

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136954.12 + phase: 0
(369 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
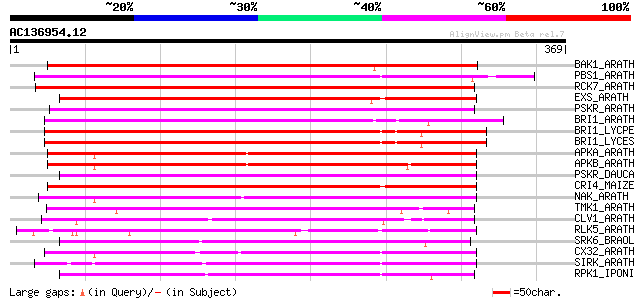
BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated rec... 256 8e-68
PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC 2.7... 249 1e-65
RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase RLC... 243 4e-64
EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase E... 236 9e-62
PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (... 223 6e-58
BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC ... 221 2e-57
BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.... 220 5e-57
BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precurso... 219 9e-57
APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor ... 218 2e-56
APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor ... 215 1e-55
PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.... 215 2e-55
CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4 pr... 212 1e-54
NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK ... 209 9e-54
TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precur... 209 1e-53
CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (... 207 3e-53
RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC... 206 8e-53
SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase rec... 206 1e-52
CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx3... 203 5e-52
SIRK_ARATH (O64483) Senescence-induced receptor-like serine/thre... 186 6e-47
RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC 2... 184 4e-46
>BAK1_ARATH (Q94F62) BRASSINOSTEROID INSENSITIVE 1-associated
receptor kinase 1 precursor (EC 2.7.1.37)
(BRI1-associated receptor kinase 1) (Somatic
embryogenesis receptor-like kinase 3)
Length = 615
Score = 256 bits (653), Expect = 8e-68
Identities = 127/289 (43%), Positives = 190/289 (64%), Gaps = 3/289 (1%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKA-EMEFAVEVE 84
F+ +EL A++ FS+ LG GGFG VY GR +DG +AVK+LK ++ E++F EVE
Sbjct: 277 FSLRELQVASDNFSNKNILGRGGFGKVYKGRLADGTLVAVKRLKEERTQGGELQFQTEVE 336
Query: 85 VLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIG 144
++ H+NLL LRG+C+ +RL+VY YM N S+ S L + + L+W KR IA+G
Sbjct: 337 MISMAVHRNLLRLRGFCMTPTERLLVYPYMANGSVASCLRERPESQPPLDWPKRQRIALG 396
Query: 145 SAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTL 204
SA G+ YLH P IIHRD+KA+N+LLD +F +V DFG AKL+ +H+TT V+GT+
Sbjct: 397 SARGLAYLHDHCDPKIIHRDVKAANILLDEEFEAVVGDFGLAKLMDYKDTHVTTAVRGTI 456
Query: 205 GYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE--KLPGGLKRTITEWAEPLITKG 262
G++APEY GK SE DV+ +G++LLEL+TG++ + +L + +W + L+ +
Sbjct: 457 GHIAPEYLSTGKSSEKTDVFGYGVMLLELITGQRAFDLARLANDDDVMLLDWVKGLLKEK 516
Query: 263 RFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKG 311
+ +VD L+GN+ + +V+Q + VA LC QS P +RP M +VV +L+G
Sbjct: 517 KLEALVDVDLQGNYKDEEVEQLIQVALLCTQSSPMERPKMSEVVRMLEG 565
>PBS1_ARATH (Q9FE20) Serine/threonine-protein kinase PBS1 (EC
2.7.1.37) (AvrPphB susceptible protein 1)
Length = 456
Score = 249 bits (635), Expect = 1e-65
Identities = 147/340 (43%), Positives = 202/340 (59%), Gaps = 13/340 (3%)
Query: 17 GVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRT-SDGLQIAVKKLKAMNSKA 75
G+ + F ++EL AT F D LGEGGFG VY GR S G +AVK+L +
Sbjct: 65 GLGQIAAHTFAFRELAAATMNFHPDTFLGEGGFGRVYKGRLDSTGQVVAVKQLDRNGLQG 124
Query: 76 EMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNW 135
