

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.7 - phase: 0
(396 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
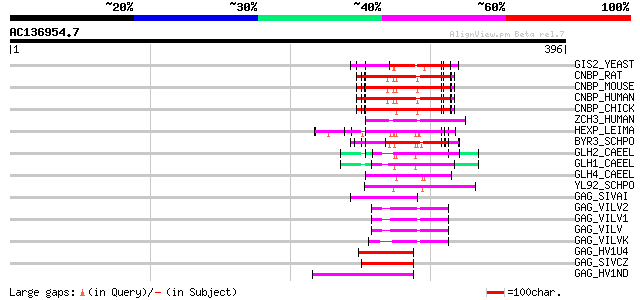
Score E
Sequences producing significant alignments: (bits) Value
GIS2_YEAST (P53849) Zinc-finger protein GIS2 71 4e-12
CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP) (... 66 1e-10
CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)... 66 1e-10
CNBP_HUMAN (P62633) Cellular nucleic acid binding protein (CNBP)... 66 1e-10
CNBP_CHICK (O42395) Cellular nucleic acid binding protein (CNBP) 66 1e-10
ZCH3_HUMAN (Q9NUD5) Zinc finger CCHC domain containing protein 3 65 2e-10
HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding p... 63 1e-09
BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog 60 1e-08
GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-... 59 2e-08
GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-... 57 8e-08
GLH4_CAEEL (O76743) ATP-dependent RNA helicase glh-4 (EC 3.6.1.-... 55 2e-07
YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I 55 3e-07
GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17; ... 51 5e-06
GAG_VILV2 (P23425) Gag polyprotein [Contains: Core protein p16; ... 50 8e-06
GAG_VILV1 (P23424) Gag polyprotein [Contains: Core protein p16; ... 50 8e-06
GAG_VILV (P03352) Gag polyprotein [Contains: Core protein p16; C... 50 8e-06
GAG_VILVK (P35955) Gag polyprotein [Contains: Core protein p16; ... 49 2e-05
GAG_HV1U4 (P24736) Gag polyprotein [Contains: Core protein p17 (... 48 4e-05
GAG_SIVCZ (P17282) Gag polyprotein [Contains: Core protein p18; ... 48 5e-05
GAG_HV1ND (P18800) Gag polyprotein [Contains: Core protein p17 (... 48 5e-05
>GIS2_YEAST (P53849) Zinc-finger protein GIS2
Length = 153
Score = 71.2 bits (173), Expect = 4e-12
Identities = 31/69 (44%), Positives = 41/69 (58%), Gaps = 5/69 (7%)
Query: 244 PSKRDAPAEIVCFKCGEKGHKSNVCTKDEK----KCFRCGQKGHMLADCKRGDVVCYNCN 299
P K +++ C+KCG H + C K++ KC+ CGQ GHM DC+ D +CYNCN
Sbjct: 83 PKKTSRFSKVSCYKCGGPNHMAKDCMKEDGISGLKCYTCGQAGHMSRDCQN-DRLCYNCN 141
Query: 300 EEGHISTQC 308
E GHIS C
Sbjct: 142 ETGHISKDC 150
Score = 66.2 bits (160), Expect = 1e-10
Identities = 29/77 (37%), Positives = 43/77 (55%), Gaps = 6/77 (7%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDE----KKCFRCGQKGHMLADCKRGDVVCYNCNEEGH 303
D +E +C+ C + GH CT K+C+ CG+ GH+ ++C C+NCN+ GH
Sbjct: 18 DCDSERLCYNCNKPGHVQTDCTMPRTVEFKQCYNCGETGHVRSECTVQR--CFNCNQTGH 75
Query: 304 ISTQCTQPKKVRVGGKV 320
IS +C +PKK KV
Sbjct: 76 ISRECPEPKKTSRFSKV 92
Score = 56.2 bits (134), Expect = 1e-07
Identities = 23/59 (38%), Positives = 33/59 (54%), Gaps = 5/59 (8%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVV----CYNCNEEGHISTQCT 309
C+ CG+ GH + C E+ C+ C + GH+ DC V CYNC E GH+ ++CT
Sbjct: 6 CYVCGKIGHLAEDC-DSERLCYNCNKPGHVQTDCTMPRTVEFKQCYNCGETGHVRSECT 63
Score = 54.3 bits (129), Expect = 5e-07
Identities = 20/43 (46%), Positives = 29/43 (66%), Gaps = 1/43 (2%)
Query: 272 EKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKV 314
+K C+ CG+ GH+ DC + +CYNCN+ GH+ T CT P+ V
Sbjct: 3 QKACYVCGKIGHLAEDCD-SERLCYNCNKPGHVQTDCTMPRTV 44
Score = 43.1 bits (100), Expect = 0.001
Identities = 16/45 (35%), Positives = 25/45 (55%), Gaps = 1/45 (2%)
Query: 246 KRDAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKR 290
K D + + C+ CG+ GH S C D + C+ C + GH+ DC +
Sbjct: 109 KEDGISGLKCYTCGQAGHMSRDCQND-RLCYNCNETGHISKDCPK 152
>CNBP_RAT (P62634) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 66.2 bits (160), Expect = 1e-10
Identities = 29/63 (46%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADC-KRGDVVCYNCNEEGHIST 306
D E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 112 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 169
Query: 307 QCT 309
