

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.9 + phase: 0
(341 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
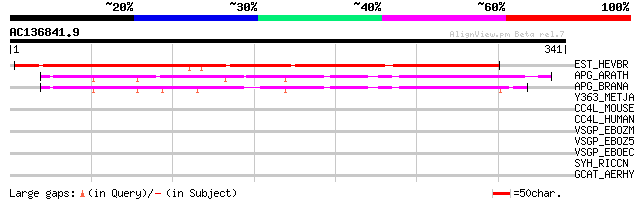
Score E
Sequences producing significant alignments: (bits) Value
EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early nodule... 249 9e-66
APG_ARATH (P40602) Anter-specific proline-rich protein APG precu... 65 2e-10
APG_BRANA (P40603) Anter-specific proline-rich protein APG (Prot... 62 2e-09
Y363_METJA (Q57809) Hypothetical MCM-type protein MJ0363 32 2.3
CC4L_MOUSE (Q8K3P5) Carbon catabolite repressor protein 4 homolo... 32 2.3
CC4L_HUMAN (Q9ULM6) Carbon catabolite repressor protein 4 homolo... 32 2.3
VSGP_EBOZM (P60170) Pre-small/secreted glycoprotein precursor (p... 32 3.0
VSGP_EBOZ5 (P60171) Pre-small/secreted glycoprotein precursor (p... 32 3.0
VSGP_EBOEC (P87670) Pre-small/secreted glycoprotein precursor (p... 32 3.0
SYH_RICCN (Q92IK8) Histidyl-tRNA synthetase (EC 6.1.1.21) (Histi... 30 6.7
GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase p... 30 8.7
>EST_HEVBR (Q7Y1X1) Esterase precursor (EC 3.1.1.-) (Early
nodule-specific protein homolog) (Latex allergen Hev b
13)
Length = 391
Score = 249 bits (635), Expect = 9e-66
Identities = 137/306 (44%), Positives = 188/306 (60%), Gaps = 18/306 (5%)
Query: 4 MTLIYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISG 63
+TL ++LC +L A S+ C +PAI+NFGDSNSDTG A + P G +FF +G
Sbjct: 12 ITLSFLLCMLSLAYA--SETCDFPAIFNFGDSNSDTGGKAAAFYPLNPPYGETFFHRSTG 69
Query: 64 RCCDGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPIR------PIIAGLT- 116
R DGRLI+DFI+E LPYLS YL+S+GSN++HGA+FA A + I+ P G +
Sbjct: 70 RYSDGRLIIDFIAESFNLPYLSPYLSSLGSNFKHGADFATAGSTIKLPTTIIPAHGGFSP 129
Query: 117 -YLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKP 175
YL Q SQF F ++ F + T VP F KA+YT DIGQND+ G
Sbjct: 130 FYLDVQYSQFRQFIPRSQ--FIRETGGIFAELVPEEYYFEKALYTFDIGQNDLTEGFL-- 185
Query: 176 NSSEEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNL 235
N + EEV ++PD+++ F+ V+K+Y+ AR FWIHNTGPI C+ + ++P +
Sbjct: 186 NLTVEEVNATVPDLVNSFSANVKKIYDLGARTFWIHNTGPIGCLSFILTYFPWAEK---- 241
Query: 236 DANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNP 295
D+ GC K +NE+AQ +N +LK+ V QLR+ PLA F +VD+Y+VKY+L S GF P
Sbjct: 242 DSAGCAKAYNEVAQHFNHKLKEIVAQLRKDLPLATFVHVDIYSVKYSLFSEPEKHGFEFP 301
Query: 296 LEFCCG 301
L CCG
Sbjct: 302 LITCCG 307
>APG_ARATH (P40602) Anter-specific proline-rich protein APG
precursor
Length = 534
Score = 65.5 bits (158), Expect = 2e-10
Identities = 76/333 (22%), Positives = 142/333 (41%), Gaps = 43/333 (12%)
Query: 20 PSKKCVYPAIYNFGDSNSDTGAGYATMAAVE---HPNGISF-FGSISGRCCDGRLILDFI 75
P K + PA++ FGDS DTG ++ P G+ F F +GR +G + D++
Sbjct: 197 PENKTI-PAVFFFGDSVFDTGNNNNLETKIKSNYRPYGMDFKFRVATGRFSNGMVASDYL 255