EF VEV +L + H NL+ L GYC DQRL+VY++MP SL HLH + L+W
Sbjct: 125 NREFLVEVLMLSLLHHPNLVNLIGYCADGDQRLLVYEFMPLGSLEDHLHDLPPDKEALDW 184
Query: 136 QKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEG-VS 194
RM IA G+A+G+ +LH + P +I+RD K+SN+LLD F P ++DFG AKL P G S
Sbjct: 185 NMRMKIAAGAAKGLEFLHDKANPPVIYRDFKSSNILLDEGFHPKLSDFGLAKLGPTGDKS 244
Query: 195 HMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE-KLPGGLKRTITE 253
H++TRV GT GY APEYAM G+++ DVYSFG++ LEL+TGRK I+ ++P G ++ +
Sbjct: 245 HVSTRVMGTYGYCAPEYAMTGQLTVKSDVYSFGVVFLELITGRKAIDSEMPHG-EQNLVA 303
Query: 254 WAEPLIT-KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVV---SLL 309
WA PL + +F + DP+L+G F + Q + VA++C+Q + RP + VV S L
Sbjct: 304 WARPLFNDRRKFIKLADPRLKGRFPTRALYQALAVASMCIQEQAATRPLIADVVTALSYL 363
Query: 310 KGQEPDQGKVTKMRIDSVKYNDELLALDQPSDDDYDGNSS 349
Q D K DS + DE A +DD G+ S
Sbjct: 364 ANQAYDPSK-----DDSRRNRDERGARLITRNDDGGGSGS 398
>RCK7_ARATH (Q9LQQ8) Putative serine/threonine-protein kinase
RLCKVII (EC 2.7.1.37)
Length = 423
Score = 243 bits (621), Expect = 4e-64
Identities = 131/295 (44%), Positives = 180/295 (60%), Gaps = 3/295 (1%)
Query: 18 VANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQI-AVKKLKAMNSKAE 76
V + FT++EL AT F D LGEGGFG V+ G Q+ A+K+L +
Sbjct: 83 VTGKKAQTFTFQELAEATGNFRSDCFLGEGGFGKVFKGTIEKLDQVVAIKQLDRNGVQGI 142
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
EF VEV L H NL+ L G+C DQRL+VY+YMP SL HLH +G+ L+W
Sbjct: 143 REFVVEVLTLSLADHPNLVKLIGFCAEGDQRLLVYEYMPQGSLEDHLHVLPSGKKPLDWN 202
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEG-VSH 195
RM IA G+A G+ YLH +TP +I+RD+K SN+LL D+ P ++DFG AK+ P G +H
Sbjct: 203 TRMKIAAGAARGLEYLHDRMTPPVIYRDLKCSNILLGEDYQPKLSDFGLAKVGPSGDKTH 262
Query: 196 MTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWA 255
++TRV GT GY AP+YAM G+++ D+YSFG++LLEL+TGRK I+ + + WA
Sbjct: 263 VSTRVMGTYGYCAPDYAMTGQLTFKSDIYSFGVVLLELITGRKAIDNTKTRKDQNLVGWA 322
Query: 256 EPLITKGR-FRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
PL R F MVDP L+G + + Q + ++A+CVQ +P RP + VV L
Sbjct: 323 RPLFKDRRNFPKMVDPLLQGQYPVRGLYQALAISAMCVQEQPTMRPVVSDVVLAL 377
>EXS_ARATH (Q9LYN8) Leucine-rich repeat receptor protein kinase EXS
precursor (EC 2.7.1.37) (Extra sporogenous cells protein)
(EXCESS MICROSPOROCYTES1 protein)
Length = 1192
Score = 236 bits (601), Expect = 9e-62
Identities = 125/281 (44%), Positives = 172/281 (60%), Gaps = 7/281 (2%)
Query: 34 ATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRVRHKN 93
AT+ FS +G+GGFG+VY +AVKKL ++ EF E+E LG+V+H N
Sbjct: 913 ATDHFSKKNIIGDGGFGTVYKACLPGEKTVAVKKLSEAKTQGNREFMAEMETLGKVKHPN 972
Query: 94 LLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSAEGILYLH 153
L+ L GYC +++L+VY+YM N SL L Q L+W KR+ IA+G+A G+ +LH
Sbjct: 973 LVSLLGYCSFSEEKLLVYEYMVNGSLDHWLRNQTGMLEVLDWSKRLKIAVGAARGLAFLH 1032
Query: 154 HEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAM 213
H PHIIHRDIKASN+LLD DF P VADFG A+LI SH++T + GT GY+ PEY
Sbjct: 1033 HGFIPHIIHRDIKASNILLDGDFEPKVADFGLARLISACESHVSTVIAGTFGYIPPEYGQ 1092
Query: 214 WGKVSESCDVYSFGILLLELVTGRKP----IEKLPGGLKRTITEWAEPLITKGRFRDMVD 269
+ + DVYSFG++LLELVTG++P ++ GG + WA I +G+ D++D
Sbjct: 1093 SARATTKGDVYSFGVILLELVTGKEPTGPDFKESEGG---NLVGWAIQKINQGKAVDVID 1149
Query: 270 PKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
P L +N + + +A LC+ P KRPNM V+ LK
Sbjct: 1150 PLLVSVALKNSQLRLLQIAMLCLAETPAKRPNMLDVLKALK 1190
>PSKR_ARATH (Q9ZVR7) Putative phytosulfokine receptor precursor (EC
