

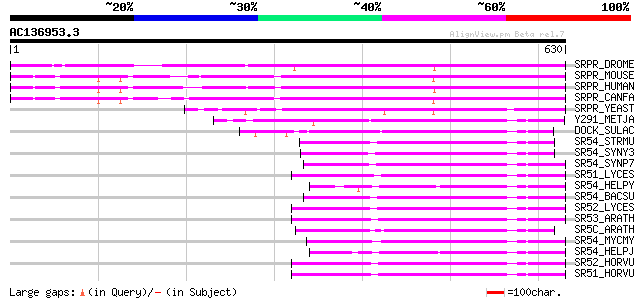
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136953.3 + phase: 0 /pseudo
(630 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
SRPR_DROME (Q9U5L1) Signal recognition particle receptor alpha s... 446 e-125
SRPR_MOUSE (Q9DBG7) Signal recognition particle receptor alpha s... 432 e-120
SRPR_HUMAN (P08240) Signal recognition particle receptor alpha s... 430 e-120
SRPR_CANFA (P06625) Signal recognition particle receptor alpha s... 429 e-120
SRPR_YEAST (P32916) Signal recognition particle receptor alpha s... 243 1e-63
Y291_METJA (Q57739) Probable signal recognition particle protein... 227 6e-59
DOCK_SULAC (P27414) Probable signal recognition particle protein... 188 4e-47
SR54_STRMU (Q54431) Signal recognition particle protein (Fifty-f... 144 7e-34
SR54_SYNY3 (P74214) Signal recognition particle protein (Fifty-f... 139 3e-32
SR54_SYNP7 (Q55311) Signal sequence binding protein 137 9e-32
SR51_LYCES (P49971) Signal recognition particle 54 kDa protein 1... 135 4e-31
SR54_HELPY (P56005) Signal recognition particle protein (Fifty-f... 134 6e-31
SR54_BACSU (P37105) Signal recognition particle protein (Fifty-f... 133 1e-30
SR52_LYCES (P49972) Signal recognition particle 54 kDa protein 2... 133 1e-30
SR53_ARATH (P49967) Signal recognition particle 54 kDa protein 3... 133 2e-30
SR5C_ARATH (P37107) Signal recognition particle 54 kDa protein, ... 132 2e-30
SR54_MYCMY (Q01442) Signal recognition particle protein (Fifty-f... 132 2e-30
SR54_HELPJ (Q9ZK62) Signal recognition particle protein (Fifty-f... 132 3e-30
SR52_HORVU (P49969) Signal recognition particle 54 kDa protein 2... 132 3e-30
SR51_HORVU (P49968) Signal recognition particle 54 kDa protein 1... 132 3e-30
>SRPR_DROME (Q9U5L1) Signal recognition particle receptor alpha
subunit homolog (SR-alpha) (Docking protein alpha)
(DP-alpha) (GTP-binding protein)
Length = 614
Score = 446 bits (1147), Expect = e-125
Identities = 269/651 (41%), Positives = 384/651 (58%), Gaps = 59/651 (9%)
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
ML+ ++IFT+GG++LW N GN+ S I++LIR +LEER+ + Y Y+ A K
Sbjct: 1 MLDFVVIFTKGGVVLWHSNASGNSF-ASCINSLIRGVILEERNTEAKY-YEEDHLAVQFK 58
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVYDPTRTVYR-DFDEIFKQLKIE 119
N+L LV+ A++Q+++ L Y+D LA ++ F + Y R DFD ++++
Sbjct: 59 --LDNELDLVYAAIFQKVIKLNYLDGFLADMQAAFKEKYGDIRLGDDYDFDREYRRVLSA 116
Query: 120 AEARAEDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGGGLKNDGDGKNGKKNSENDR 179
AE + K+ + N Q K+ +
Sbjct: 117 AEEASAKQVKAPKTMRSYNESQ-------------------------------KSKKTVA 145
Query: 180 SAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFD--VNRLQKKVRNKGGNGKKTDAV 237
S I ++ R N + + D + ++K+R K KKT
Sbjct: 146 SMIQDDKKPVEKRVNIQEAPPPSKSQPSSPPTGSPMDKIIMEKRRKLREKLTPTKKTSPS 205
Query: 238 VTKA-EPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESMMDKDE 296
+K+ +P+K KK RVWD D D+ D + + E +M
Sbjct: 206 DSKSSKPEKAGKKPRVWDLGGNSKDAALLDR---SRDSPDDVQYQNINSELVGTMQGVIR 262
Query: 297 IFSSDSEDEEDDDDDNAGKKSKPDAK----KKGWFSSMFQSIAGKANLEKSDLEPALKAL 352
+SEDE D++D ++ +++ + K+G S F+ I G + +DL+PAL+ +
Sbjct: 263 DLDVESEDEADNEDASSEGEAEEQVQSKKGKRGGLLSYFKGIVGAKTMSLADLQPALEKM 322
Query: 353 KDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDI 412
+D L++KNVA EIA KLC+SVAASL+GK++ +F I+S V+ A+ ++LVRIL+P+R IDI
Sbjct: 323 RDHLISKNVASEIAAKLCDSVAASLDGKQMGTFDSIASQVKEALTESLVRILSPKRRIDI 382
Query: 413 LRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQL 472
+RD +K +PY ++F GVNGVGKSTNLAK+ +WL +++ NV++AACDTFR+GAVEQL
Sbjct: 383 IRDALESKRNGRPYTIIFCGVNGVGKSTNLAKICFWLIENDFNVLIAACDTFRAGAVEQL 442
Query: 473 RTHARRLQ-------------IPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRM 519
RTH R L + ++EKGY KD A +A EAI+ A DVVLVDTAGRM
Sbjct: 443 RTHTRHLNALHPAAKHDGRNMVQLYEKGYGKDAAGIAMEAIKFAHDTRVDVVLVDTAGRM 502
Query: 520 QDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGI 579
QDNEPLMR+LSKLI +NNPDLVLFVGEALVGN+AVDQL KFNQ LAD S++ P +IDGI
Sbjct: 503 QDNEPLMRSLSKLIKVNNPDLVLFVGEALVGNEAVDQLVKFNQSLADYSSNENPHIIDGI 562
Query: 580 LLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