+CT
Sbjct: 170 ECT 172
Score = 63.5 bits (153), Expect = 9e-10
Identities = 23/59 (38%), Positives = 34/59 (56%), Gaps = 4/59 (6%)
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCK----RGDVVCYNCNEEGHISTQC 308
+C++CGE GH + C E C+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 53 ICYRCGESGHLAKDCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 111
Score = 63.5 bits (153), Expect = 9e-10
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query: 252 EIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
E C+ CG+ GH + C DE+KC+ CG+ GH+ DC + V CY C E GH++ C++
Sbjct: 95 EQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSK 152
Query: 311 PKKV 314
+V
Sbjct: 153 TSEV 156
Score = 61.2 bits (147), Expect = 4e-09
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query: 252 EIVCFKCGEKGHKSNVCTKDEKK----CFRCGQKGHMLADCKRGDVV-CYNCNEEGHIST 306
E C+ CG GH + C + +++ C+ CG+ GH+ DC D CY+C E GHI
Sbjct: 71 EDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQK 130
Query: 307 QCTQPKKVRVG 317
CT+ K R G
Sbjct: 131 DCTKVKCYRCG 141
Score = 59.3 bits (142), Expect = 2e-08
Identities = 29/89 (32%), Positives = 37/89 (40%), Gaps = 28/89 (31%)
Query: 255 CFKCGEKGHKSNVC------------------TKDE----------KKCFRCGQKGHMLA 286
CFKCG GH + C T D C+RCG+ GH+
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 287 DCKRGDVVCYNCNEEGHISTQCTQPKKVR 315
DC + CYNC GHI+ C +PK+ R
Sbjct: 66 DCDLQEDACYNCGRGGHIAKDCKEPKRER 94
>CNBP_MOUSE (P53996) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 178
Score = 66.2 bits (160), Expect = 1e-10
Identities = 29/63 (46%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADC-KRGDVVCYNCNEEGHIST 306
D E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 113 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 170
Query: 307 QCT 309
+CT
Sbjct: 171 ECT 173
Score = 63.5 bits (153), Expect = 9e-10
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query: 252 EIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
E C+ CG+ GH + C DE+KC+ CG+ GH+ DC + V CY C E GH++ C++
Sbjct: 96 EQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSK 153
Query: 311 PKKV 314
+V
Sbjct: 154 TSEV 157
Score = 61.2 bits (147), Expect = 4e-09
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 5/75 (6%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKK----CFRCGQKGHMLADCKRGDVV-CYNCNEEG 302
D + C+ CG GH + C + +++ C+ CG+ GH+ DC D CY+C E G
Sbjct: 68 DLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFG 127
Query: 303 HISTQCTQPKKVRVG 317
HI CT+ K R G
Sbjct: 128 HIQKDCTKVKCYRCG 142
Score = 60.5 bits (145), Expect = 7e-09
Identities = 22/60 (36%), Positives = 37/60 (61%), Gaps = 5/60 (8%)
Query: 254 VCFKCGEKGHKSNVC-TKDEKKCFRCGQKGHMLADCK----RGDVVCYNCNEEGHISTQC 308
+C++CGE GH + C ++++ C+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 53 ICYRCGESGHLAKDCDLQEDEACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 112
Score = 57.8 bits (138), Expect = 5e-08
Identities = 30/90 (33%), Positives = 38/90 (41%), Gaps = 29/90 (32%)
Query: 255 CFKCGEKGHKSNVC------------------TKDE----------KKCFRCGQKGHMLA 286
CFKCG GH + C T D C+RCG+ GH+
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 287 DCK-RGDVVCYNCNEEGHISTQCTQPKKVR 315
DC + D CYNC GHI+ C +PK+ R
Sbjct: 66 DCDLQEDEACYNCGRGGHIAKDCKEPKRER 95
>CNBP_HUMAN (P62633) Cellular nucleic acid binding protein (CNBP)
(Zinc finger protein 9)
Length = 177
Score = 66.2 bits (160), Expect = 1e-10
Identities = 29/63 (46%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADC-KRGDVVCYNCNEEGHIST 306
D E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 112 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 169
Query: 307 QCT 309
+CT
Sbjct: 170 ECT 172
Score = 63.5 bits (153), Expect = 9e-10