2.7.1.37) (Phytosulfokine LRR receptor kinase)
Length = 1008
Score = 223 bits (568), Expect = 6e-58
Identities = 113/283 (39%), Positives = 171/283 (59%)
Query: 27 TYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVL 86
+Y +L +TN F +G GGFG VY DG ++A+KKL + E EF EVE L
Sbjct: 723 SYDDLLDSTNSFDQANIIGCGGFGMVYKATLPDGKKVAIKKLSGDCGQIEREFEAEVETL 782
Query: 87 GRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSA 146
R +H NL+ LRG+C + RL++Y YM N SL LH + G L W+ R+ IA G+A
Sbjct: 783 SRAQHPNLVLLRGFCFYKNDRLLIYSYMENGSLDYWLHERNDGPALLKWKTRLRIAQGAA 842
Query: 147 EGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLGY 206
+G+LYLH PHI+HRDIK+SN+LLD +F +ADFG A+L+ +H++T + GTLGY
Sbjct: 843 KGLLYLHEGCDPHILHRDIKSSNILLDENFNSHLADFGLARLMSPYETHVSTDLVGTLGY 902
Query: 207 LAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKGRFRD 266
+ PEY + DVYSFG++LLEL+T ++P++ R + W + + R +
Sbjct: 903 IPPEYGQASVATYKGDVYSFGVVLLELLTDKRPVDMCKPKGCRDLISWVVKMKHESRASE 962
Query: 267 MVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
+ DP + ++ ++ + + +A LC+ P++RP +Q+VS L
Sbjct: 963 VFDPLIYSKENDKEMFRVLEIACLCLSENPKQRPTTQQLVSWL 1005
>BRI1_ARATH (O22476) BRASSINOSTEROID INSENSITIVE 1 precursor (EC
2.7.1.37) (AtBRI1) (Brassinosteroid LRR receptor kinase)
Length = 1196
Score = 221 bits (564), Expect = 2e-57
Identities = 122/308 (39%), Positives = 184/308 (59%), Gaps = 5/308 (1%)
Query: 24 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 83
R T+ +L ATNGF +D +G GGFG VY DG +A+KKL ++ + + EF E+
Sbjct: 869 RKLTFADLLQATNGFHNDSLIGSGGFGDVYKAILKDGSAVAIKKLIHVSGQGDREFMAEM 928
Query: 84 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAI 143
E +G+++H+NL+ L GYC D+RL+VY++M SL LH V+LNW R IAI
Sbjct: 929 ETIGKIKHRNLVPLLGYCKVGDERLLVYEFMKYGSLEDVLHDPKKAGVKLNWSTRRKIAI 988
Query: 144 GSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMT-TRVKG 202
GSA G+ +LHH +PHIIHRD+K+SNVLLD + V+DFG A+L+ +H++ + + G
Sbjct: 989 GSARGLAFLHHNCSPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAG 1048
Query: 203 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKG 262
T GY+ PEY + S DVYS+G++LLEL+TG++P + P + W + K
Sbjct: 1049 TPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKRPTDS-PDFGDNNLVGWVKQ-HAKL 1106
Query: 263 RFRDMVDPKLRGNFD--ENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQGKVT 320
R D+ DP+L E ++ Q + VA C+ +RP M QV+++ K + G +
Sbjct: 1107 RISDVFDPELMKEDPALEIELLQHLKVAVACLDDRAWRRPTMVQVMAMFKEIQAGSGIDS 1166
Query: 321 KMRIDSVK 328
+ I S++
Sbjct: 1167 QSTIRSIE 1174
>BRI1_LYCPE (Q8L899) Systemin receptor SR160 precursor (EC 2.7.1.37)
(Brassinosteroid LRR receptor kinase)
Length = 1207
Score = 220 bits (560), Expect = 5e-57
Identities = 119/297 (40%), Positives = 180/297 (60%), Gaps = 5/297 (1%)
Query: 24 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 83
R T+ +L ATNGF +D +G GGFG VY + DG +A+KKL ++ + + EF E+
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEM 933
Query: 84 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAI 143
E +G+++H+NL+ L GYC ++RL+VY+YM SL LH + ++LNW R IAI
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKTGIKLNWPARRKIAI 993
Query: 144 GSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMT-TRVKG 202
G+A G+ +LHH PHIIHRD+K+SNVLLD + V+DFG A+L+ +H++ + + G
Sbjct: 994 GAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAG 1053
Query: 203 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKG 262
T GY+ PEY + S DVYS+G++LLEL+TG++P + G + W + L KG
Sbjct: 1054 TPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFG-DNNLVGWVK-LHAKG 1111