Query: 76 SE-----ELELPYLSSYLNSVGSNYRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKS 130
++ E+ YL + ++ G +FA A P + + Q F+
Sbjct: 256 AKYMGVKEIVPAYLDPKIQP--NDLLTGVSFASGGAGYNPTTSEAANAIPMLDQLTYFQD 313
Query: 131 H----TKILFDQRTEPPLRSGVPRTEDF-SKAIYTIDIGQNDI-----GYGLQKPNSSEE 180
+ +++ ++++ L +G+ +T SK + + G ND+ G G Q+ + +
Sbjct: 314 YIEKVNRLVRQEKSQYKL-AGLEKTNQLISKGVAIVVGGSNDLIITYFGSGAQRLKNDID 372
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGC 240
I D + F V +LY AR + T P+ C+P + +K + C
Sbjct: 373 SYTTIIADSAASF---VLQLYGYGARRIGVIGTPPLGCVP------SQRLKKKKI----C 419
Query: 241 VKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEFCC 300
+ N +Q +N +L + QL + P + F Y+D+YT+ ++ GF + CC
Sbjct: 420 NEELNYASQLFNSKLLLILGQLSKTLPNSTFVYMDIYTIISQMLETPAAYGFEETKKPCC 479
Query: 301 GSYQGNEIHYCGKKSIKNGTVYGIACDDSSTYI 333
+ + C K + K C ++S+Y+
Sbjct: 480 KTGLLSAGALCKKSTSK-------ICPNTSSYL 505
>APG_BRANA (P40603) Anter-specific proline-rich protein APG (Protein
CEX) (Fragment)
Length = 449
Score = 62.0 bits (149), Expect = 2e-09
Identities = 79/322 (24%), Positives = 134/322 (41%), Gaps = 48/322 (14%)
Query: 20 PSKKCVYPAIYNFGDSNSDTGAGYATMAAVE---HPNGISF-FGSISGRCCDGRLILDFI 75
P K + PA++ FGDS DTG ++ P G+ F G +GR +GR+ D+I
Sbjct: 118 PQNKTI-PAVFFFGDSIFDTGNNNNLDTKLKCNYRPYGMDFPMGVATGRFSNGRVASDYI 176
Query: 76 SE-----ELELPYLSSYLNSVG----SNYRHGANFAVASAPIRPIIAG---LTYLGFQVS 123
S+ E+ Y+ L S+ G +FA A P + +T + Q++
Sbjct: 177 SKYLGVKEIVPAYVDKKLQQNNELQQSDLLTGVSFASGGAGYLPQTSESWKVTTMLDQLT 236
Query: 124 QFILFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDI-----GYGLQKPNSS 178
F +K K L ++ + SK + G ND+ G G Q +
Sbjct: 237 YFQDYKKRMKKLVGKKKTKKI---------VSKGAAIVVAGSNDLIYTYFGNGAQHLKND 287
Query: 179 EEEVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDAN 238
+ + D + F V +LY AR + T PI C P + +K +
Sbjct: 288 VDSFTTMMADSAASF---VLQLYGYGARRIGVIGTPPIGCTP------SQRVKKKKI--- 335
Query: 239 GCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVDVYTVKYTLISNARNQGFVNPLEF 298
C + N AQ +N +L + QL + P + Y D+Y++ ++ + + GF +
Sbjct: 336 -CNEDLNYAAQLFNSKLVIILGQLSKTLPNSTIVYGDIYSIFSKMLESPEDYGFEEIKKP 394
Query: 299 CC--GSYQGNEIHYCGKKSIKN 318
CC G +G +C ++++KN
Sbjct: 395 CCKIGLTKGGV--FCKERTLKN 414
>Y363_METJA (Q57809) Hypothetical MCM-type protein MJ0363
Length = 759
Score = 32.0 bits (71), Expect = 2.3
Identities = 15/38 (39%), Positives = 25/38 (65%), Gaps = 3/38 (7%)
Query: 181 EVRRSIPDILSQFTQAVQKLYNEEARVFWIHNTGPIEC 218
EVR+SI ++ + +A ++L+NE V +IH PI+C
Sbjct: 136 EVRKSITNV---YIEAYEELFNETPNVEFIHIRNPIDC 170
>CC4L_MOUSE (Q8K3P5) Carbon catabolite repressor protein 4 homolog
(EC 3.1.-.-) (Cytoplasmic deadenylase) (CCR4 carbon
catabolite repression 4-like) (Nocturnin homolog)
Length = 557