Identities = 23/59 (38%), Positives = 34/59 (56%), Gaps = 4/59 (6%)
Query: 254 VCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCK----RGDVVCYNCNEEGHISTQC 308
+C++CGE GH + C E C+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 53 ICYRCGESGHLAKDCDLQEDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 111
Score = 63.5 bits (153), Expect = 9e-10
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query: 252 EIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
E C+ CG+ GH + C DE+KC+ CG+ GH+ DC + V CY C E GH++ C++
Sbjct: 95 EQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSK 152
Query: 311 PKKV 314
+V
Sbjct: 153 TSEV 156
Score = 61.2 bits (147), Expect = 4e-09
Identities = 27/71 (38%), Positives = 38/71 (53%), Gaps = 5/71 (7%)
Query: 252 EIVCFKCGEKGHKSNVCTKDEKK----CFRCGQKGHMLADCKRGDVV-CYNCNEEGHIST 306
E C+ CG GH + C + +++ C+ CG+ GH+ DC D CY+C E GHI
Sbjct: 71 EDACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQK 130
Query: 307 QCTQPKKVRVG 317
CT+ K R G
Sbjct: 131 DCTKVKCYRCG 141
Score = 59.3 bits (142), Expect = 2e-08
Identities = 29/89 (32%), Positives = 37/89 (40%), Gaps = 28/89 (31%)
Query: 255 CFKCGEKGHKSNVC------------------TKDE----------KKCFRCGQKGHMLA 286
CFKCG GH + C T D C+RCG+ GH+
Sbjct: 6 CFKCGRSGHWARECPTGGGRGRGMRSRGRGGFTSDRGFQFVSSSLPDICYRCGESGHLAK 65
Query: 287 DCKRGDVVCYNCNEEGHISTQCTQPKKVR 315
DC + CYNC GHI+ C +PK+ R
Sbjct: 66 DCDLQEDACYNCGRGGHIAKDCKEPKRER 94
>CNBP_CHICK (O42395) Cellular nucleic acid binding protein (CNBP)
Length = 172
Score = 66.2 bits (160), Expect = 1e-10
Identities = 29/63 (46%), Positives = 38/63 (60%), Gaps = 3/63 (4%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADC-KRGDVVCYNCNEEGHIST 306
D E C+ CGE GH CTK KC+RCG+ GH+ +C K +V CY C E GH++
Sbjct: 107 DHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSKTSEVNCYRCGESGHLAR 164
Query: 307 QCT 309
+CT
Sbjct: 165 ECT 167
Score = 63.5 bits (153), Expect = 9e-10
Identities = 26/64 (40%), Positives = 39/64 (60%), Gaps = 3/64 (4%)
Query: 252 EIVCFKCGEKGHKSNVCTK-DEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
E C+ CG+ GH + C DE+KC+ CG+ GH+ DC + V CY C E GH++ C++
Sbjct: 90 EQCCYNCGKPGHLARDCDHADEQKCYSCGEFGHIQKDCTK--VKCYRCGETGHVAINCSK 147
Query: 311 PKKV 314
+V
Sbjct: 148 TSEV 151
Score = 62.0 bits (149), Expect = 3e-09
Identities = 23/60 (38%), Positives = 37/60 (61%), Gaps = 5/60 (8%)
Query: 254 VCFKCGEKGHKSNVC-TKDEKKCFRCGQKGHMLADCK----RGDVVCYNCNEEGHISTQC 308
+C++CGE GH + C +++K C+ CG+ GH+ DCK + CYNC + GH++ C
Sbjct: 47 ICYRCGESGHLAKDCDLQEDKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDC 106
Score = 61.2 bits (147), Expect = 4e-09
Identities = 27/75 (36%), Positives = 39/75 (52%), Gaps = 5/75 (6%)
Query: 248 DAPAEIVCFKCGEKGHKSNVCTKDEKK----CFRCGQKGHMLADCKRGDVV-CYNCNEEG 302
D + C+ CG GH + C + +++ C+ CG+ GH+ DC D CY+C E G
Sbjct: 62 DLQEDKACYNCGRGGHIAKDCKEPKREREQCCYNCGKPGHLARDCDHADEQKCYSCGEFG 121
Query: 303 HISTQCTQPKKVRVG 317
HI CT+ K R G
Sbjct: 122 HIQKDCTKVKCYRCG 136
Score = 58.9 bits (141), Expect = 2e-08
Identities = 28/84 (33%), Positives = 37/84 (43%), Gaps = 23/84 (27%)
Query: 255 CFKCGEKGHKSNVCTKDEKK----------------------CFRCGQKGHMLADCK-RG 291
CFKCG GH + C + C+RCG+ GH+ DC +
Sbjct: 6 CFKCGRTGHWARECPTGIGRGRGMRSRGRAGFQFMSSSLPDICYRCGESGHLAKDCDLQE 65
Query: 292 DVVCYNCNEEGHISTQCTQPKKVR 315
D CYNC GHI+ C +PK+ R
Sbjct: 66 DKACYNCGRGGHIAKDCKEPKRER 89
>ZCH3_HUMAN (Q9NUD5) Zinc finger CCHC domain containing protein 3
Length = 404
Score = 65.5 bits (158), Expect = 2e-10
Identities = 28/71 (39%), Positives = 42/71 (58%), Gaps = 3/71 (4%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKV 314
CFKCG + H S CT+D +CFRCG++GH+ C++G +VC C + GH QC +
Sbjct: 336 CFKCGSRTHMSGSCTQD--RCFRCGEEGHLSPYCRKG-IVCNLCGKRGHAFAQCPKAVHN 392
Query: 315 RVGGKVFALTG 325
