LMKVKEARVHAEEASALS---------AAAVNHSLELWNQLDKHKNSGFMSD-----IEA 347
+ K+A V A A A + + A+++S N+++K K++ ++ ++A
Sbjct: 619 ALDPKKAAVAAAIARAKAKKLAQTNSTSEAISNSQTAENEVEKTKSAVEKTEENSTALDA 678
Query: 348 KLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEG 407
K A+ A AIA A A A AN AS A ++ +E + +Q N+T L
Sbjct: 679 KKAAIAAAIARAKA---KKLAQANSASEAISNSQTAENEVEKTKSAVEKTQQNSTALDPK 735
Query: 408 TSNLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATKRAENMDAI--LKAAE 465
+ + A + K NS I ++ A + SA K EN A+ KAA
Sbjct: 736 KAAVAAAIARAKAKKLAQANSTSEAISNSQTAENEVEKTKSAVEKTQENSTALDPKKAAV 795
Query: 466 LAAEAVSQAGKI 477
AA A ++A K+
Sbjct: 796 AAAIARAKAKKL 807
>ESC1_SCHPO (Q04635) Protein esc1
Length = 413
Score = 42.7 bits (99), Expect = 0.005
Identities = 61/237 (25%), Positives = 88/237 (36%), Gaps = 53/237 (22%)
Query: 68 SSKATPTIANPLIPLSSPLWSLPTLSADSLQSSALARGSVVDYSQALTPLHPYQSPSPRN 127
S + TPT + PL +S P S P+ SA S TP P Q P N
Sbjct: 8 SMQPTPTSSIPLRQMSQPTTSAPSNSASS------------------TPYSPQQVPLTHN 49
Query: 128 FLGHST-SWISQAPLRGP---WIGSPTPAPDNNTHLSASPSSDTIKLASVKGSLPPSSSI 183
ST S R P + P P SA+P+S + A++ + P +
Sbjct: 50 SYPLSTPSSFQHGQTRLPPINCLAEPFNRPQPWHSNSAAPASSSPTSATLSTAAHPVHT- 108
Query: 184 KDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLL 243
A +G S++V +VPP ++ + K + H
Sbjct: 109 ------NAAQVAGSSSSYV----------YSVPPTNSTTSQASAKHS----AVPHRSSQF 148
Query: 244 QS--LTPAVASRASTSVSAATPVGNVPMSSVEKSVVSV--------SPLADQPKNDQ 290
QS LTP+ +ST VS++ V SS + VSV +PL +QP Q
Sbjct: 149 QSTTLTPSTTDSSSTDVSSSDSVSTSASSSNASNTVSVTSPASSSATPLPNQPSQQQ 205
>VTA2_XENLA (P18709) Vitellogenin A2 precursor (VTG A2) [Contains:
Lipovitellin I; Lipovitellin II; Phosvitin]
Length = 1807
Score = 42.0 bits (97), Expect = 0.009
Identities = 49/237 (20%), Positives = 107/237 (44%), Gaps = 26/237 (10%)
Query: 152 APDNNTHLSASPSSDTIKLASVKGSLPPSSSIKDVTPGPPASSSGLQSTFVGTDSQLDAN 211
A + LS+S SS + +S S SSS P+SSS S++ + + N
Sbjct: 1121 ARKQQSSLSSSSSSSSSSSSSSSSSSSSSSS------SSPSSSSS--SSYSKRSKRREHN 1172
Query: 212 NVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLLQSLTPAVASRASTSVSAATPVGNVPMSS 271
+ SS + K++ + HGQK + S + + +S +S+S S+++ + SS
Sbjct: 1173 PHHQRESSSSSSQEQNKKRNLQENRKHGQKGMSSSSSSSSSSSSSSSSSSSSSSSSSSSS 1232
Query: 272 VEKSVVSVSPLADQPKNDQTVEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWN 331
E ++P ++ + + +S ++ +E +S+ S+++ S E+WN
Sbjct: 1233 EE----------NRPHKNRQHDNKQAKMQSNQHQQKKNKFSESSSSSSSSS---SSEMWN 1279
Query: 332 QLDKHKNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAF--QAKLMADE 386