Query: 263 RFRDMVDPKL--RGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQG 317
+ D+ D +L E ++ Q + VA C+ KRP M QV+++ K + G
Sbjct: 1112 KITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSG 1168
>BRI1_LYCES (Q8GUQ5) Brassinosteroid LRR receptor kinase precursor (EC
2.7.1.37) (tBRI1) (Altered brassinolide sensitivity 1)
(Systemin receptor SR160)
Length = 1207
Score = 219 bits (558), Expect = 9e-57
Identities = 119/297 (40%), Positives = 180/297 (60%), Gaps = 5/297 (1%)
Query: 24 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEV 83
R T+ +L ATNGF +D +G GGFG VY + DG +A+KKL ++ + + EF E+
Sbjct: 874 RKLTFADLLEATNGFHNDSLVGSGGFGDVYKAQLKDGSVVAIKKLIHVSGQGDREFTAEM 933
Query: 84 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAI 143
E +G+++H+NL+ L GYC ++RL+VY+YM SL LH + ++LNW R IAI
Sbjct: 934 ETIGKIKHRNLVPLLGYCKVGEERLLVYEYMKYGSLEDVLHDRKKIGIKLNWPARRKIAI 993
Query: 144 GSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMT-TRVKG 202
G+A G+ +LHH PHIIHRD+K+SNVLLD + V+DFG A+L+ +H++ + + G
Sbjct: 994 GAARGLAFLHHNCIPHIIHRDMKSSNVLLDENLEARVSDFGMARLMSAMDTHLSVSTLAG 1053
Query: 203 TLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKG 262
T GY+ PEY + S DVYS+G++LLEL+TG++P + G + W + L KG
Sbjct: 1054 TPGYVPPEYYQSFRCSTKGDVYSYGVVLLELLTGKQPTDSADFG-DNNLVGWVK-LHAKG 1111
Query: 263 RFRDMVDPKL--RGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLKGQEPDQG 317
+ D+ D +L E ++ Q + VA C+ KRP M QV+++ K + G
Sbjct: 1112 KITDVFDRELLKEDASIEIELLQHLKVACACLDDRHWKRPTMIQVMAMFKEIQAGSG 1168
>APKA_ARATH (Q06548) Protein kinase APK1A, chloroplast precursor (EC
2.7.1.-)
Length = 410
Score = 218 bits (555), Expect = 2e-56
Identities = 120/297 (40%), Positives = 183/297 (61%), Gaps = 13/297 (4%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWG----------RTSDGLQIAVKKLKAMNSKA 75
F++ EL +AT F D LGEGGFG V+ G R GL IAVKKL +
Sbjct: 56 FSFAELKSATRNFRPDSVLGEGGFGCVFKGWIDEKSLTASRPGTGLVIAVKKLNQDGWQG 115
Query: 76 EMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNW 135
E+ EV LG+ H++L+ L GYC+ D+ RL+VY++MP SL +HL + L+W
Sbjct: 116 HQEWLAEVNYLGQFSHRHLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGLYFQPLSW 175
Query: 136 QKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEG-VS 194
+ R+ +A+G+A+G+ +LH T +I+RD K SN+LLDS++ ++DFG AK P G S
Sbjct: 176 KLRLKVALGAAKGLAFLHSSET-RVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPIGDKS 234
Query: 195 HMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEW 254
H++TRV GT GY APEY G ++ DVYSFG++LLEL++GR+ ++K +R + EW
Sbjct: 235 HVSTRVMGTHGYAAPEYLATGHLTTKSDVYSFGVVLLELLSGRRAVDKNRPSGERNLVEW 294
Query: 255 AEP-LITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
A+P L+ K + ++D +L+ + + + ++ C+ +E + RPNM +VVS L+
Sbjct: 295 AKPYLVNKRKIFRVIDNRLQDQYSMEEACKVATLSLRCLTTEIKLRPNMSEVVSHLE 351
>APKB_ARATH (P46573) Protein kinase APK1B, chloroplast precursor (EC
2.7.1.-)
Length = 412
Score = 215 bits (548), Expect = 1e-55
Identities = 120/298 (40%), Positives = 182/298 (60%), Gaps = 15/298 (5%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWG----------RTSDGLQIAVKKLKAMNSKA 75
FT+ EL AT F D LGEGGFGSV+ G + G+ IAVKKL +
Sbjct: 57 FTFAELKAATRNFRPDSVLGEGGFGSVFKGWIDEQTLTASKPGTGVVIAVKKLNQDGWQG 116
Query: 76 EMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNW 135
E+ EV LG+ H NL+ L GYC+ D+ RL+VY++MP SL +HL + + L+W
Sbjct: 117 HQEWLAEVNYLGQFSHPNLVKLIGYCLEDEHRLLVYEFMPRGSLENHLFRRGSYFQPLSW 176
Query: 136 QKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEG-VS 194
