

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136841.7 + phase: 0 /pseudo
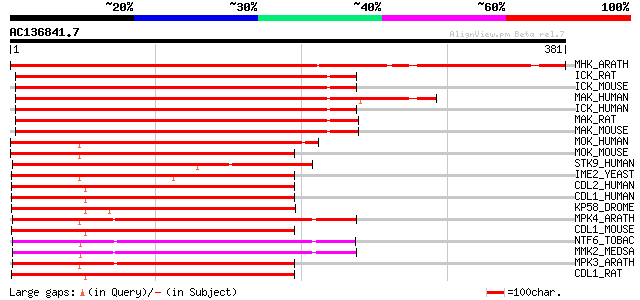
(381 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
MHK_ARATH (P43294) Serine/threonine-protein kinase MHK (EC 2.7.1... 417 e-116
ICK_RAT (Q62726) Serine/threonine kinase ICK (EC 2.7.1.37) (Inte... 281 3e-75
ICK_MOUSE (Q9JKV2) Serine/threonine kinase ICK (EC 2.7.1.37) (In... 280 4e-75
MAK_HUMAN (P20794) Serine/threonine-protein kinase MAK (EC 2.7.1... 279 1e-74
ICK_HUMAN (Q9UPZ9) Serine/threonine kinase ICK (EC 2.7.1.37) (In... 278 2e-74
MAK_RAT (P20793) Serine/threonine-protein kinase MAK (EC 2.7.1.3... 277 4e-74
MAK_MOUSE (Q04859) Serine/threonine-protein kinase MAK (EC 2.7.1... 276 5e-74
MOK_HUMAN (Q9UQ07) MAPK/MAK/MRK overlapping kinase (EC 2.7.1.37)... 196 8e-50
MOK_MOUSE (Q9WVS4) MAPK/MAK/MRK overlapping kinase (EC 2.7.1.37)... 195 1e-49
STK9_HUMAN (O76039) Serine/threonine-protein kinase 9 (EC 2.7.1.... 173 6e-43
IME2_YEAST (P32581) Meiosis induction protein kinase IME2/SME1 (... 169 1e-41
CDL2_HUMAN (Q9UQ88) PITSLRE serine/threonine-protein kinase CDC2... 165 2e-40
CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2... 164 3e-40
KP58_DROME (Q9VPC0) Serine/threonine-protein kinase PITSLRE (EC ... 163 6e-40
MPK4_ARATH (Q39024) Mitogen-activated protein kinase homolog 4 (... 163 8e-40
CDL1_MOUSE (P24788) PITSLRE serine/threonine-protein kinase CDC2... 162 1e-39
NTF6_TOBAC (Q40531) Mitogen-activated protein kinase homolog NTF... 162 1e-39
MMK2_MEDSA (Q40353) Mitogen-activated protein kinase homolog MMK... 161 2e-39
MPK3_ARATH (Q39023) Mitogen-activated protein kinase homolog 3 (... 161 3e-39
CDL1_RAT (P46892) PITSLRE serine/threonine-protein kinase CDC2L1... 161 3e-39
>MHK_ARATH (P43294) Serine/threonine-protein kinase MHK (EC
2.7.1.37)
Length = 443
Score = 417 bits (1072), Expect = e-116
Identities = 218/382 (57%), Positives = 275/382 (71%), Gaps = 15/382 (3%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMD 60
+ T+E+VAVK++KRKF +WEE NLRE+KALRK+NH +IIKL+E+VRE+NELFFIFE MD
Sbjct: 32 LETYEVVAVKKMKRKFYYWEECVNLREVKALRKLNHPHIIKLKEIVREHNELFFIFECMD 91
Query: 61 CNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKIADF 120
NLY ++KERE+PFSE EIR FM QMLQGL+HMHK G+FHRDLKPENLLVTN++LKIADF
Sbjct: 92 HNLYHIMKERERPFSEGEIRSFMSQMLQGLAHMHKNGYFHRDLKPENLLVTNNILKIADF 151
Query: 121 GLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGES 180
GLAREV+SMPPYT+YVSTRWYRAPEVLLQS YTPAVDMWA+GAILAEL+ LTP+FPGES
Sbjct: 152 GLAREVASMPPYTEYVSTRWYRAPEVLLQSSLYTPAVDMWAVGAILAELYALTPLFPGES 211
Query: 181 EIDQMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLITA 240
EIDQ+YKI C+LG PD T F + SR++ + H +++D++PNA+ EAIDLI