+ H+N D+ + + + +++ +++ +S +A+ +AK + D+
Sbjct: 1280 KKKHHRN---FYDLNFRRTARTKGTEHRGSRLSSSSESSSSSSESAYRHKAKFLGDK 1333
>TOLA_HAEIN (P44678) TolA protein
Length = 372
Score = 42.0 bits (97), Expect = 0.009
Identities = 48/178 (26%), Positives = 77/178 (42%), Gaps = 13/178 (7%)
Query: 283 ADQPKNDQTVEK-RILSDESLMKVKEARVHAEE-----ASALSAAAVNHSLELWNQLDKH 336
A++ K Q E ++ +D ++ A AEE A+ ++A E +L+
Sbjct: 138 AEEAKAKQAAEAAKLKADAEAKRLAAAAKQAEEEAKAKAAEIAAQKAKQEAEAKAKLEAE 197
Query: 337 KNSGFMSDIEAKLASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENT 396
+ +++ +AK + A A AAA A AKA A A AA +AK AD+A +
Sbjct: 198 AKAKAVAEAKAKAEAEAKAKAAAEAKAKADAEA-----KAATEAKRKADQASLDDFLNGG 252
Query: 397 SQGNNTFLPEGTSNLG--QATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASAATK 452
G + G +N G Q + A++ G G V KE RR ++ + A K
Sbjct: 253 DIGGGSASKGGNTNKGGTQGSGAALGSGDGGKVGDQYAGVIKKEIQRRFLKDPNFAGK 310
>SSRP_HUMAN (Q08945) Structure-specific recognition protein 1
(SSRP1) (Recombination signal sequence recognition
protein) (T160) (Chromatin-specific transcription
elongation factor 80 kDa subunit) (FACT 80 kDa subunit)
Length = 709
Score = 42.0 bits (97), Expect = 0.009
Identities = 68/317 (21%), Positives = 118/317 (36%), Gaps = 52/317 (16%)
Query: 668 SLQHEGTRKRRRAAMGDYAWSVGDRVDAWIQESWREGVITEKNKKD------ETTLTVHI 721
++++ G ++ + +YA S D+ DA+++ EG I E+N D E T
Sbjct: 424 NIKNRGLKEGMNPSYDEYADSDEDQHDAYLERMKEEGKIREENANDSSDDSGEETDESFN 483
Query: 722 PASGETSVLRAWNLRPSLIWKDGQWLDFSKVGANDSSTHKGDTPH-EKRPKLGSNAVEVK 780
P E V +F + SS+++GD+ EK+ K A K
Sbjct: 484 PGEEEEDVAE----------------EFDSNASASSSSNEGDSDRDEKKRKQLKKAKMAK 527
Query: 781 GKDKMSKNIDAAESANPDE--------MRSLNLTENEIVF-NIGKSSTNESKQDPQRQVR 831
+ K ++ + +P+ M LN + +I + G S T+ SK+ ++
Sbjct: 528 DRKSRKKPVEVKKGKDPNAPKRPMSAYMLWLNASREKIKSDHPGISITDLSKK--AGEIW 585
Query: 832 SGLQKEGSKVIFGVPKPGKKRKFMEVSKHYV-AHGSSKVNDKNDSVKIANFSMPQGSELR 890
G+ KE K + +R + + K Y G S DK+ K M + S
Sbjct: 586 KGMSKE-KKEEWDRKAEDARRDYEKAMKEYEGGRGESSKRDKSKKKKKVKVKMEKKSTPS 644
Query: 891 GWRNSSKNDSKEKLGADSKPKTKFGKPPGVLGRVNPPRNTSVSNTEMNKDSSNHTKNASQ 950
R SS S +L K K ++ S++ NK ++
Sbjct: 645 --RGSSSKSSSRQLSESFKSKEFV--------------SSDESSSGENKSKKKRRRSEDS 688
Query: 951 SESRVERAPYSTTDGAT 967
E + P S+ D A+
Sbjct: 689 EEEELASTPPSSEDSAS 705
>FSH_DROME (P13709) Female sterile homeotic protein (Fragile-chorion
membrane protein)
Length = 2038
Score = 42.0 bits (97), Expect = 0.009
Identities = 76/294 (25%), Positives = 114/294 (37%), Gaps = 39/294 (13%)