R+ +A+G+A+G+ +LH+ T +I+RD K SN+LLDS++ ++DFG AK P G S
Sbjct: 177 TLRLKVALGAAKGLAFLHNAET-SVIYRDFKTSNILLDSEYNAKLSDFGLAKDGPTGDKS 235
Query: 195 HMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEW 254
H++TR+ GT GY APEY G ++ DVYS+G++LLE+++GR+ ++K ++ + EW
Sbjct: 236 HVSTRIMGTYGYAAPEYLATGHLTTKSDVYSYGVVLLEVLSGRRAVDKNRPPGEQKLVEW 295
Query: 255 AEPLITKGR--FRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
A PL+ R FR ++D +L+ + + + +A C+ E + RPNM +VVS L+
Sbjct: 296 ARPLLANKRKLFR-VIDNRLQDQYSMEEACKVATLALRCLTFEIKLRPNMNEVVSHLE 352
>PSKR_DAUCA (Q8LPB4) Phytosulfokine receptor precursor (EC 2.7.1.37)
(Phytosulfokine LRR receptor kinase)
Length = 1021
Score = 215 bits (547), Expect = 2e-55
Identities = 111/277 (40%), Positives = 164/277 (59%)
Query: 34 ATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRVRHKN 93
+T+ F+ +G GGFG VY DG ++A+K+L + + EF EVE L R +H N
Sbjct: 739 STSSFNQANIIGCGGFGLVYKATLPDGTKVAIKRLSGDTGQMDREFQAEVETLSRAQHPN 798
Query: 94 LLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSAEGILYLH 153
L+ L GYC + +L++Y YM N SL LH + G L+W+ R+ IA G+AEG+ YLH
Sbjct: 799 LVHLLGYCNYKNDKLLIYSYMDNGSLDYWLHEKVDGPPSLDWKTRLRIARGAAEGLAYLH 858
Query: 154 HEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVKGTLGYLAPEYAM 213
PHI+HRDIK+SN+LL FV +ADFG A+LI +H+TT + GTLGY+ PEY
Sbjct: 859 QSCEPHILHRDIKSSNILLSDTFVAHLADFGLARLILPYDTHVTTDLVGTLGYIPPEYGQ 918
Query: 214 WGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKGRFRDMVDPKLR 273
+ DVYSFG++LLEL+TGR+P++ R + W + T+ R ++ DP +
Sbjct: 919 ASVATYKGDVYSFGVVLLELLTGRRPMDVCKPRGSRDLISWVLQMKTEKRESEIFDPFIY 978
Query: 274 GNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
++ + +A C+ P+ RP +Q+VS L+
Sbjct: 979 DKDHAEEMLLVLEIACRCLGENPKTRPTTQQLVSWLE 1015
>CRI4_MAIZE (O24585) Putative receptor protein kinase CRINKLY4
precursor (EC 2.7.1.-)
Length = 901
Score = 212 bits (540), Expect = 1e-54
Identities = 122/290 (42%), Positives = 181/290 (62%), Gaps = 8/290 (2%)
Query: 26 FTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKK-LKAMN-SKAEMEFAVEV 83
F+Y+EL AT GFS+D ++G+G F V+ G DG +AVK+ +KA + K+ EF E+
Sbjct: 493 FSYEELEQATGGFSEDSQVGKGSFSCVFKGILRDGTVVAVKRAIKASDVKKSSKEFHNEL 552
Query: 84 EVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAG-EVQLNWQKRMSIA 142
++L R+ H +LL L GYC +RL+VY++M + SL HLHG+ + +LNW +R++IA
Sbjct: 553 DLLSRLNHAHLLNLLGYCEDGSERLLVYEFMAHGSLYQHLHGKDPNLKKRLNWARRVTIA 612
Query: 143 IGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EGVSHMTTRVK 201
+ +A GI YLH P +IHRDIK+SN+L+D D VADFG + L P + + ++
Sbjct: 613 VQAARGIEYLHGYACPPVIHRDIKSSNILIDEDHNARVADFGLSILGPADSGTPLSELPA 672
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE-KLPGGLKRTITEWAEPLIT 260
GTLGYL PEY ++ DVYSFG++LLE+++GRK I+ + G I EWA PLI
Sbjct: 673 GTLGYLDPEYYRLHYLTTKSDVYSFGVVLLEILSGRKAIDMQFEEG---NIVEWAVPLIK 729
Query: 261 KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
G ++DP L D +K+ +VA CV+ + RP+M +V + L+
Sbjct: 730 AGDIFAILDPVLSPPSDLEALKKIASVACKCVRMRGKDRPSMDKVTTALE 779
>NAK_ARATH (P43293) Probable serine/threonine-protein kinase NAK (EC
2.7.1.37)
Length = 389
Score = 209 bits (532), Expect = 9e-54
Identities = 114/303 (37%), Positives = 181/303 (59%), Gaps = 13/303 (4%)
Query: 20 NNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWG----------RTSDGLQIAVKKLK 69
N + + F+ EL +AT F D +GEGGFG V+ G + G+ IAVK+L
Sbjct: 50 NANLKNFSLSELKSATRNFRPDSVVGEGGFGCVFKGWIDESSLAPSKPGTGIVIAVKRLN 109