+LTKFDTIDDKVGAA+SM YI+G P++FVG GQ+Y DLK +NV ++V +L+
Sbjct: 563 VLTKFDTIDDKVGAAISMTYITGQPIVFVGTGQTYADLKAINVNAVVNSLM 613
>SRPR_MOUSE (Q9DBG7) Signal recognition particle receptor alpha
subunit (SR-alpha) (Docking protein alpha) (DP-alpha)
Length = 636
Score = 432 bits (1112), Expect = e-120
Identities = 276/672 (41%), Positives = 390/672 (57%), Gaps = 80/672 (11%)
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
ML+ IF++GGL+LW + ++ G P++ LIRS LL+ER G +S+ ++A +LK
Sbjct: 1 MLDFFTIFSKGGLVLWCFQGVSDSCTG-PVNALIRSVLLQERGGNNSFTHEA----LTLK 55
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVY-------------DPTRTVYR 107
+ N LVFV +Q+IL L YVD L+ V R F Y + T
Sbjct: 56 YKLDNQFELVFVVGFQKILTLTYVDKLIDDVHRLFRDKYRTEIQQQSALSLLNGTFDFQN 115
Query: 108 DFDEIFKQLKIEAEARA-------EDLKKSNPVIVGGNRKQQVTWKGDGSDGK-KNGSAG 159
DF + ++ + ++ RA ED +K+ + + + +G+ + K KN
Sbjct: 116 DFLRLLREAEESSKIRAPTTMKKFEDSEKAKKPV-----RSMIETRGEKTKEKAKNNKKR 170
Query: 160 GGLKNDGDGKNG-KKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVN 218
G K DG K + ++S + G EN ++ +
Sbjct: 171 GAKKEGSDGTLATSKTAPAEKSGL-------------------SAGPENGELSKEEL-IR 210
Query: 219 RLQKKVRNKGGNGKKTDAVVTKAE-PKKVVKK-NRVWDEKPVETK--LDFTDHVDIDGDA 274
R +++ K G G + TK++ PK+ KK RVW+ K LD++ +G
Sbjct: 211 RKREEFIQKHGKGLDKSSKSTKSDTPKEKGKKAPRVWELGGCANKEVLDYSTPT-TNGTP 269
Query: 275 DKDRKVDY-LAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKK--KGWFSSMF 331
+ D L + G +D SS DD+ A + +KP A K G M
Sbjct: 270 EAALSEDINLIRGTGPGGQLQDLDCSS-------SDDEGATQNTKPSATKGTLGGMFGML 322
Query: 332 QSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISST 391
+ + G +L + D+E L ++D L+ KNVA +IA +LCESVA LEGK + +F+ ++ST
Sbjct: 323 KGLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKLEGKVMGTFSTVTST 382
Query: 392 VQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQ 451
V+ A++++LV+IL P+R +D+LRD+ A+ +++PYVV F GVNGVGKSTNLAK+++WL +
Sbjct: 383 VKQALQESLVQILQPQRRVDMLRDIMDAQRRQRPYVVTFCGVNGVGKSTNLAKISFWLLE 442
Query: 452 HNVNVMMAACDTFRSGAVEQLRTHARRL-------------QIPIFEKGYEKDPAVVAKE 498
+ +V++AACDTFR+GAVEQLRTH RRL + +FEKGY KD A +A E
Sbjct: 443 NGFSVLIAACDTFRAGAVEQLRTHTRRLTALHPPEKHGGRTMVQLFEKGYGKDAAGIAME 502
Query: 499 AIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLS 558
AI A G DVVLVDTAGRMQDN PLM AL+KLI +N PDLVLFVGEALVGN+AVDQL
Sbjct: 503 AIAFARNQGFDVVLVDTAGRMQDNAPLMTALAKLITVNTPDLVLFVGEALVGNEAVDQLV 562
Query: 559 KFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 618
KFN+ LAD S + TPRLIDGI+LTKFDTIDDKVGAA+SM YI+ P++FVG GQ+Y DL+
Sbjct: 563 KFNRALADHSMAQTPRLIDGIVLTKFDTIDDKVGAAISMTYITSKPIVFVGTGQTYCDLR 622
Query: 619 KLNVKSIVKTLL 630
LN K++V L+
Sbjct: 623 SLNAKAVVAALM 634
>SRPR_HUMAN (P08240) Signal recognition particle receptor alpha
subunit (SR-alpha) (Docking protein alpha) (DP-alpha)
Length = 638
Score = 430 bits (1105), Expect = e-120
Identities = 277/673 (41%), Positives = 388/673 (57%), Gaps = 80/673 (11%)
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
ML+ IF++GGL+LW + ++ G P++ LIRS LL+ER G +S+ ++A +LK
Sbjct: 1 MLDFFTIFSKGGLVLWCFQGVSDSCTG-PVNALIRSVLLQERGGNNSFTHEA----LTLK 55
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVY-------------DPTRTVYR 107
+ N LVFV +Q+IL L YVD L+ V R F Y + T
Sbjct: 56 YKLDNQFELVFVVGFQKILTLTYVDKLIDDVHRLFRDKYRTEIQQQSALSLLNGTFDFQN 115
Query: 108 DFDEIFKQLKIEAEARA-------EDLKKSNPVIVGGNRKQQVTWKGDGSDGKKNGSAGG 160
DF + ++ + ++ RA ED +K+ + + + +G+ K S
Sbjct: 116 DFLRLLREAEESSKIRAPTTMKKFEDSEKAKKPV-----RSMIETRGEKPKEKAKNSKKK 170
Query: 161 GLKNDG-DGKNG-KKNSENDRSAI-VNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDV 217
G K +G DG K ++S + V NG L ++
Sbjct: 171 GAKKEGSDGPLATSKPVPAEKSGLPVGPENGVELSKEELI-------------------- 210
Query: 218 NRLQKKVRNKGGNGKKTDAVVTKAE-PKKVVKKN-RVWDEKPVETK--LDFTDHVDIDGD 273
R +++ K G G + TK++ PK+ KK RVW+ K LD++ +G
Sbjct: 211 RRKREEFIQKHGRGMEKSNKSTKSDAPKEKGKKAPRVWELGGCANKEVLDYSTPTT-NGT 269
Query: 274 ADKDRKVDY-LAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKKK--GWFSSM 330
+ D L + G +D SS DD+ A +KP A K G M
Sbjct: 270 PEAALSEDINLIRGTGSGGQLQDLDCSSS------DDEGAAQNSTKPSATKGTLGGMFGM 323