V ++ + G
Sbjct: 393 SVAAQLTGVAG 403
Score = 42.7 bits (99), Expect = 0.002
Identities = 18/47 (38%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Query: 273 KKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKVRVGGK 319
K CF+CG + HM C + C+ C EEGH+S C + + GK
Sbjct: 334 KTCFKCGSRTHMSGSCTQDR--CFRCGEEGHLSPYCRKGIVCNLCGK 378
>HEXP_LEIMA (Q04832) DNA-binding protein HEXBP (Hexamer-binding
protein)
Length = 271
Score = 63.2 bits (152), Expect = 1e-09
Identities = 29/78 (37%), Positives = 37/78 (47%), Gaps = 14/78 (17%)
Query: 255 CFKCGEKGHKSNVCTKDEKK-------CFRCGQKGHMLADCKRG-------DVVCYNCNE 300
CF+CGE+GH S C + + CFRCG+ GHM DC CY C +
Sbjct: 45 CFRCGEEGHMSRECPNEARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQ 104
Query: 301 EGHISTQCTQPKKVRVGG 318
EGH+S C + GG
Sbjct: 105 EGHLSRDCPSSQGGSRGG 122
Score = 62.4 bits (150), Expect = 2e-09
Identities = 32/107 (29%), Positives = 47/107 (43%), Gaps = 16/107 (14%)
Query: 218 KGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTK------- 270
+GH +R P S +G R ++ + C+KCG+ GH S C
Sbjct: 105 EGHLSRDCPSSQGGSRGGYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDCPNGQGGYSG 164
Query: 271 -DEKKCFRCGQKGHMLADCKR--------GDVVCYNCNEEGHISTQC 308
++ C++CG GH+ DC GD CY C E GH+S +C
Sbjct: 165 AGDRTCYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSREC 211
Score = 61.6 bits (148), Expect = 3e-09
Identities = 26/70 (37%), Positives = 37/70 (52%), Gaps = 14/70 (20%)
Query: 255 CFKCGEKGHKSNVCTKDE--------KKCFRCGQKGHMLADCKR------GDVVCYNCNE 300
C+KCG+ GH S C + +KC++CG+ GHM +C GD CY C +
Sbjct: 170 CYKCGDAGHISRDCPNGQGGYSGAGDRKCYKCGESGHMSRECPSAGSTGSGDRACYKCGK 229
Query: 301 EGHISTQCTQ 310
GHIS +C +
Sbjct: 230 PGHISRECPE 239
Score = 60.1 bits (144), Expect = 1e-08
Identities = 38/121 (31%), Positives = 50/121 (40%), Gaps = 32/121 (26%)
Query: 219 GHQNRPKP-----YSAPTDKGKQRLNDERRPSKRDAP---------AEIVCFKCGEKGHK 264
GH +R P YS D+ + D S RD P + C+KCGE GH
Sbjct: 149 GHISRDCPNGQGGYSGAGDRTCYKCGDAGHIS-RDCPNGQGGYSGAGDRKCYKCGESGHM 207
Query: 265 SNVCTK------DEKKCFRCGQKGHMLADCKR-----------GDVVCYNCNEEGHISTQ 307
S C ++ C++CG+ GH+ +C GD CY C E GHIS
Sbjct: 208 SRECPSAGSTGSGDRACYKCGKPGHISRECPEAGGSYGGSRGGGDRTCYKCGEAGHISRD 267
Query: 308 C 308
C
Sbjct: 268 C 268
Score = 59.3 bits (142), Expect = 2e-08
Identities = 31/94 (32%), Positives = 41/94 (42%), Gaps = 30/94 (31%)
Query: 245 SKRDAPAEIVCFKCGEKGHKSNVCTKDEK-------KCFRCGQKGHMLADC--------- 288
++ A + CF+CGE GH S C K +C++CGQ+GH+ DC
Sbjct: 62 ARSGAAGAMTCFRCGEAGHMSRDCPNSAKPGAAKGFECYKCGQEGHLSRDCPSSQGGSRG 121
Query: 289 ----KR----------GDVVCYNCNEEGHISTQC 308
KR GD CY C + GHIS C
Sbjct: 122 GYGQKRGRSGAQGGYSGDRTCYKCGDAGHISRDC 155
Score = 57.8 bits (138), Expect = 5e-08
Identities = 28/88 (31%), Positives = 42/88 (46%), Gaps = 17/88 (19%)
Query: 240 DERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEKK-------CFRCGQKGHMLADCKR-- 290
D +RP + + C CG++GH + C + + K CFRCG++GHM +C
Sbjct: 6 DVKRPRTESSTS---CRNCGKEGHYARECPEADSKGDERSTTCFRCGEEGHMSRECPNEA 62
Query: 291 -----GDVVCYNCNEEGHISTQCTQPKK 313
G + C+ C E GH+S C K
Sbjct: 63 RSGAAGAMTCFRCGEAGHMSRDCPNSAK 90
Score = 43.5 bits (101), Expect = 0.001
Identities = 19/56 (33%), Positives = 27/56 (47%), Gaps = 9/56 (16%)
Query: 269 TKDEKKCFRCGQKGHMLADCKRGD-------VVCYNCNEEGHISTQCTQPKKVRVG 317
T+ C CG++GH +C D C+ C EEGH+S +C P + R G
Sbjct: 12 TESSTSCRNCGKEGHYARECPEADSKGDERSTTCFRCGEEGHMSREC--PNEARSG 65
>BYR3_SCHPO (P36627) Cellular nucleic acid binding protein homolog
Length = 179
Score = 60.1 bits (144), Expect = 1e-08
Identities = 25/69 (36%), Positives = 36/69 (51%), Gaps = 7/69 (10%)
Query: 247 RDAPAEIVCFKCGEKGHKSNVCT--KDEKKCFRCGQKGHMLADC-----KRGDVVCYNCN 299
R+ +C+ C + GHK++ CT + EK C+ CG GH++ DC R CY C
Sbjct: 30 RECTKGSICYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDCPSSPNPRQGAECYKCG 89
Query: 300 EEGHISTQC 308
GHI+ C
Sbjct: 90 RVGHIARDC 98
Score = 60.1 bits (144), Expect = 1e-08
Identities = 24/59 (40%), Positives = 35/59 (58%), Gaps = 3/59 (5%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGD--VVCYNCNEEGHISTQCTQP 311
C+ CG GH++ CT K C+ CG+ GH +C++ +CY CN+ GHI+ CT P
Sbjct: 118 CYACGSYGHQARDCTMGVK-CYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNCTSP 175
Score = 55.8 bits (133), Expect = 2e-07
Identities = 23/69 (33%), Positives = 34/69 (48%), Gaps = 3/69 (4%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADC--KRGDVVCYNCNEEGHISTQCTQPK 312
C+ CGE GH++ CTK C+ C Q GH ++C + + CY C GH+ C
Sbjct: 19 CYNCGENGHQARECTKG-SICYNCNQTGHKASECTEPQQEKTCYACGTAGHLVRDCPSSP 77
Query: 313 KVRVGGKVF 321
R G + +
Sbjct: 78 NPRQGAECY 86
Score = 55.8 bits (133), Expect = 2e-07
Identities = 18/45 (40%), Positives = 32/45 (71%), Gaps = 1/45 (2%)
Query: 269 TKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
T+ +C+ CG+ GH +C +G + CYNCN+ GH +++CT+P++
Sbjct: 13 TRPGPRCYNCGENGHQARECTKGSI-CYNCNQTGHKASECTEPQQ 56
Score = 51.6 bits (122), Expect = 3e-06
Identities = 26/80 (32%), Positives = 34/80 (42%), Gaps = 14/80 (17%)
Query: 244 PSKRDAPAEIVCFKCGEKGHKSNVCT-------------KDEKKCFRCGQKGHMLADCKR 290
PS + C+KCG GH + C + C+ CG GH DC
Sbjct: 74 PSSPNPRQGAECYKCGRVGHIARDCRTNGQQSGGRFGGHRSNMNCYACGSYGHQARDCTM 133
Query: 291 GDVVCYNCNEEGHISTQCTQ 310
G V CY+C + GH S +C Q
Sbjct: 134 G-VKCYSCGKIGHRSFECQQ 152
Score = 49.3 bits (116), Expect = 2e-05
Identities = 25/87 (28%), Positives = 36/87 (40%), Gaps = 18/87 (20%)
Query: 252 EIVCFKCGEKGHKSNVCTKDEK-----KCFRCGQKGHMLADCK-------------RGDV 293
E C+ CG GH C +C++CG+ GH+ DC+ R ++
Sbjct: 57 EKTCYACGTAGHLVRDCPSSPNPRQGAECYKCGRVGHIARDCRTNGQQSGGRFGGHRSNM 116
Query: 294 VCYNCNEEGHISTQCTQPKKVRVGGKV 320
CY C GH + CT K GK+
Sbjct: 117 NCYACGSYGHQARDCTMGVKCYSCGKI 143
Score = 42.4 bits (98), Expect = 0.002
Identities = 16/44 (36%), Positives = 25/44 (56%), Gaps = 2/44 (4%)
Query: 247 RDAPAEIVCFKCGEKGHKSNVC--TKDEKKCFRCGQKGHMLADC 288
RD + C+ CG+ GH+S C D + C++C Q GH+ +C
Sbjct: 129 RDCTMGVKCYSCGKIGHRSFECQQASDGQLCYKCNQPGHIAVNC 172
>GLH2_CAEEL (Q966L9) ATP-dependent RNA helicase glh-2 (EC 3.6.1.-)
(Germline helicase-2)
Length = 974
Score = 58.9 bits (141), Expect = 2e-08
Identities = 37/135 (27%), Positives = 48/135 (35%), Gaps = 39/135 (28%)
Query: 237 RLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEK----------------------- 273
R ND P K P VC+ C + GH S C ++ K
Sbjct: 382 RSNDCPEPKKEREPR--VCYNCQQPGHNSRDCPEERKPREGRNGFTSGFGGGNDGGFGGG 439
Query: 274 --------------KCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKVRVGGK 319
KCF C +GH A+C C+NC E+GH S +C P K R G +
Sbjct: 440 NAEGFGNNEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREGAE 499
Query: 320 VFALTGTQTANEDRL 334
T ED +
Sbjct: 500 GEGPKATYVPVEDNM 514
Score = 50.4 bits (119), Expect = 8e-06
Identities = 27/101 (26%), Positives = 39/101 (37%), Gaps = 42/101 (41%)
Query: 255 CFKCGEKGHKSNVCTKDEKK-----CFRCGQKGHMLADC--------------------- 288
CF C + GH+SN C + +K+ C+ C Q GH DC
Sbjct: 373 CFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKPREGRNGFTSGFGGGN 432
Query: 289 ----------------KRGDVVCYNCNEEGHISTQCTQPKK 313
+RG + C+NC EGH S +C +P +
Sbjct: 433 DGGFGGGNAEGFGNNEERGPMKCFNCKGEGHRSAECPEPPR 473
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/67 (35%), Positives = 32/67 (46%), Gaps = 12/67 (17%)
Query: 260 EKGHKSNVCTKDEKKCFRCGQKGHMLADC-----KRGDVVCYNCNEEGHISTQCTQPKKV 314
++G ++N CF C Q GH DC +R VCYNC + GH S C + +K
Sbjct: 251 DRGERNN-------NCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKP 303
Query: 315 RVGGKVF 321
R G F
Sbjct: 304 REGRNGF 310
Score = 46.6 bits (109), Expect = 1e-04