Sbjct: 212 EIDQLYKICCVLGKPDWTTFPEAKSISRIMS-ISHTEFPQTRIADLLPNAAPEAIDLINR 270
Query: 241 RCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKPNLELKLH 300
C ++ H F + Q + + LEL+L N PNLEL L
Sbjct: 271 LCSWDPLKRPTADEALNHP---FFSMATQASYPI-----HDLELRLDNMAALPNLELNLW 322
Query: 301 DFGPDPDDCFLGLTLAVKPSVSNLDVVQNARQGMGENLV*LDAL*KCRICYFARILMTIQ 360
DF +P++CFLGLTLAVKPS L++++N Q M EN + + R L++
Sbjct: 323 DFNREPEECFLGLTLAVKPSAPKLEMLRNVSQDMSENFLFCPGVNNDREPSVFWSLLS-- 380
Query: 361 TNQPDQNGVHSSAETS-LSLSF 381
PD+NG+H+ E+S LSLSF
Sbjct: 381 ---PDENGLHAPVESSPLSLSF 399
>ICK_RAT (Q62726) Serine/threonine kinase ICK (EC 2.7.1.37)
(Intestinal cell kinase) (MAK-related kinase) (MRK)
Length = 629
Score = 281 bits (718), Expect = 3e-75
Identities = 138/235 (58%), Positives = 179/235 (75%), Gaps = 2/235 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E++A+K++KRKF WEE NLRE+K+L+K+NH NI+KL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELIAIKKMKRKFYSWEECMNLREVKSLKKLNHANIVKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QLIKER K F E IR M Q+LQGL+ +HK GFFHRDLKPENLL +++KIADFGLA
Sbjct: 88 QLIKERNKLFPESAIRNIMYQILQGLAFIHKHGFFHRDLKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G I+AE++TL P+FPG SEID
Sbjct: 148 REIRSRPPYTDYVSTRWYRAPEVLLRSTNYSSPIDVWAVGCIMAEVYTLRPLFPGASEID 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
++KI +LG P T + G S ++F+ + + P L +IPNAS EAI L+
Sbjct: 208 TIFKICQVLGTPKKTDWPEGYQLSSAMNFIWPQCI-PNNLKTLIPNASSEAIQLL 261
>ICK_MOUSE (Q9JKV2) Serine/threonine kinase ICK (EC 2.7.1.37)
(Intestinal cell kinase) (mICK) (MAK-related kinase)
(MRK)
Length = 629
Score = 280 bits (716), Expect = 4e-75
Identities = 138/235 (58%), Positives = 179/235 (75%), Gaps = 2/235 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E++A+K++KRKF WEE NLRE+K+L+K+NH NI+KL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELIAIKKMKRKFYSWEECMNLREVKSLKKLNHANIVKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QLIKER K F E IR M Q+LQGL+ +HK GFFHRDLKPENLL +++KIADFGLA
Sbjct: 88 QLIKERNKLFPESAIRNIMYQILQGLAFIHKHGFFHRDLKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G I+AE++TL P+FPG SEID
Sbjct: 148 REIRSRPPYTDYVSTRWYRAPEVLLRSTNYSSPIDIWAVGCIMAEVYTLRPLFPGASEID 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
++KI +LG P T + G S ++F+ + + P L +IPNAS EAI L+
Sbjct: 208 TIFKICQVLGTPKKTDWPEGYQLSSAMNFLWPQCI-PNNLKTLIPNASSEAIQLL 261
>MAK_HUMAN (P20794) Serine/threonine-protein kinase MAK (EC
2.7.1.37) (Male germ cell-associated kinase)
Length = 623
Score = 279 bits (713), Expect = 1e-74
Identities = 148/300 (49%), Positives = 201/300 (66%), Gaps = 17/300 (5%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNIMYQILQGLAFIHKHGFFHRDMKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+ L P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSVYSSPIDVWAVGSIMAELYMLRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT---- 239