Query: 331 FQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISS 390
+ + G +L + D+E L ++D L+ KNVA +IA +LCESVA LEGK + +F+ ++S
Sbjct: 324 LKGLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKLEGKVMGTFSTVTS 383
Query: 391 TVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQ 450
TV+ A++++LV+IL P+R +D+LRD+ A+ +++PYVV F GVNGVGKSTNLAK+++WL
Sbjct: 384 TVKQALQESLVQILQPQRRVDMLRDIMDAQRRQRPYVVTFCGVNGVGKSTNLAKISFWLL 443
Query: 451 QHNVNVMMAACDTFRSGAVEQLRTHARRLQ-------------IPIFEKGYEKDPAVVAK 497
++ +V++AACDTFR+GAVEQLRTH RRL + +FEKGY KD A +A
Sbjct: 444 ENGFSVLIAACDTFRAGAVEQLRTHTRRLSALHPPEKHGGRTMVQLFEKGYGKDAAGIAM 503
Query: 498 EAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQL 557
EAI A G DVVLVDTAGRMQDN PLM AL+KLI +N PDLVLFVGEALVGN+AVDQL
Sbjct: 504 EAIAFARNQGFDVVLVDTAGRMQDNAPLMTALAKLITVNTPDLVLFVGEALVGNEAVDQL 563
Query: 558 SKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDL 617
KFN+ LAD S + TPRLIDGI+LTKFDTIDDKVGAA+SM YI+ P++FVG GQ+Y DL
Sbjct: 564 VKFNRALADHSMAQTPRLIDGIVLTKFDTIDDKVGAAISMTYITSKPIVFVGTGQTYCDL 623
Query: 618 KKLNVKSIVKTLL 630
+ LN K++V L+
Sbjct: 624 RSLNAKAVVAALM 636
>SRPR_CANFA (P06625) Signal recognition particle receptor alpha
subunit (SR-alpha) (Docking protein alpha) (DP-alpha)
Length = 638
Score = 429 bits (1104), Expect = e-120
Identities = 276/671 (41%), Positives = 385/671 (57%), Gaps = 76/671 (11%)
Query: 1 MLEQLLIFTRGGLILWSCNEIGNALKGSPIDTLIRSCLLEERSGASSYNYDAPGAAYSLK 60
ML+ IF++GGL+LW + ++ G P++ LIRS LL+ER G +S+ ++A +LK
Sbjct: 1 MLDFFTIFSKGGLVLWCFQGVSDSCTG-PVNALIRSVLLQERGGNNSFTHEA----LTLK 55
Query: 61 WTFHNDLGLVFVAVYQRILHLLYVDDLLAAVKREFSQVY-------------DPTRTVYR 107
+ N LVFV +Q+IL L YVD L+ V R F Y + T
Sbjct: 56 YKLDNQFELVFVVGFQKILTLTYVDKLIDDVHRLFRDKYRTEIQQQSALSLLNGTFDFQN 115
Query: 108 DFDEIFKQLKIEAEARA-------EDLKKSNPVIVGGNRKQQVTWKGDGSDGK-KNGSAG 159
DF + ++ + ++ RA ED +K+ + + + +G+ K KN
Sbjct: 116 DFLRLLREREESSKIRAPTTMKKFEDSEKAKKPV-----RSMIETRGEKPKEKAKNSKKK 170
Query: 160 GGLKNDGDGKNGKKNSENDRSAIVNNGNGYNLRSNGVVGNVSVNGKENDSVNNGAFDVNR 219
G K DG + G +G+ G EN + + R
Sbjct: 171 GAKKESSDGP-------------LATGKAVPAEKSGLPA-----GPENGVELSKEELIRR 212
Query: 220 LQKKVRNKGGNGKKTDAVVTKAE-PKKVVKK-NRVWDEKPVETK--LDFTDHVDIDGDAD 275
+++ K G G + + TK++ PK+ KK RVW K LD++ +G D
Sbjct: 213 KREEFIQKHGRGLEKSSKSTKSDAPKEKGKKAPRVWALGGCANKEVLDYSAPT-TNGAPD 271
Query: 276 KDRKVDY-LAKEQGESMMDKDEIFSSDSEDEEDDDDDNAGKKSKPDAKK--KGWFSSMFQ 332
D L + G +D SS DD++ A SKP A K G M +
Sbjct: 272 AAPPEDINLIRGTGPGGQLQDLDCSS------SDDEETAQNASKPSATKGTLGGMFGMLK 325
Query: 333 SIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTV 392
+ G +L + D+E L ++D L+ KNVA +IA +LCESVA LEGK + +F+ ++STV
Sbjct: 326 GLVGSKSLSREDMESVLDKMRDHLIAKNVAADIAVQLCESVANKLEGKVMGTFSTVTSTV 385
Query: 393 QAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQH 452
+ A++++LV+IL P+R +D+LRD+ A+ ++PYVV F GVNGVGKSTNLAK+++WL ++
Sbjct: 386 KQALQESLVQILQPQRRVDMLRDIMDAQRHQRPYVVTFCGVNGVGKSTNLAKISFWLLEN 445
Query: 453 NVNVMMAACDTFRSGAVEQLRTHARRL-------------QIPIFEKGYEKDPAVVAKEA 499
+V++AACDTFR+GAVE +RTH RRL + +FEKGY KD A +A EA
Sbjct: 446 GFSVLIAACDTFRAGAVEHVRTHTRRLSALHPPEKHAGPTMVQLFEKGYGKDAAGIAMEA 505
Query: 500 IQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSK 559
I A G DVVLVDTAGRMQDN PLM AL+KLI +N PDLVLFVGEALVGN+AVDQL K
Sbjct: 506 IAFARNQGFDVVLVDTAGRMQDNAPLMTALAKLITVNTPDLVLFVGEALVGNEAVDQLVK 565
Query: 560 FNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKK 619
FN+ LAD S + TPRLIDGI+LTKFDTIDDKVGAA+SM YI+ P++FVG GQ+Y DL+
Sbjct: 566 FNRALADHSMAQTPRLIDGIVLTKFDTIDDKVGAAISMTYITSKPIVFVGTGQTYCDLRS 625
Query: 620 LNVKSIVKTLL 630
LN K++V L+
Sbjct: 626 LNAKAVVAALM 636
>SRPR_YEAST (P32916) Signal recognition particle receptor alpha
subunit homolog (SR-alpha) (Docking protein alpha)
(DP-alpha)
Length = 621
Score = 243 bits (620), Expect = 1e-63
Identities = 163/472 (34%), Positives = 247/472 (51%), Gaps = 63/472 (13%)
Query: 199 NVSVNGKENDSVNNGAFDVNRLQKKVRNKGGNGKKTDAVVTKAEPKKVVKKNRVWDEKPV 258
N + NG S N +KK+R+ G + T V +K + + DE
Sbjct: 172 NFTKNGSVPQSHNKNT------KKKLRDTKGKKQSTGNV---GSGRKWGRDGGMLDEMNH 222
Query: 259 E--TKLDFTD-------HVDIDGDADKDRKVDYLAKEQGESMMDK-DEIFSSDSEDEEDD 308