Score = 32.0 bits (71), Expect = 2.3
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 4/76 (5%)
Query: 216 IECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVD 275
+E YY FF E+G NG P + A+ + Q + V F KFT V
Sbjct: 241 VETEQYYSFFLVELKERGY---NGFFSPKSR-ARTMSEQERKHVDGCAIFFKTEKFTLVQ 296
Query: 276 VYTVKYTLISNARNQG 291
+TV++ ++ A ++G
Sbjct: 297 KHTVEFNQLAMANSEG 312
>CC4L_HUMAN (Q9ULM6) Carbon catabolite repressor protein 4 homolog
(EC 3.1.-.-) (Cytoplasmic deadenylase) (CCR4 carbon
catabolite repression 4-like) (Nocturnin homolog)
Length = 557
Score = 32.0 bits (71), Expect = 2.3
Identities = 22/76 (28%), Positives = 35/76 (45%), Gaps = 4/76 (5%)
Query: 216 IECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLRRMFPLAKFTYVD 275
+E YY FF E+G NG P + A+ + Q + V F KFT V
Sbjct: 241 VETEQYYSFFLVELKERGY---NGFFSPKSR-ARTMSEQERKHVDGCAIFFKTEKFTLVQ 296
Query: 276 VYTVKYTLISNARNQG 291
+TV++ ++ A ++G
Sbjct: 297 KHTVEFNQLAMANSEG 312
>VSGP_EBOZM (P60170) Pre-small/secreted glycoprotein precursor
(pre-sGP) [Contains: Small/secreted glycoprotein (sGP);
Delta-peptide]
Length = 364
Score = 31.6 bits (70), Expect = 3.0
Identities = 43/181 (23%), Positives = 65/181 (35%), Gaps = 21/181 (11%)
Query: 95 YRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDF 154
++ GA F I G T+ V+ F++ K F PLR V TED
Sbjct: 154 HKEGAFFLYDRLASTVIYRGTTFAEGVVA-FLILPQAKKDFFSSH---PLREPVNATEDP 209
Query: 155 SKAIYTIDI-------GQNDIGYGLQKPNSSEEEVR-RSIPDILSQFTQAVQ---KLYNE 203
S Y+ I G N+ Y + N + ++ R P L Q + + K N
Sbjct: 210 SSGYYSTTIRYQATGFGTNETEYLFEVDNLTYVQLESRFTPQFLLQLNETIYTSGKRSNT 269
Query: 204 EARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLR 263
++ W N I + F+ K C L+Q Y + K V ++R
Sbjct: 270 TGKLIWKVNPEIDTTIGEWAFWETKKTSLEKFAVKSC------LSQLYQTEPKTSVVRVR 323
Query: 264 R 264
R
Sbjct: 324 R 324
>VSGP_EBOZ5 (P60171) Pre-small/secreted glycoprotein precursor
(pre-sGP) [Contains: Small/secreted glycoprotein (sGP);
Delta-peptide]
Length = 364
Score = 31.6 bits (70), Expect = 3.0
Identities = 43/181 (23%), Positives = 65/181 (35%), Gaps = 21/181 (11%)
Query: 95 YRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDF 154
++ GA F I G T+ V+ F++ K F PLR V TED
Sbjct: 154 HKEGAFFLYDRLASTVIYRGTTFAEGVVA-FLILPQAKKDFFSSH---PLREPVNATEDP 209
Query: 155 SKAIYTIDI-------GQNDIGYGLQKPNSSEEEVR-RSIPDILSQFTQAVQ---KLYNE 203
S Y+ I G N+ Y + N + ++ R P L Q + + K N
Sbjct: 210 SSGYYSTTIRYQATGFGTNETEYLFEVDNLTYVQLESRFTPQFLLQLNETIYTSGKRSNT 269
Query: 204 EARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLR 263
++ W N I + F+ K C L+Q Y + K V ++R
Sbjct: 270 TGKLIWKVNPEIDTTIGEWAFWETKKTSLEKFAVKSC------LSQLYQTEPKTSVVRVR 323
Query: 264 R 264
R
Sbjct: 324 R 324
>VSGP_EBOEC (P87670) Pre-small/secreted glycoprotein precursor
(pre-sGP) [Contains: Small/secreted glycoprotein (sGP);