Identities = 24/67 (35%), Positives = 32/67 (46%), Gaps = 12/67 (17%)
Query: 260 EKGHKSNVCTKDEKKCFRCGQKGHMLADC-----KRGDVVCYNCNEEGHISTQCTQPKKV 314
++G ++N CF C Q GH DC +R VCYNC + GH S C + +K
Sbjct: 365 DRGERNN-------NCFNCQQPGHRSNDCPEPKKEREPRVCYNCQQPGHNSRDCPEERKP 417
Query: 315 RVGGKVF 321
R G F
Sbjct: 418 REGRNGF 424
>GLH1_CAEEL (P34689) ATP-dependent RNA helicase glh-1 (EC 3.6.1.-)
(Germline helicase-1)
Length = 763
Score = 57.0 bits (136), Expect = 8e-08
Identities = 37/137 (27%), Positives = 49/137 (35%), Gaps = 41/137 (29%)
Query: 237 RLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEK----------------------- 273
R +D P K P VC+ C + GH S CT++ K
Sbjct: 169 RSSDCPEPRKEREPR--VCYNCQQPGHTSRECTEERKPREGRTGGFGGGAGFGNNGGNDG 226
Query: 274 ----------------KCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKKVRVG 317
KCF C +GH A+C C+NC E+GH S +C P K R G
Sbjct: 227 FGGDGGFGGGEERGPMKCFNCKGEGHRSAECPEPPRGCFNCGEQGHRSNECPNPAKPREG 286
Query: 318 GKVFALTGTQTANEDRL 334
+ T ED +
Sbjct: 287 VEGEGPKATYVPVEDNM 303
Score = 53.1 bits (126), Expect = 1e-06
Identities = 26/64 (40%), Positives = 33/64 (50%), Gaps = 8/64 (12%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHMLADC-----KRGDVVCYNCNEEGHISTQCTQPKK 313
GE GH + CF C Q GH +DC +R VCYNC + GH S +CT+ +K
Sbjct: 147 GEGGHGGG---ERNNNCFNCQQPGHRSSDCPEPRKEREPRVCYNCQQPGHTSRECTEERK 203
Query: 314 VRVG 317
R G
Sbjct: 204 PREG 207
>GLH4_CAEEL (O76743) ATP-dependent RNA helicase glh-4 (EC 3.6.1.-)
(Germline helicase-4)
Length = 1156
Score = 55.5 bits (132), Expect = 2e-07
Identities = 28/67 (41%), Positives = 33/67 (48%), Gaps = 6/67 (8%)
Query: 255 CFKCGEKGHKSNVCTKDEKK---CFRCGQKGHMLADCKRGDVV---CYNCNEEGHISTQC 308
C CGE+GH S C K + C C Q GH +DC + V C NC EGH + C
Sbjct: 572 CHNCGEEGHISKECDKPKVPRFPCRNCEQLGHFASDCDQPRVPRGPCRNCGIEGHFAVDC 631
Query: 309 TQPKKVR 315
QPK R
Sbjct: 632 DQPKVPR 638
Score = 44.7 bits (104), Expect = 4e-04
Identities = 25/71 (35%), Positives = 35/71 (49%), Gaps = 10/71 (14%)
Query: 255 CFKCGEKGHKSNVCTKDEKK---CFRCGQKGHMLADCKRGDV------VCYNCNEEGHIS 305
C CG +GH + C + + C CGQ+GH DC+ V C C EEGH
Sbjct: 618 CRNCGIEGHFAVDCDQPKVPRGPCRNCGQEGHFAKDCQNERVRMEPTEPCRRCAEEGHWG 677
Query: 306 TQC-TQPKKVR 315
+C T+PK ++
Sbjct: 678 YECPTRPKDLQ 688
Score = 37.0 bits (84), Expect = 0.088
Identities = 18/47 (38%), Positives = 25/47 (52%), Gaps = 2/47 (4%)
Query: 295 CYNCNEEGHISTQCTQPKKVRVGGKVFALTG--TQTANEDRLIRGTC 339
C+NC EEGHIS +C +PK R + G ++ R+ RG C
Sbjct: 572 CHNCGEEGHISKECDKPKVPRFPCRNCEQLGHFASDCDQPRVPRGPC 618
>YL92_SCHPO (Q9HFF2) Hypothetical protein C683.02c in chromosome I
Length = 218
Score = 55.1 bits (131), Expect = 3e-07
Identities = 30/91 (32%), Positives = 42/91 (45%), Gaps = 12/91 (13%)
Query: 254 VCFKCGEKGHKSNVCTKDE----KKCFRCGQKGHMLADCKRGDV-------VCYNCNEEG 302
+CF+CG K H N C+K KCF C + GH+ C++ C C+
Sbjct: 101 ICFRCGSKEHSLNACSKKGPLKFAKCFICHENGHLSGQCEQNPKGLYPKGGCCKFCSSVH 160
Query: 303 HISTQCTQPKKVRVG-GKVFALTGTQTANED 332
H++ C Q K V G V + GT A+ED
Sbjct: 161 HLAKDCDQVNKDDVSFGHVVGVAGTTGADED 191
Score = 38.5 bits (88), Expect = 0.030
Identities = 34/140 (24%), Positives = 52/140 (36%), Gaps = 39/140 (27%)
Query: 203 EDTKAHYKVMSERRGKGHQNRPKPY--SAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGE 260
E T KV+ + + + N + G + DER+ KR +
Sbjct: 15 EATPYESKVLEQPKNSSNTNEESSSQDNMKASFGSSKRYDERQKKKRSEYRRL------- 67
Query: 261 KGHKSNVCTKDEKKCFRCGQKGHMLADC------------------------KRGDV--- 293
+ N +D K CF C Q+GH++ DC K+G +
Sbjct: 68 --RRINQRNRD-KFCFACRQQGHIVQDCPEAKDNVSICFRCGSKEHSLNACSKKGPLKFA 124
Query: 294 VCYNCNEEGHISTQCTQPKK 313
C+ C+E GH+S QC Q K
Sbjct: 125 KCFICHENGHLSGQCEQNPK 144
>GAG_SIVAI (Q02843) Gag polyprotein [Contains: Core protein p17;
Core protein p24; Core protein p15]
Length = 513
Score = 51.2 bits (121), Expect = 5e-06