E KLDF+ V +D +KD D E G+ ++ + D++ SS
Sbjct: 223 EDAAKLDFSSSNSHNSSQVALDSTINKDSFGD--RTEGGDFLIKEIDDLLSSHK------ 274
Query: 309 DDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEK 368
D+ +G ++K F + + + G + +SDL+ L+ L +L+TKNVA E A+
Sbjct: 275 DEITSGNEAKNSGYVSTAFGFLQKHVLGNKTINESDLKSVLEKLTQQLITKNVAPEAADY 334
Query: 369 LCESVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRK---- 424
L + V+ L G K A++T + +T + ++ AL +ILTP S+D+LR++ + + ++
Sbjct: 335 LTQQVSHDLVGSKTANWTSVENTARESLTKALTQILTPGVSVDLLREIQSKRSKKDEEGK 394
Query: 425 --PYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRL--- 479
PYV VGVNGVGKSTNL+K+A+WL Q+N V++ ACDTFRSGAVEQLR H L
Sbjct: 395 CDPYVFSIVGVNGVGKSTNLSKLAFWLLQNNFKVLIVACDTFRSGAVEQLRVHVENLAQL 454
Query: 480 -------------------QIPIFEKGYEKDPAV--VAKEAIQEASRNGSDVVLVDTAGR 518
+ +FE GY V +AK+AI+ + D+VL+DTAGR
Sbjct: 455 MDDSHVRGSKNKRGKTGNDYVELFEAGYGGSDLVTKIAKQAIKYSRDQNFDIVLMDTAGR 514
Query: 519 MQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDG 578
++ LM L PD ++ VGEALVG D+V Q FN R +D
Sbjct: 515 RHNDPTLMSPLKSFADQAKPDKIIMVGEALVGTDSVQQAKNFNDAFG------KGRNLDF 568
Query: 579 ILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
+++K DT+ + +G ++MVY +G P++FVG GQ+YTDL+ L+VK V TL+
Sbjct: 569 FIISKCDTVGEMLGTMVNMVYATGIPILFVGVGQTYTDLRTLSVKWAVNTLM 620
>Y291_METJA (Q57739) Probable signal recognition particle protein
MJ0291
Length = 409
Score = 227 bits (579), Expect = 6e-59
Identities = 148/405 (36%), Positives = 228/405 (55%), Gaps = 31/405 (7%)
Query: 232 KKTDAVVTKAEPKKVVKKNRVWDEKPVETKLDFTDHVDIDGDADKDRKVDYLAKEQGESM 291
K T+ + +K E ++V +EK ++K+ FT K+ K + + KE+ E
Sbjct: 15 KITEKIYSKGEAEEVK------EEKEEKSKISFTSLFK------KEPKKEEVKKEKAEEE 62
Query: 292 MDKDEIFSSD-SEDEEDDDDDNAGKKSKPDAKKKGWFSSMFQSIAGKANLEKS------D 344
+ K EI ++ SE E+ ++ K + + KK +F + A K L+K D
Sbjct: 63 IKKSEIVKTEPSEKVEEAKEEIKEIKEEKEEKKITFFDRFGLTRAIKKVLKKEVVILEED 122
Query: 345 LEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDALVRIL 404
+E L+ L+ L+ +VA E+ EKL E++ L G+K++ + A+++A+ IL
Sbjct: 123 IEDVLEELEIALLEADVALEVVEKLIENIKNELVGRKISPDDNVEEITINAVKNAIKNIL 182
Query: 405 TPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTF 464
+ + IDI + K + KP V+VFVG+NG GK+T +AK+AY L+Q +V++AA DTF
Sbjct: 183 SQEK-IDIEEIIKKNKAEGKPTVIVFVGINGTGKTTTIAKLAYKLKQKGYSVVLAAGDTF 241
Query: 465 RSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEP 524
R+GA+EQL HA+ + + + + D A V +AIQ A G DVVL DTAGR N
Sbjct: 242 RAGAIEQLEQHAKNVGVKVIKHKPGADSAAVIYDAIQHAKARGIDVVLADTAGRQATNVN 301
Query: 525 LMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGILLTKF 584
LM + K++ + PDLV+FVG+AL GNDAV Q +FN+ + IDGI+LTK
Sbjct: 302 LMEEIKKVVRVTKPDLVIFVGDALTGNDAVYQAEEFNRAVN----------IDGIILTKV 351
Query: 585 DTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTL 629
D D K GAALS+ Y G P++++G GQ Y DL + + +V+ L
Sbjct: 352 DA-DAKGGAALSIGYAIGKPILYLGVGQRYQDLIEFDADWMVRKL 395
>DOCK_SULAC (P27414) Probable signal recognition particle protein
(Docking protein) (P41)
Length = 369
Score = 188 bits (477), Expect = 4e-47
Identities = 114/366 (31%), Positives = 205/366 (55%), Gaps = 28/366 (7%)
Query: 261 KLDFTDHVD-IDGDADKD----RKVDYLAKEQGESMMDKDEIFSSDSEDEEDDDDDN--- 312
K F++ +D I G+ +K R+ D L ++ E++ + + +E++ + +
Sbjct: 8 KKAFSNFLDKISGEENKKEPETRQTDQLESKKEETIQQQQNVQQPQAENKIEQKQEKISV 67
Query: 313 -AGKKSKPDAKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCE 371
G+++K + K+ S F + K +++ DL ++ L+ +L+ +V+ E+ EK+ E
Sbjct: 68 QTGQENKQENKR-----SFFDFLKYKT-IKEDDLNDVIEELRFQLLDSDVSYEVTEKILE 121
Query: 372 SVAASLEGKKLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFV 431
+ +L GKK++ + V ++ ++ ILT + D++ + ++ +KP+V++F
Sbjct: 122 DLKNNLIGKKVSRREEVEEIVINTLKKSITEILTKNQKTDLIEKIRSSG--KKPFVIIFF 179
Query: 432 GVNGVGKSTNLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKD 491
GVNGVGK+T +AKV L+++N++ ++AA DTFR+ A EQL HA +L++ + Y D
Sbjct: 180 GVNGVGKTTTIAKVVNMLKKNNLSTIIAASDTFRAAAQEQLAYHASKLEVQLIRGKYGAD 239
Query: 492 PAVVAKEAIQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGN 551
PA VA +AI A DVVL+DTAGRM + L+ L +++ + PD + + ++L G+