Query: 70 AMNSKAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAG 129
+ E+ E+ LG++ H NL+ L GYC+ ++ RL+VY++M SL +HL +
Sbjct: 110 QEGFQGHREWLAEINYLGQLDHPNLVKLIGYCLEEEHRLLVYEFMTRGSLENHLFRRGTF 169
Query: 130 EVQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLI 189
L+W R+ +A+G+A G+ +LH+ P +I+RD KASN+LLDS++ ++DFG A+
Sbjct: 170 YQPLSWNTRVRMALGAARGLAFLHN-AQPQVIYRDFKASNILLDSNYNAKLSDFGLARDG 228
Query: 190 PEG-VSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLK 248
P G SH++TRV GT GY APEY G +S DVYSFG++LLEL++GR+ I+K +
Sbjct: 229 PMGDNSHVSTRVMGTQGYAAPEYLATGHLSVKSDVYSFGVVLLELLSGRRAIDKNQPVGE 288
Query: 249 RTITEWAEPLIT-KGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVS 307
+ +WA P +T K R ++DP+L+G + + + +A C+ + + RP M ++V
Sbjct: 289 HNLVDWARPYLTNKRRLLRVMDPRLQGQYSLTRALKIAVLALDCISIDAKSRPTMNEIVK 348
Query: 308 LLK 310
++
Sbjct: 349 TME 351
>TMK1_ARATH (P43298) Putative receptor protein kinase TMK1 precursor
(EC 2.7.1.-)
Length = 942
Score = 209 bits (531), Expect = 1e-53
Identities = 116/293 (39%), Positives = 172/293 (58%), Gaps = 10/293 (3%)
Query: 25 IFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLK--AMNSKAEMEFAVE 82
+ + + L + TN FS D LG GGFG VY G DG +IAVK+++ + K EF E
Sbjct: 575 LISIQVLRSVTNNFSSDNILGSGGFGVVYKGELHDGTKIAVKRMENGVIAGKGFAEFKSE 634
Query: 83 VEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHL-HGQYAGEVQLNWQKRMSI 141
+ VL +VRH++L+ L GYC+ +++L+VY+YMP +L HL G L W++R+++
Sbjct: 635 IAVLTKVRHRHLVTLLGYCLDGNEKLLVYEYMPQGTLSRHLFEWSEEGLKPLLWKQRLTL 694
Query: 142 AIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTTRVK 201
A+ A G+ YLH IHRD+K SN+LL D VADFG +L PEG + TR+
Sbjct: 695 ALDVARGVEYLHGLAHQSFIHRDLKPSNILLGDDMRAKVADFGLVRLAPEGKGSIETRIA 754
Query: 202 GTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLI-- 259
GT GYLAPEYA+ G+V+ DVYSFG++L+EL+TGRK +++ + W + +
Sbjct: 755 GTFGYLAPEYAVTGRVTTKVDVYSFGVILMELITGRKSLDESQPEESIHLVSWFKRMYIN 814
Query: 260 TKGRFRDMVDPKLRGNFDENQVKQTVNVAAL---CVQSEPEKRPNMKQVVSLL 309
+ F+ +D + + DE + VA L C EP +RP+M V++L
Sbjct: 815 KEASFKKAIDTTI--DLDEETLASVHTVAELAGHCCAREPYQRPDMGHAVNIL 865
>CLV1_ARATH (Q9SYQ8) Receptor protein kinase CLAVATA1 precursor (EC
2.7.1.-)
Length = 980
Score = 207 bits (527), Expect = 3e-53
Identities = 112/303 (36%), Positives = 181/303 (58%), Gaps = 22/303 (7%)
Query: 22 SWRIFTYKELHTATNGFSDDYK----LGEGGFGSVYWGRTSDGLQIAVKKLKAMNS-KAE 76
+W++ +++L + + K +G+GG G VY G + + +A+K+L + +++
Sbjct: 672 AWKLTAFQKLDFKSEDVLECLKEENIIGKGGAGIVYRGSMPNNVDVAIKRLVGRGTGRSD 731
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
F E++ LGR+RH++++ L GY D L++Y+YMPN SL LHG G +Q W+
Sbjct: 732 HGFTAEIQTLGRIRHRHIVRLLGYVANKDTNLLLYEYMPNGSLGELLHGSKGGHLQ--WE 789
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEG-VSH 195
R +A+ +A+G+ YLHH+ +P I+HRD+K++N+LLDSDF VADFG AK + +G S
Sbjct: 790 TRHRVAVEAAKGLCYLHHDCSPLILHRDVKSNNILLDSDFEAHVADFGLAKFLVDGAASE 849
Query: 196 MTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGL-------- 247
+ + G+ GY+APEYA KV E DVYSFG++LLEL+ G+KP+ + G+
Sbjct: 850 CMSSIAGSYGYIAPEYAYTLKVDEKSDVYSFGVVLLELIAGKKPVGEFGEGVDIVRWVRN 909
Query: 248 -KRTITEWAEPLITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVV 306
+ IT+ ++ I +VDP+L G + V +A +CV+ E RP M++VV
Sbjct: 910 TEEEITQPSDAAIVVA----IVDPRLTG-YPLTSVIHVFKIAMMCVEEEAAARPTMREVV 964
Query: 307 SLL 309
+L
Sbjct: 965 HML 967
>RLK5_ARATH (P47735) Receptor-like protein kinase 5 precursor (EC
2.7.1.37)
Length = 999
Score = 206 bits (524), Expect = 8e-53
Identities = 127/331 (38%), Positives = 186/331 (55%), Gaps = 34/331 (10%)