Query: 184 KDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQKLL 243
K P P SSSG G+ + A +SGP + K V + Q L
Sbjct: 158 KQRAPATPKSSSGGAGASTGSGTS---------SAAVTSGPGSGSTKVSVAASSAQQSGL 208
Query: 244 QSLTPAVASRAST----------SVSAATPVGNV--PMSSVEKSVVSVSPLADQPKNDQT 291
Q T A +ST AA PV + +SS S+ P++ P + T
Sbjct: 209 QGATGAGGGSSSTPGTQPGSGAGGAIAARPVSAMGGTVSSTAGGAPSIPPISTMPPH--T 266
Query: 292 VEKRILSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAK--L 349
V + + M A A+ +AAA+ SL Q + + + + + L
Sbjct: 267 VPGSTNTTTTAMAGGVGGPGAAGANP-NAAALMASLLNAGQTGAYPGAPGQTAVNSSSLL 325
Query: 350 ASAAVAIAAAAAVAKAAAAAANVASNAAFQAKLMADEALISSGYENTSQGNNTFLPEGTS 409
+ A+AAAAA A AAAAAA A+ AA A + A+ N + ++ G S
Sbjct: 326 DGSTAAVAAAAAAAAAAAAAAGGAAGAAGGAGTIPAVAV------NAANAVQAYVNAGVS 379
Query: 410 -----NLGQATPASILKGANGPNSPGSFIVAAKEAIRRRVEAASA--ATKRAEN 456
+ PA I KG + A E+ ++++ SA AT+R N
Sbjct: 380 VGVDAVIPPQQPAKIKKGVKRKADTTTPTANAFESPYTQMDSKSAKIATRRESN 433
Score = 37.0 bits (84), Expect = 0.29
Identities = 54/203 (26%), Positives = 73/203 (35%), Gaps = 35/203 (17%)
Query: 183 IKDVTPGPPASSSGLQSTFVGTDSQLDANNVTVPPAQQSSGPKAKKRKKDVLSEDHGQ-K 241
I+ + P PP QL + +PP Q S K + Q K
Sbjct: 1580 IESMMPSPPDKQ------------QLQQHQKVLPPQQSPSDMKLHPNAAAAAAVASAQAK 1627
Query: 242 LLQSLTPAV-----ASRASTSVSAATPVGNVPMSSVEKSVVSVSPLADQPKNDQTVEKRI 296
L+Q+ AS S+ SA +P + SS Q +N R+
Sbjct: 1628 LVQTFKANEQNLKNASSWSSLASANSPQSHTSSSSSSSKAKPAMDSFQQFRNKAKERDRL 1687
Query: 297 LSDESLMKVKEARVHAEEASALSAAAVNHSLELWNQLDKHKNSGFMSDIEAKLASAAV-- 354
E+ K K+ + A E Q KH S S A +A AA
Sbjct: 1688 KLLEAAEKEKKNQKEAAEKE---------------QQRKHHKSSSSSLTSAAVAQAAAIA 1732
Query: 355 AIAAAAAVAKAAAAAANVASNAA 377
A AAAAV AAAAA +AS+A+
Sbjct: 1733 AATAAAAVTLGAAAAAALASSAS 1755
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.308 0.125 0.347
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 125,110,360
Number of Sequences: 164201
Number of extensions: 5346909
Number of successful extensions: 18576
Number of sequences better than 10.0: 311
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 302
Number of HSP's that attempted gapping in prelim test: 17546
Number of HSP's gapped (non-prelim): 894
length of query: 1091
length of database: 59,974,054
effective HSP length: 121
effective length of query: 970
effective length of database: 40,105,733
effective search space: 38902561010
effective search space used: 38902561010
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.7 bits)
S2: 71 (32.0 bits)
Medicago: description of AC137077.8