Sbjct: 240 PASVAFDAISFAKSRNIDVVLIDTAGRMHIDSDLVEELKRVLRIAKPDFRILILDSLAGS 299
Query: 552 DAVDQLSKFNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCG 611
DA++Q F + D ++LTK D D K G ALS+ Y PV+++G G
Sbjct: 300 DALEQARHFENNVG----------YDAVILTKVDA-DAKGGIALSLAYELKKPVVYMGVG 348
Query: 612 QSYTDL 617
Q+Y DL
Sbjct: 349 QNYDDL 354
>SR54_STRMU (Q54431) Signal recognition particle protein (Fifty-four
homolog)
Length = 516
Score = 144 bits (363), Expect = 7e-34
Identities = 83/290 (28%), Positives = 154/290 (52%), Gaps = 19/290 (6%)
Query: 330 MFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRIS 389
+F+++ GK L + D++ K ++ L+ +VA + ++ + V G ++ S
Sbjct: 13 VFKNLRGKRKLSEKDVQEVTKEIRLALLEADVALPVVKEFIKRVRKRAVGHEVIDTLDPS 72
Query: 390 STVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWL 449
+ + + L +L + K + P +++ VG+ G GK+T K+A L
Sbjct: 73 QQIIKIVNEELTAVLGSETA-------EIEKSSKIPTIIMMVGLQGAGKTTFAGKLANKL 125
Query: 450 -QQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGS 508
++ N +M A D +R A++QL+T +++ +P+F+ G E + + + +A N +
Sbjct: 126 VKEENARPLMIAADIYRPAAIDQLKTLGQQINVPVFDMGTEHSAVEIVSQGLAQAKENRN 185
Query: 509 DVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLS 568
D VL+DTAGR+Q +E LM L + L NP+ +L V ++++G +A + +FNQ+L
Sbjct: 186 DYVLIDTAGRLQIDEKLMTELRDIKALANPNEILLVVDSMIGQEAANVAREFNQQLE--- 242
Query: 569 TSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 618
+ G++LTK D D + GAALS+ I+G P+ F G G+ TD++
Sbjct: 243 -------VTGVILTKIDG-DTRGGAALSVRQITGKPIKFTGTGEKITDIE 284
>SR54_SYNY3 (P74214) Signal recognition particle protein (Fifty-four
homolog)
Length = 482
Score = 139 bits (349), Expect = 3e-32
Identities = 80/288 (27%), Positives = 156/288 (53%), Gaps = 18/288 (6%)
Query: 331 FQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISS 390
++ + G+ + +S+++ AL+ ++ L+ +V ++ + + V G + S
Sbjct: 13 WKKLRGQDKISESNIKEALQEVRRALLAADVNLQVVKGFIKDVEQKALGADVISGVNPGQ 72
Query: 391 TVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQ 450
+ D LV ++ +V A+ ++ P V++ G+ G GK+T AK+A +L+
Sbjct: 73 QFIKIVYDELVNLMGES-------NVPLAQAEQAPTVILMAGLQGTGKTTATAKLALYLR 125
Query: 451 QHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDV 510
+ + +M A D +R A++QL+T +++ +P+F+ G + +P +A++ +++A G D
Sbjct: 126 KQKRSALMVATDVYRPAAIDQLKTLGQQIDVPVFDLGSDANPVEIARQGVEKAKELGVDT 185
Query: 511 VLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTS 570
VL+DTAGR+Q + +M L+++ + PD L V +A+ G +A + F++++
Sbjct: 186 VLIDTAGRLQIDPQMMAELAEIKQVVKPDDTLLVVDAMTGQEAANLTHTFHEQIG----- 240
Query: 571 PTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 618
I G +LTK D D + GAALS+ ISG P+ FVG G+ L+
Sbjct: 241 -----ITGAILTKLDG-DTRGGAALSVRQISGQPIKFVGVGEKVEALQ 282
>SR54_SYNP7 (Q55311) Signal sequence binding protein
Length = 485
Score = 137 bits (345), Expect = 9e-32
Identities = 81/297 (27%), Positives = 157/297 (52%), Gaps = 18/297 (6%)
Query: 334 IAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQ 393
+ G+ + +S+++ ALK ++ L+ +V + ++ V G ++ S R
Sbjct: 16 LRGQDKISESNVQEALKEVRRALLEADVNLGVVKEFIAQVQEKAVGTQVISGIRPDQQFI 75
Query: 394 AAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHN 453
+ D LV+++ V A+ + P V++ G+ G GK+T AK+A L++
Sbjct: 76 KVVYDELVQVMGETH-------VPLAQAAKAPTVILMAGLQGAGKTTATAKLALHLRKEG 128
Query: 454 VNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLV 513
+ ++ A D +R A++QL T +++ +P+FE G + +P +A++ +++A + G D V+V
Sbjct: 129 RSTLLVATDVYRPAAIDQLITLGKQIDVPVFELGSDANPVEIARQGVEKARQEGIDTVIV 188
Query: 514 DTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTP 573
DTAGR+Q + +M L+++ P VL V ++++G +A F++++
Sbjct: 189 DTAGRLQIDTEMMEELAQVKEAIAPHEVLLVVDSMIGQEAASLTRSFHERIG-------- 240
Query: 574 RLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
I G +LTK D D + GAALS+ +SG P+ FVG G+ L+ + + + +L
Sbjct: 241 --ITGAILTKLDG-DSRGGAALSIRRVSGQPIKFVGLGEKVEALQPFHPERMASRIL 294
>SR51_LYCES (P49971) Signal recognition particle 54 kDa protein 1
(SRP54)
Length = 496
Score = 135 bits (339), Expect = 4e-31
Identities = 87/311 (27%), Positives = 153/311 (48%), Gaps = 19/311 (6%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G S Q ++ +++ L L + L+ +V ++ + ++ + +