Query: 5 VLIVHVCRPT----SYGVANNSWRIFTYKELHTATNGFSD--DYK--LGEGGFGSVYWGR 56
V+ + CR S +A + WR F +LH + + +D D K +G G G VY
Sbjct: 644 VMFIAKCRKLRALKSSTLAASKWRSF--HKLHFSEHEIADCLDEKNVIGFGSSGKVYKVE 701
Query: 57 TSDGLQIAVKKLKAMNSKAEME----------FAVEVEVLGRVRHKNLLGLRGYCVGDDQ 106
G +AVKKL + E FA EVE LG +RHK+++ L C D
Sbjct: 702 LRGGEVVAVKKLNKSVKGGDDEYSSDSLNRDVFAAEVETLGTIRHKSIVRLWCCCSSGDC 761
Query: 107 RLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIK 166
+L+VY+YMPN SL LHG G V L W +R+ IA+ +AEG+ YLHH+ P I+HRD+K
Sbjct: 762 KLLVYEYMPNGSLADVLHGDRKGGVVLGWPERLRIALDAAEGLSYLHHDCVPPIVHRDVK 821
Query: 167 ASNVLLDSDFVPLVADFGFAKL-------IPEGVSHMTTRVKGTLGYLAPEYAMWGKVSE 219
+SN+LLDSD+ VADFG AK+ PE +S + G+ GY+APEY +V+E
Sbjct: 822 SSNILLDSDYGAKVADFGIAKVGQMSGSKTPEAMS----GIAGSCGYIAPEYVYTLRVNE 877
Query: 220 SCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKGRFRDMVDPKLRGNFDEN 279
D+YSFG++LLELVTG++P + G + + +W + K ++DPKL F E
Sbjct: 878 KSDIYSFGVVLLELVTGKQPTDSELG--DKDMAKWVCTALDKCGLEPVIDPKLDLKFKE- 934
Query: 280 QVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
++ + +++ LC P RP+M++VV +L+
Sbjct: 935 EISKVIHIGLLCTSPLPLNRPSMRKVVIMLQ 965
>SRK6_BRAOL (Q09092) Putative serine/threonine-protein kinase
receptor precursor (EC 2.7.1.37) (S-receptor kinase)
(SRK)
Length = 849
Score = 206 bits (523), Expect = 1e-52
Identities = 111/281 (39%), Positives = 168/281 (59%), Gaps = 9/281 (3%)
Query: 34 ATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAEMEFAVEVEVLGRVRHKN 93
AT FS KLG+GGFG VY GR DG +IAVK+L + + EF EV ++ R++H N
Sbjct: 524 ATENFSSCNKLGQGGFGIVYKGRLLDGKEIAVKRLSKTSVQGTDEFMNEVTLIARLQHIN 583
Query: 94 LLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSAEGILYLH 153
L+ + G C+ D+++++Y+Y+ NLSL S+L G+ +LNW +R I G A G+LYLH
Sbjct: 584 LVQVLGCCIEGDEKMLIYEYLENLSLDSYLFGK-TRRSKLNWNERFDITNGVARGLLYLH 642
Query: 154 HEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHMTT-RVKGTLGYLAPEYA 212
+ IIHRD+K SN+LLD + +P ++DFG A++ + T +V GT GY++PEYA
Sbjct: 643 QDSRFRIIHRDLKVSNILLDKNMIPKISDFGMARIFERDETEANTMKVVGTYGYMSPEYA 702
Query: 213 MWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITKGRFRDMVDPKL 272
M+G SE DV+SFG+++LE+V+G+K + + + +GR ++VDP +
Sbjct: 703 MYGIFSEKSDVFSFGVIVLEIVSGKKNRGFYNLDYENDLLSYVWSRWKEGRALEIVDPVI 762
Query: 273 RGN-------FDENQVKQTVNVAALCVQSEPEKRPNMKQVV 306
+ F +V + + + LCVQ E RP M VV
Sbjct: 763 VDSLSSQPSIFQPQEVLKCIQIGLLCVQELAEHRPAMSSVV 803
>CX32_ARATH (P27450) Probable serine/threonine-protein kinase Cx32,
chloroplast precursor (EC 2.7.1.37)
Length = 419
Score = 203 bits (517), Expect = 5e-52
Identities = 112/300 (37%), Positives = 176/300 (58%), Gaps = 18/300 (6%)
Query: 24 RIFTYKELHTATNGFSDDYKLGEGGFGSVYWG----------RTSDGLQIAVKKLKAMNS 73
+++ + +L TAT F D LG+GGFG VY G R G+ +A+K+L + +
Sbjct: 72 KVYNFLDLKTATKNFKPDSMLGQGGFGKVYRGWVDATTLAPSRVGSGMIVAIKRLNSESV 131
Query: 74 KAEMEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQL 133
+ E+ EV LG + H+NL+ L GYC D + L+VY++MP SL SHL +
Sbjct: 132 QGFAEWRSEVNFLGMLSHRNLVKLLGYCREDKELLLVYEFMPKGSLESHL---FRRNDPF 188
Query: 134 NWQKRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EG 192
W R+ I IG+A G+ +L H + +I+RD KASN+LLDS++ ++DFG AKL P +
Sbjct: 189 PWDLRIKIVIGAARGLAFL-HSLQREVIYRDFKASNILLDSNYDAKLSDFGLAKLGPADE 247
Query: 193 VSHMTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIE-KLPGGLKRTI 251
SH+TTR+ GT GY APEY G + DV++FG++LLE++TG K P G + ++
Sbjct: 248 KSHVTTRIMGTYGYAAPEYMATGHLYVKSDVFAFGVVLLEIMTGLTAHNTKRPRG-QESL 306