Identities = 21/49 (42%), Positives = 27/49 (54%), Gaps = 1/49 (2%)
Query: 244 PSKRDAPAEIVCFKCGEKGHKSNVCTKDEK-KCFRCGQKGHMLADCKRG 291
P K+ + CF CG+ GH C + KCF+CG+ GHM DCK G
Sbjct: 381 PQKKGPRGPLKCFNCGKFGHMQRECKAPRQIKCFKCGKIGHMAKDCKNG 429
Score = 40.8 bits (94), Expect = 0.006
Identities = 15/36 (41%), Positives = 22/36 (60%), Gaps = 1/36 (2%)
Query: 274 KCFRCGQKGHMLADCKRG-DVVCYNCNEEGHISTQC 308
KCF CG+ GHM +CK + C+ C + GH++ C
Sbjct: 391 KCFNCGKFGHMQRECKAPRQIKCFKCGKIGHMAKDC 426
Score = 32.3 bits (72), Expect = 2.2
Identities = 9/26 (34%), Positives = 18/26 (68%)
Query: 290 RGDVVCYNCNEEGHISTQCTQPKKVR 315
RG + C+NC + GH+ +C P++++
Sbjct: 387 RGPLKCFNCGKFGHMQRECKAPRQIK 412
Score = 31.2 bits (69), Expect = 4.8
Identities = 13/34 (38%), Positives = 17/34 (49%)
Query: 249 APAEIVCFKCGEKGHKSNVCTKDEKKCFRCGQKG 282
AP +I CFKCG+ GH + C + G G
Sbjct: 407 APRQIKCFKCGKIGHMAKDCKNGQANFLGYGHWG 440
>GAG_VILV2 (P23425) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 50.4 bits (119), Expect = 8e-06
Identities = 19/55 (34%), Positives = 33/55 (59%), Gaps = 6/55 (10%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
G+ GHK +KC+ CG+ GH+ C++G ++C++C + GH+ C Q K+
Sbjct: 376 GKAGHKGV-----NQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 31.6 bits (70), Expect = 3.7
Identities = 16/56 (28%), Positives = 25/56 (44%), Gaps = 8/56 (14%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
C G +G K + + + + G KG + CYNC + GH++ QC Q
Sbjct: 355 CRDVGSEGFKMQLLAQALRPQGKAGHKGV--------NQKCYNCGKPGHLARQCRQ 402
>GAG_VILV1 (P23424) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 50.4 bits (119), Expect = 8e-06
Identities = 19/55 (34%), Positives = 33/55 (59%), Gaps = 6/55 (10%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
G+ GHK +KC+ CG+ GH+ C++G ++C++C + GH+ C Q K+
Sbjct: 376 GKAGHKGV-----NQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 31.6 bits (70), Expect = 3.7
Identities = 16/56 (28%), Positives = 25/56 (44%), Gaps = 8/56 (14%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
C G +G K + + + + G KG + CYNC + GH++ QC Q
Sbjct: 355 CRDVGSEGFKMQLLAQALRPQGKAGHKGV--------NQKCYNCGKPGHLARQCRQ 402
>GAG_VILV (P03352) Gag polyprotein [Contains: Core protein p16; Core
protein p25; Core protein p14]
Length = 442
Score = 50.4 bits (119), Expect = 8e-06
Identities = 19/55 (34%), Positives = 33/55 (59%), Gaps = 6/55 (10%)
Query: 259 GEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
G+ GHK +KC+ CG+ GH+ C++G ++C++C + GH+ C Q K+
Sbjct: 376 GKAGHKGV-----NQKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 31.6 bits (70), Expect = 3.7
Identities = 16/56 (28%), Positives = 25/56 (44%), Gaps = 8/56 (14%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
C G +G K + + + + G KG + CYNC + GH++ QC Q
Sbjct: 355 CRDVGSEGFKMQLLAQALRPQGKAGHKGV--------NQKCYNCGKPGHLARQCRQ 402
>GAG_VILVK (P35955) Gag polyprotein [Contains: Core protein p16;
Core protein p25; Core protein p14]
Length = 442
Score = 48.9 bits (115), Expect = 2e-05
Identities = 19/57 (33%), Positives = 33/57 (57%), Gaps = 9/57 (15%)
Query: 257 KCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQPKK 313
K G+KG +KC+ CG+ GH+ C++G ++C++C + GH+ C Q K+
Sbjct: 377 KAGQKGVN--------QKCYNCGKPGHLARQCRQG-IICHHCGKRGHMQKDCRQKKQ 424
Score = 33.5 bits (75), Expect = 0.97
Identities = 17/56 (30%), Positives = 26/56 (46%), Gaps = 8/56 (14%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCGQKGHMLADCKRGDVVCYNCNEEGHISTQCTQ 310
C G +G K + + + + GQKG + CYNC + GH++ QC Q
Sbjct: 355 CRDVGSEGFKMQLLAQALRPQGKAGQKGV--------NQKCYNCGKPGHLARQCRQ 402
>GAG_HV1U4 (P24736) Gag polyprotein [Contains: Core protein p17
(Matrix protein); Core protein p24 (Core antigen); Core
protein p2; Core protein p7 (Nucleocapsid protein); Core
protein p1; Core protein p6]