Sbjct: 4 AQLGGSISRALQQMSNATIIDEKVLNECLNEITRALLQADVQFKLVRDMSTNIKKIVNLE 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ +Q A+ + L +IL P + L+ + KP VV+FVG+ G GK+T
Sbjct: 64 DLAAGHNKRRIIQQAVYNELCKILDPGKPAFTLK-------KGKPSVVMFVGLQGSGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGY-EKDPAVVAKEA 499
K AY Q+ + DTFR+GA +QL+ +A + +IP + Y E DP +A +
Sbjct: 117 TCTKYAYHHQKRGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSSYTESDPVKIAVDG 176
Query: 500 IQEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSK 559
++ + D+++VDT+GR + L + ++ PDLV+FV ++ +G A DQ
Sbjct: 177 VETFKKENCDLIIVDTSGRHKQEAALFEEMRQVSEAQKPDLVIFVMDSSIGQAAFDQAQA 236
Query: 560 FNQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKK 619
F Q +A + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 FRQSVA----------VGAVIVTKMDG-HAKGGGALSRVAATKSPVIFIGTGEHMDEFEV 285
Query: 620 LNVKSIVKTLL 630
+VK V LL
Sbjct: 286 FDVKPFVSRLL 296
>SR54_HELPY (P56005) Signal recognition particle protein (Fifty-four
homolog)
Length = 448
Score = 134 bits (338), Expect = 6e-31
Identities = 94/296 (31%), Positives = 158/296 (52%), Gaps = 35/296 (11%)
Query: 341 EKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQA----AM 396
++ L+ AL LK L+ +V ++A +L KK+ S T+++ + A+
Sbjct: 21 DEKALDRALDELKKTLLKNDVHHKVARELL---------KKVESQTKLNGIGKQQFLDAL 71
Query: 397 EDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNV 456
E +L+ IL+ + S Q P VV+ G+ G GK+T AK+A++L+ N V
Sbjct: 72 EKSLLEILSAKGSSGF------TFAQTPPTVVLMAGLQGSGKTTTTAKLAHYLKTKNKKV 125
Query: 457 MMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAV--VAKEAIQEASRNGSDVVLVD 514
++ ACD R AVEQL+ ++ + +F YE++ +V +A A++ A DV+LVD
Sbjct: 126 LLCACDLQRLAAVEQLKVLGEQVGVEVF---YEENKSVKEIASNALKRAKEAQFDVLLVD 182
Query: 515 TAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPR 574
+AGR+ ++ LM+ L ++ + NP VL+V +AL G D V + FN+++
Sbjct: 183 SAGRLAIDKELMQELKEVKEILNPHEVLYVADALSGQDGVKSANTFNEEIG--------- 233
Query: 575 LIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
+ G++L+KFD+ D K G AL + Y G P+ F+G G+ DL + IV L+
Sbjct: 234 -VSGVVLSKFDS-DSKGGIALGITYQLGLPLRFIGSGEKIPDLDVFVPERIVGRLM 287
>SR54_BACSU (P37105) Signal recognition particle protein (Fifty-four
homolog)
Length = 446
Score = 133 bits (335), Expect = 1e-30
Identities = 81/298 (27%), Positives = 153/298 (51%), Gaps = 19/298 (6%)
Query: 334 IAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQ 393
I GK + + D++ ++ ++ L+ +V ++ + + V+ G+ + V
Sbjct: 17 IRGKGKVSEQDVKEMMREVRLALLEADVNFKVVKDFVKKVSERAVGQDVMKSLTPGQQVI 76
Query: 394 AAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQ-H 452
+++ L ++ S A +R P V++ VG+ G GK+T K+A L++ H
Sbjct: 77 KVVQEELTELMGGEES-------KIAVAKRPPTVIMMVGLQGAGKTTTSGKLANLLRKKH 129
Query: 453 NVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVL 512
N M+ A D +R A++QL T ++L +P+F G + P +AK+AI++A D V+
Sbjct: 130 NRKPMLVAADIYRPAAIKQLETLGKQLDMPVFSLGDQVSPVEIAKQAIEKAKEEHYDYVI 189
Query: 513 VDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPT 572
+DTAGR+ + LM L+ + + NP+ + V +++ G DAV+ FN++L
Sbjct: 190 LDTAGRLHIDHELMDELTNVKEIANPEEIFLVVDSMTGQDAVNVAKSFNEQLG------- 242
Query: 573 PRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
+ G++LTK D D + GAALS+ ++ P+ F G G+ L+ + + + +L
Sbjct: 243 ---LTGVVLTKLDG-DTRGGAALSIRAVTNTPIKFAGLGEKLDALEPFHPERMASRIL 296
>SR52_LYCES (P49972) Signal recognition particle 54 kDa protein 2
(SRP54)
Length = 499
Score = 133 bits (335), Expect = 1e-30
Identities = 84/310 (27%), Positives = 151/310 (48%), Gaps = 18/310 (5%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G S Q ++ +++ L L + L+ +V ++ + ++ +
Sbjct: 4 AELGGSISRALQQMSNATIIDEKVLNECLNEITRALLQADVQFKLVRDMTTNIKKIVNLD 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ +Q A+ + L +IL P + ++ KP +V+FVG+ G GK+T
Sbjct: 64 DLAAGHNKRRIIQQAVFNELCKILDPGKP-------SFTPKKGKPSIVMFVGLQGSGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A + +
Sbjct: 117 TCTKYAYYHQKKGWKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVDGV 176
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ + D+++VDT+GR + L + ++ PDLV+FV ++ +G A DQ F