+++KI +LG P + + G + ++F + V P+ L +IPNAS EAI L+T
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYQLASSMNFRFPQCV-PINLKTLIPNASNEAIQLMTEMLN 266
Query: 240 ------ARCGSVTATSFFPSCSILHKVSNFLTYTEQVNTRVPRSLSDPLELKLSNKRVKP 293
+F +L SN L + +N ++ PLE K S V+P
Sbjct: 267 WDPKKRPTASQALKHPYFQVGQVLGPSSNHLESKQSLNKQL-----QPLESKPSLVEVEP 321
>ICK_HUMAN (Q9UPZ9) Serine/threonine kinase ICK (EC 2.7.1.37)
(Intestinal cell kinase) (hICK) (MAK-related kinase)
(MRK) (Laryngeal cancer kinase 2) (LCK2)
Length = 632
Score = 278 bits (711), Expect = 2e-74
Identities = 137/235 (58%), Positives = 178/235 (75%), Gaps = 2/235 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E++A+K++KRKF WEE NLRE+K+L+K+NH N++KL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELIAIKKMKRKFYSWEECMNLREVKSLKKLNHANVVKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QLIKER K F E IR M Q+LQGL+ +HK GFFHRDLKPENLL +++KIADFGLA
Sbjct: 88 QLIKERNKLFPESAIRNIMYQILQGLAFIHKHGFFHRDLKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G I+AE++TL P+FPG SEID
Sbjct: 148 REIRSKPPYTDYVSTRWYRAPEVLLRSTNYSSPIDVWAVGCIMAEVYTLRPLFPGASEID 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLI 238
++KI +LG P T + G S ++F + V P L +IPNAS EA+ L+
Sbjct: 208 TIFKICQVLGTPKKTDWPEGYQLSSAMNFRWPQCV-PNNLKTLIPNASSEAVQLL 261
>MAK_RAT (P20793) Serine/threonine-protein kinase MAK (EC 2.7.1.37)
(Male germ cell-associated kinase)
Length = 622
Score = 277 bits (708), Expect = 4e-74
Identities = 134/236 (56%), Positives = 181/236 (75%), Gaps = 2/236 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+FIFEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFIFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNIMYQILQGLAFIHKHGFFHRDMKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+T P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSVYSSPIDVWAVGSIMAELYTFRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT 239
+++KI +LG P + + G + ++F + + P+ L +IPNAS EAI L+T
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYQLASSMNFRFPQCI-PINLKTLIPNASSEAIQLMT 262
>MAK_MOUSE (Q04859) Serine/threonine-protein kinase MAK (EC
2.7.1.37) (Male germ cell-associated kinase) (Protein
kinase RCK)
Length = 622
Score = 276 bits (707), Expect = 5e-74
Identities = 133/236 (56%), Positives = 181/236 (76%), Gaps = 2/236 (0%)
Query: 5 EIVAVKRLKRKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDCNLY 64
E+VA+KR+KRKF W+E NLRE+K+L+K+NH N+IKL+EV+REN+ L+F+FEYM NLY
Sbjct: 28 ELVAIKRMKRKFYSWDECMNLREVKSLKKLNHANVIKLKEVIRENDHLYFVFEYMKENLY 87
Query: 65 QLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKIADFGLA 123