Length = 492
Score = 48.1 bits (113), Expect = 4e-05
Identities = 18/40 (45%), Positives = 26/40 (65%), Gaps = 1/40 (2%)
Query: 250 PAEIVCFKCGEKGHKSNVCTKDEKK-CFRCGQKGHMLADC 288
P I CF CG++GH + C KK C++CG++GH + DC
Sbjct: 380 PRRIKCFNCGKEGHLAKNCRAPRKKGCWKCGKEGHQMKDC 419
Score = 42.0 bits (97), Expect = 0.003
Identities = 19/52 (36%), Positives = 31/52 (59%), Gaps = 8/52 (15%)
Query: 274 KCFRCGQKGHMLADC----KRGDVVCYNCNEEGHISTQCTQPKKVRVGGKVF 321
KCF CG++GH+ +C K+G C+ C +EGH CT+ ++ GK++
Sbjct: 384 KCFNCGKEGHLAKNCRAPRKKG---CWKCGKEGHQMKDCTE-RQANFLGKIW 431
Score = 33.9 bits (76), Expect = 0.75
Identities = 17/62 (27%), Positives = 30/62 (47%), Gaps = 6/62 (9%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCG---QKGHMLADCKRGDVVCYNCNEEGHISTQCTQP 311
C G GHK+ V + + + Q+G+ + + C+NC +EGH++ C P
Sbjct: 345 CQGVGGPGHKARVLAEAMSQVQQTSIMMQRGNFRGPRR---IKCFNCGKEGHLAKNCRAP 401
Query: 312 KK 313
+K
Sbjct: 402 RK 403
>GAG_SIVCZ (P17282) Gag polyprotein [Contains: Core protein p18;
Core protein p25; Core protein p16]
Length = 508
Score = 47.8 bits (112), Expect = 5e-05
Identities = 18/38 (47%), Positives = 26/38 (68%), Gaps = 1/38 (2%)
Query: 252 EIVCFKCGEKGHKSNVCTKDEKK-CFRCGQKGHMLADC 288
+I CF CG++GH + C +K C+RCGQ+GH + DC
Sbjct: 399 KIKCFNCGKEGHLARNCKAPRRKGCWRCGQEGHQMKDC 436
Score = 41.6 bits (96), Expect = 0.004
Identities = 18/44 (40%), Positives = 26/44 (58%), Gaps = 7/44 (15%)
Query: 270 KDEKKCFRCGQKGHMLADCK----RGDVVCYNCNEEGHISTQCT 309
K + KCF CG++GH+ +CK +G C+ C +EGH CT
Sbjct: 397 KRKIKCFNCGKEGHLARNCKAPRRKG---CWRCGQEGHQMKDCT 437
Score = 30.8 bits (68), Expect = 6.3
Identities = 8/21 (38%), Positives = 15/21 (71%)
Query: 293 VVCYNCNEEGHISTQCTQPKK 313
+ C+NC +EGH++ C P++
Sbjct: 400 IKCFNCGKEGHLARNCKAPRR 420
>GAG_HV1ND (P18800) Gag polyprotein [Contains: Core protein p17
(Matrix protein); Core protein p24 (Core antigen); Core
protein p2; Core protein p7 (Nucleocapsid protein); Core
protein p1; Core protein p6]
Length = 496
Score = 47.8 bits (112), Expect = 5e-05
Identities = 23/73 (31%), Positives = 36/73 (48%), Gaps = 1/73 (1%)
Query: 217 GKGHQNRPKPYSAPTDKGKQRLNDERRPSKRDAPAEIVCFKCGEKGHKSNVCTKDEKK-C 275
G GH+ R + G +R + + I CF CG++GH + C KK C
Sbjct: 351 GPGHKARVLAEAMSQVTGSATAVMMQRGNFKGPRKSIKCFNCGKEGHTAKNCRAPRKKGC 410
Query: 276 FRCGQKGHMLADC 288
++CG++GH + DC
Sbjct: 411 WKCGREGHQMKDC 423
Score = 40.4 bits (93), Expect = 0.008
Identities = 19/52 (36%), Positives = 29/52 (55%), Gaps = 8/52 (15%)
Query: 274 KCFRCGQKGHMLADC----KRGDVVCYNCNEEGHISTQCTQPKKVRVGGKVF 321
KCF CG++GH +C K+G C+ C EGH CT+ ++ GK++
Sbjct: 388 KCFNCGKEGHTAKNCRAPRKKG---CWKCGREGHQMKDCTE-RQANFLGKIW 435
Score = 33.1 bits (74), Expect = 1.3
Identities = 18/64 (28%), Positives = 28/64 (43%), Gaps = 7/64 (10%)
Query: 255 CFKCGEKGHKSNVCTKDEKKCFRCG-----QKGHMLADCKRGDVVCYNCNEEGHISTQCT 309
C G GHK+ V + + Q+G+ R + C+NC +EGH + C
Sbjct: 346 CQGVGGPGHKARVLAEAMSQVTGSATAVMMQRGNFKGP--RKSIKCFNCGKEGHTAKNCR 403
Query: 310 QPKK 313
P+K
Sbjct: 404 APRK 407
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.321 0.136 0.413
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 47,762,690
Number of Sequences: 164201
Number of extensions: 2095295
Number of successful extensions: 5838
Number of sequences better than 10.0: 121
Number of HSP's better than 10.0 without gapping: 68
Number of HSP's successfully gapped in prelim test: 53
Number of HSP's that attempted gapping in prelim test: 5223
Number of HSP's gapped (non-prelim): 378
length of query: 396
length of database: 59,974,054
effective HSP length: 112
effective length of query: 284
effective length of database: 41,583,542
effective search space: 11809725928
effective search space used: 11809725928
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC136954.7