Sbjct: 177 ETFKKENCDLIIVDTSGRHKQEAALFEEMRQVAEATKPDLVIFVMDSSIGQAAFDQAQAF 236
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
Q +A + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 KQSVA----------VGAVIVTKMDG-HAKGGGALSAVAATKSPVIFIGTGEHMDEFEVF 285
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 286 DVKPFVSRLL 295
>SR53_ARATH (P49967) Signal recognition particle 54 kDa protein 3
(SRP54)
Length = 495
Score = 133 bits (334), Expect = 2e-30
Identities = 83/310 (26%), Positives = 152/310 (48%), Gaps = 18/310 (5%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G + Q ++ +++ L L + L+ +V+ + +++ ++ + +
Sbjct: 4 AELGGRITRAIQQMSNVTIIDEKALNECLNEITRALLQSDVSFPLVKEMQSNIKKIVNLE 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ ++ A+ L ++L P + A ++ K VV+FVG+ G GK+T
Sbjct: 64 DLAAGHNKRRIIEQAIFSELCKMLDPGKPA-------FAPKKAKASVVMFVGLQGAGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E +
Sbjct: 117 TCTKYAYYHQKKGYKPALVCADTFRAGAFDQLKQNATKAKIPFYGSYTESDPVKIAVEGV 176
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ D+++VDT+GR + L + ++ PDLV+FV ++ +G A DQ F
Sbjct: 177 DTFKKENCDLIIVDTSGRHKQEASLFEEMRQVAEATKPDLVIFVMDSSIGQAAFDQAQAF 236
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
Q +A + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 KQSVA----------VGAVIITKMDG-HAKGGGALSAVAATKSPVIFIGTGEHMDEFEVF 285
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 286 DVKPFVSRLL 295
>SR5C_ARATH (P37107) Signal recognition particle 54 kDa protein,
chloroplast precursor (SRP54) (54 chloroplast protein)
(54CP) (FFC)
Length = 564
Score = 132 bits (333), Expect = 2e-30
Identities = 84/294 (28%), Positives = 155/294 (52%), Gaps = 18/294 (6%)
Query: 325 GWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLAS 384
G + + + G+ L K ++ ++ ++ L+ +V+ + + +SV+ G +
Sbjct: 83 GGLEAAWSKLKGEEVLTKDNIAEPMRDIRRALLEADVSLPVVRRFVQSVSDQAVGMGVIR 142
Query: 385 FTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAK 444
+ + + D LV+++ S ++ AK P V++ G+ GVGK+T AK
Sbjct: 143 GVKPDQQLVKIVHDELVKLMGGEVS-----ELQFAKSG--PTVILLAGLQGVGKTTVCAK 195
Query: 445 VAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEAS 504
+A +L++ + M+ A D +R A++QL ++ +P++ G + PA +AK+ ++EA
Sbjct: 196 LACYLKKQGKSCMLIAGDVYRPAAIDQLVILGEQVGVPVYTAGTDVKPADIAKQGLKEAK 255
Query: 505 RNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKL 564
+N DVV++DTAGR+Q ++ +M L + NP VL V +A+ G +A ++ FN ++
Sbjct: 256 KNNVDVVIMDTAGRLQIDKGMMDELKDVKKFLNPTEVLLVVDAMTGQEAAALVTTFNVEI 315
Query: 565 ADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLK 618
I G +LTK D D + GAALS+ +SG P+ VG G+ DL+
Sbjct: 316 G----------ITGAILTKLDG-DSRGGAALSVKEVSGKPIKLVGRGERMEDLE 358
>SR54_MYCMY (Q01442) Signal recognition particle protein (Fifty-four
homolog)
Length = 447
Score = 132 bits (333), Expect = 2e-30
Identities = 87/295 (29%), Positives = 155/295 (52%), Gaps = 23/295 (7%)
Query: 338 ANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAME 397
+ L + +++ LK ++ L+ +V E A+++ +V G ++ + +
Sbjct: 22 STLNEENIKETLKEIRLSLLEADVNIEAAKEIINNVKQKALGGYISEGASAHQQMIKIVH 81
Query: 398 DALVRILTPRRS-IDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVN- 455
+ LV IL + +DI +KP VV+ VG+ G GK+T K+AY L + N
Sbjct: 82 EELVNILGKENAPLDI---------NKKPSVVMMVGLQGSGKTTTANKLAYLLNKKNKKK 132
Query: 456 VMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDT 515
V++ D +R GA+EQL ++ +FEKG ++DP A++A++ A N DVV++DT
Sbjct: 133 VLLVGLDIYRPGAIEQLVQLGQKTNTQVFEKG-KQDPVKTAEQALEYAKENNFDVVILDT 191
Query: 516 AGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRL 575
AGR+Q ++ LM+ L L +P+ +L V + + G + ++ ++FN KL
Sbjct: 192 AGRLQVDQVLMKELDNLKKKTSPNEILLVVDGMSGQEIINVTNEFNDKLK---------- 241
Query: 576 IDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
+ G+++TK D D + GA LS+ Y++ P+ F+G G+ Y L K + L+
Sbjct: 242 LSGVVVTKLDG-DARGGATLSISYLTKLPIKFIGEGEGYNALAAFYPKRMADRLM 295
>SR54_HELPJ (Q9ZK62) Signal recognition particle protein (Fifty-four
homolog)
Length = 448
Score = 132 bits (332), Expect = 3e-30
Identities = 88/290 (30%), Positives = 151/290 (51%), Gaps = 23/290 (7%)
Query: 341 EKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGKKLASFTRISSTVQAAMEDAL 400