Query: 252 TEWAEP-LITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
+W P L K R + ++D ++G + + + C++ +P+ RP+MK+VV +L+
Sbjct: 307 VDWLRPELSNKHRVKQIMDKGIKGQYTTKVATEMARITLSCIEPDPKNRPHMKEVVEVLE 366
>SIRK_ARATH (O64483) Senescence-induced receptor-like
serine/threonine kinase precursor (FLG22-induced
receptor-like kinase 1)
Length = 876
Score = 186 bits (473), Expect = 6e-47
Identities = 107/295 (36%), Positives = 169/295 (57%), Gaps = 7/295 (2%)
Query: 17 GVANNSWRIFTYKELHTATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSKAE 76
G + R F Y E+ TN F + +G+GGFG VY G +G Q+AVK L +++
Sbjct: 555 GPLKTAKRYFKYSEVVNITNNF--ERVIGKGGFGKVYHG-VINGEQVAVKVLSEESAQGY 611
Query: 77 MEFAVEVEVLGRVRHKNLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQ 136
EF EV++L RV H NL L GYC + +++Y+YM N +L +L G+ + L+W+
Sbjct: 612 KEFRAEVDLLMRVHHTNLTSLVGYCNEINHMVLIYEYMANENLGDYLAGKRS--FILSWE 669
Query: 137 KRMSIAIGSAEGILYLHHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIP-EGVSH 195
+R+ I++ +A+G+ YLH+ P I+HRD+K +N+LL+ +ADFG ++ EG
Sbjct: 670 ERLKISLDAAQGLEYLHNGCKPPIVHRDVKPTNILLNEKLQAKMADFGLSRSFSVEGSGQ 729
Query: 196 MTTRVKGTLGYLAPEYAMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWA 255
++T V G++GYL PEY +++E DVYS G++LLE++TG+ I K I++
Sbjct: 730 ISTVVAGSIGYLDPEYYSTRQMNEKSDVYSLGVVLLEVITGQPAIASSKTE-KVHISDHV 788
Query: 256 EPLITKGRFRDMVDPKLRGNFDENQVKQTVNVAALCVQSEPEKRPNMKQVVSLLK 310
++ G R +VD +LR +D + +A C + +RP M QVV LK
Sbjct: 789 RSILANGDIRGIVDQRLRERYDVGSAWKMSEIALACTEHTSAQRPTMSQVVMELK 843
>RPK1_IPONI (P93194) Receptor-like protein kinase precursor (EC
2.7.1.37)
Length = 1109
Score = 184 bits (466), Expect = 4e-46
Identities = 107/283 (37%), Positives = 162/283 (56%), Gaps = 9/283 (3%)
Query: 34 ATNGFSDDYKLGEGGFGSVYWGRTSDGLQIAVKKLKAMNSK-AEMEFAVEVEVLGRVRHK 92
AT +D Y +G+G G++Y S AVKKL K + E+E +G+VRH+
Sbjct: 812 ATENLNDKYVIGKGAHGTIYKATLSPDKVYAVKKLVFTGIKNGSVSMVREIETIGKVRHR 871
Query: 93 NLLGLRGYCVGDDQRLIVYDYMPNLSLLSHLHGQYAGEVQLNWQKRMSIAIGSAEGILYL 152
NL+ L + + + LI+Y YM N SL LH + L+W R +IA+G+A G+ YL
Sbjct: 872 NLIKLEEFWLRKEYGLILYTYMENGSLHDILHETNPPK-PLDWSTRHNIAVGTAHGLAYL 930
Query: 153 HHEVTPHIIHRDIKASNVLLDSDFVPLVADFGFAKLIPEGVSHM-TTRVKGTLGYLAPEY 211
H + P I+HRDIK N+LLDSD P ++DFG AKL+ + + + + V+GT+GY+APE
Sbjct: 931 HFDCDPAIVHRDIKPMNILLDSDLEPHISDFGIAKLLDQSATSIPSNTVQGTIGYMAPEN 990
Query: 212 AMWGKVSESCDVYSFGILLLELVTGRKPIEKLPGGLKRTITEWAEPLITK-GRFRDMVDP 270
A S DVYS+G++LLEL+T +K ++ G + I W + T+ G + +VDP
Sbjct: 991 AFTTVKSRESDVYSYGVVLLELITRKKALDPSFNG-ETDIVGWVRSVWTQTGEIQKIVDP 1049
Query: 271 KLRGNFDEN----QVKQTVNVAALCVQSEPEKRPNMKQVVSLL 309
L ++ QV + +++A C + E +KRP M+ VV L
Sbjct: 1050 SLLDELIDSSVMEQVTEALSLALRCAEKEVDKRPTMRDVVKQL 1092
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.319 0.138 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 45,479,268
Number of Sequences: 164201
Number of extensions: 1971538
Number of successful extensions: 8736
Number of sequences better than 10.0: 1631
Number of HSP's better than 10.0 without gapping: 1051
Number of HSP's successfully gapped in prelim test: 582
Number of HSP's that attempted gapping in prelim test: 5400
Number of HSP's gapped (non-prelim): 1859
length of query: 369
length of database: 59,974,054
effective HSP length: 112
effective length of query: 257
effective length of database: 41,583,542
effective search space: 10686970294
effective search space used: 10686970294
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC136954.12