Delta-peptide]
Length = 364
Score = 31.6 bits (70), Expect = 3.0
Identities = 43/181 (23%), Positives = 65/181 (35%), Gaps = 21/181 (11%)
Query: 95 YRHGANFAVASAPIRPIIAGLTYLGFQVSQFILFKSHTKILFDQRTEPPLRSGVPRTEDF 154
++ GA F I G T+ V+ F++ K F PLR V TED
Sbjct: 154 HKEGAFFLYDRLASTVIYRGTTFAEGVVA-FLILPQAKKDFFSSH---PLREPVNATEDP 209
Query: 155 SKAIYTIDI-------GQNDIGYGLQKPNSSEEEVR-RSIPDILSQFTQAVQ---KLYNE 203
S Y+ I G N+ Y + N + ++ R P L Q + + K N
Sbjct: 210 SSGYYSTTIRYQATGFGTNETEYLFEVDNLTYVQLESRFTPQFLLQLNETIYTSGKRSNT 269
Query: 204 EARVFWIHNTGPIECIPYYYFFYPHKNEKGNLDANGCVKPHNELAQEYNRQLKDQVFQLR 263
++ W N I + F+ K C L+Q Y + K V ++R
Sbjct: 270 TGKLIWKVNPEIDTTIGEWAFWETKKTSLEKFAVKSC------LSQLYQTEPKTSVVRVR 323
Query: 264 R 264
R
Sbjct: 324 R 324
>SYH_RICCN (Q92IK8) Histidyl-tRNA synthetase (EC 6.1.1.21)
(Histidine--tRNA ligase) (HisRS)
Length = 414
Score = 30.4 bits (67), Expect = 6.7
Identities = 14/51 (27%), Positives = 29/51 (56%)
Query: 156 KAIYTIDIGQNDIGYGLQKPNSSEEEVRRSIPDILSQFTQAVQKLYNEEAR 206
K ++ + IG+N+I Y L+ + E I + L + + +Q+++NE A+
Sbjct: 325 KPVFVLPIGKNNICYALEIVDKLRTENIAIIIESLGKIAKRMQRIFNENAQ 375
>GCAT_AERHY (P10480) Phosphatidylcholine-sterol acyltransferase
precursor (EC 2.3.1.43) (Glycerophospholipid-cholesterol
acyltransferase) (GCAT)
Length = 335
Score = 30.0 bits (66), Expect = 8.7
Identities = 49/211 (23%), Positives = 83/211 (39%), Gaps = 22/211 (10%)
Query: 7 IYILCFFNLCVACPSKKCVYPAIYNFGDSNSDTGAGYATMAAVEHPNGISFFGSISGRCC 66
+ +L L V + + I FGDS SDTG Y+ M P+ ++ GR
Sbjct: 6 VCLLGLVALTVQAADSRPAFSRIVMFGDSLSDTGKMYSKMRGY-LPSSPPYY---EGRFS 61
Query: 67 DGRLILDFISEELELPYLSSYLNSVGSNYRHGANFAVASAPI--RPIIAGLTYLGFQVSQ 124
+G + L+ ++ E P L+ +N G AVA I P + L ++V+Q
Sbjct: 62 NGPVWLEQLTN--EFPGLTI------ANEAEGGPTAVAYNKISWNPKYQVINNLDYEVTQ 113
Query: 125 FI---LFKSHTKILFDQRTEPPLRSGVPRTEDFSKAIYTIDIGQNDIGYGLQKPNSSEEE 181
F+ FK ++ L G +D + I N + N ++E
Sbjct: 114 FLQKDSFKPDDLVILWVGANDYLAYGWNTEQDAKRVRDAISDAANRMVL-----NGAKEI 168
Query: 182 VRRSIPDILSQFTQAVQKLYNEEARVFWIHN 212
+ ++PD+ + QK+ + V HN
Sbjct: 169 LLFNLPDLGQNPSARSQKVVEAASHVSAYHN 199
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.139 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,161,173
Number of Sequences: 164201
Number of extensions: 1833183
Number of successful extensions: 3760
Number of sequences better than 10.0: 11
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 3747
Number of HSP's gapped (non-prelim): 12
length of query: 341
length of database: 59,974,054
effective HSP length: 111
effective length of query: 230
effective length of database: 41,747,743
effective search space: 9601980890
effective search space used: 9601980890
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC136841.9