++ L+ AL LK L+ +V ++A +L + V + + + + A+E +L
Sbjct: 21 DEKALDRALDELKKTLLKNDVHHKVARELLKKVESQTKAGGIGKQQFLD-----ALEKSL 75
Query: 401 VRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKSTNLAKVAYWLQQHNVNVMMAA 460
+ IL+ + S Q P VV+ G+ G GK+T AK+A++L+ N V++ A
Sbjct: 76 LEILSAKGSSGF------TFAQTPPTVVLMAGLQGSGKTTTTAKLAHYLKTKNKKVLLCA 129
Query: 461 CDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAIQEASRNGSDVVLVDTAGRMQ 520
CD R AVEQL+ ++ + +F + K +A A++ A DV++VD+AGR+
Sbjct: 130 CDLQRLAAVEQLKVLGEQVGVEVFHE-ENKSVKEIANNALKRAKEAQFDVLIVDSAGRLA 188
Query: 521 DNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKFNQKLADLSTSPTPRLIDGIL 580
++ LM+ L ++ + NP VL+V +AL G D V + FN+++ + G++
Sbjct: 189 IDKELMQELKEVKEVLNPHEVLYVADALSGQDGVKSANTFNEEIG----------VSGVV 238
Query: 581 LTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKLNVKSIVKTLL 630
L+KFD+ D K G AL + Y G P+ F+G G+ DL + IV L+
Sbjct: 239 LSKFDS-DSKGGIALGITYQLGLPLRFIGSGEKIPDLDVFMPERIVGRLM 287
>SR52_HORVU (P49969) Signal recognition particle 54 kDa protein 2
(SRP54)
Length = 497
Score = 132 bits (332), Expect = 3e-30
Identities = 84/310 (27%), Positives = 149/310 (47%), Gaps = 18/310 (5%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G S ++ +++ L L + L+ +V ++ + ++ + +
Sbjct: 4 AQLGGSISRALAQMSNATVIDEKVLGECLNEISRALLQSDVQFKMVRDMQTNIRKIVNLE 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ T +Q A+ L +L P + ++ KP VV+FVG+ G GK+T
Sbjct: 64 TLAAGTNKRRIIQQAVFTELCNMLDPGKPA-------FTPKKGKPSVVMFVGLQGSGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E +
Sbjct: 117 TCTKYAYYHQRKGFKPSLVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGL 176
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ + SD++++DT+GR + L + ++ PDLV+FV + +G A DQ F
Sbjct: 177 ERFRKENSDLIIIDTSGRHKQEAALFEEMRQVAEATKPDLVIFVMDGSIGQAAFDQAQAF 236
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
Q + + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 KQSAS----------VGAVIITKLDG-HAKGGGALSAVAATKSPVIFIGTGEHIDEFEIF 285
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 286 DVKPFVSRLL 295
>SR51_HORVU (P49968) Signal recognition particle 54 kDa protein 1
(SRP54)
Length = 497
Score = 132 bits (332), Expect = 3e-30
Identities = 84/310 (27%), Positives = 149/310 (47%), Gaps = 18/310 (5%)
Query: 321 AKKKGWFSSMFQSIAGKANLEKSDLEPALKALKDRLMTKNVAEEIAEKLCESVAASLEGK 380
A+ G S ++ +++ L L + L+ +V ++ + ++ + +
Sbjct: 4 AQLGGSISRALAQMSNATVIDEKVLGECLNEISRALLQSDVQFKMVRDMQTNIRKIVNLE 63
Query: 381 KLASFTRISSTVQAAMEDALVRILTPRRSIDILRDVHAAKEQRKPYVVVFVGVNGVGKST 440
LA+ T +Q A+ L +L P + ++ KP VV+FVG+ G GK+T
Sbjct: 64 TLAAGTNKRRIIQQAVFTELCNMLDPGKPA-------FTTKKGKPSVVMFVGLQGSGKTT 116
Query: 441 NLAKVAYWLQQHNVNVMMAACDTFRSGAVEQLRTHARRLQIPIFEKGYEKDPAVVAKEAI 500
K AY+ Q+ + DTFR+GA +QL+ +A + +IP + E DP +A E +
Sbjct: 117 TCTKYAYYHQRKGFKPSLVCADTFRAGAFDQLKQNATKAKIPFYGSYMESDPVKIAVEGL 176
Query: 501 QEASRNGSDVVLVDTAGRMQDNEPLMRALSKLIYLNNPDLVLFVGEALVGNDAVDQLSKF 560
+ + SD++++DT+GR + L + ++ PDLV+FV + +G A DQ F
Sbjct: 177 ERFRKENSDLIIIDTSGRHKQEAALFEEMRQVAEATKPDLVIFVMDGSIGQAAFDQAQAF 236
Query: 561 NQKLADLSTSPTPRLIDGILLTKFDTIDDKVGAALSMVYISGAPVMFVGCGQSYTDLKKL 620
Q + + +++TK D K G ALS V + +PV+F+G G+ + +
Sbjct: 237 KQSAS----------VGAVIITKLDG-HAKGGGALSAVAATKSPVIFIGTGEHIDEFEIF 285
Query: 621 NVKSIVKTLL 630
+VK V LL
Sbjct: 286 DVKPFVSRLL 295
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.314 0.133 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 75,121,449
Number of Sequences: 164201
Number of extensions: 3468557
Number of successful extensions: 18819
Number of sequences better than 10.0: 503
Number of HSP's better than 10.0 without gapping: 188
Number of HSP's successfully gapped in prelim test: 331
Number of HSP's that attempted gapping in prelim test: 15545
Number of HSP's gapped (non-prelim): 1597
length of query: 630
length of database: 59,974,054
effective HSP length: 116
effective length of query: 514
effective length of database: 40,926,738
effective search space: 21036343332
effective search space used: 21036343332
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC136953.3