QL+K+R K F E IR M Q+LQGL+ +HK GFFHRD+KPENLL +++KIADFGLA
Sbjct: 88 QLMKDRNKLFPESVIRNIMYQILQGLAFIHKHGFFHRDMKPENLLCMGPELVKIADFGLA 147
Query: 124 REVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPGESEID 183
RE+ S PPYT YVSTRWYRAPEVLL+S Y+ +D+WA+G+I+AEL+T P+FPG SE+D
Sbjct: 148 RELRSQPPYTDYVSTRWYRAPEVLLRSSVYSSPIDVWAVGSIMAELYTFRPLFPGTSEVD 207
Query: 184 QMYKIYCILGMPDSTCFTIGANNSRLLDFVGHEVVAPVKLSDIIPNASMEAIDLIT 239
+++KI +LG P + + G + ++F + + P+ L +IPNAS EAI L+T
Sbjct: 208 EIFKICQVLGTPKKSDWPEGYQLASSMNFRFPQCI-PINLKTLIPNASSEAIQLMT 262
>MOK_HUMAN (Q9UQ07) MAPK/MAK/MRK overlapping kinase (EC 2.7.1.37)
(MOK protein kinase) (Renal tumor antigen 1) (RAGE-1)
Length = 419
Score = 196 bits (498), Expect = 8e-50
Identities = 99/215 (46%), Positives = 142/215 (66%), Gaps = 4/215 (1%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMN-HQNIIKLREVV--RENNELFFIFE 57
+R A K++K++F E+ NLREI+ALR++N H NI+ L EVV R++ L I E
Sbjct: 24 LRDGNYYACKQMKQRFESIEQVNNLREIQALRRLNPHPNILMLHEVVFDRKSGSLALICE 83
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKI 117
MD N+Y+LI+ R P SE++I +M Q+ + L H+H+ G FHRD+KPEN+L+ DVLK+
Sbjct: 84 LMDMNIYELIRGRRYPLSEKKIMHYMYQLCKSLDHIHRNGIFHRDVKPENILIKQDVLKL 143
Query: 118 ADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFP 177
DFG R V S PYT+Y+STRWYRAPE LL YT +D+W+ G + E+ +L P+FP
Sbjct: 144 GDFGSCRSVYSKQPYTEYISTRWYRAPECLLTDGFYTYKMDLWSAGCVFYEIASLQPLFP 203
Query: 178 GESEIDQMYKIYCILGMPDSTCFTIGANNSRLLDF 212
G +E+DQ+ KI+ ++G P T SR ++F
Sbjct: 204 GVNELDQISKIHDVIGTPAQKILT-KFKQSRAMNF 237
>MOK_MOUSE (Q9WVS4) MAPK/MAK/MRK overlapping kinase (EC 2.7.1.37)
(MOK protein kinase)
Length = 420
Score = 195 bits (496), Expect = 1e-49
Identities = 96/198 (48%), Positives = 135/198 (67%), Gaps = 3/198 (1%)
Query: 1 MRTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMN-HQNIIKLREVV--RENNELFFIFE 57
+R A K++K+ F E+ +LREI+ALR++N H NI+ L EVV R++ L I E
Sbjct: 24 LRDGNYYACKQMKQHFESIEQVNSLREIQALRRLNPHPNILALHEVVFDRKSGSLALICE 83
Query: 58 YMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTNDVLKI 117
MD N+Y+LI+ R P SE++I +M Q+ + L HMH+ G FHRD+KPEN+LV DVLK+
Sbjct: 84 LMDMNIYELIRGRRHPLSEKKIMLYMYQLCKSLDHMHRNGIFHRDVKPENILVKQDVLKL 143
Query: 118 ADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFP 177
DFG R V S PYT+Y+STRWYRAPE LL YT +D+W+ G + E+ +L P+FP
Sbjct: 144 GDFGSCRSVYSKQPYTEYISTRWYRAPECLLTDGFYTYKMDLWSAGCVFYEIASLQPLFP 203
Query: 178 GESEIDQMYKIYCILGMP 195
G +E+DQ+ KI+ ++G P
Sbjct: 204 GVNELDQISKIHDVIGTP 221
>STK9_HUMAN (O76039) Serine/threonine-protein kinase 9 (EC 2.7.1.37)
(Cyclin-dependent kinase-like 5)
Length = 1030
Score = 173 bits (439), Expect = 6e-43
Identities = 91/210 (43%), Positives = 138/210 (65%), Gaps = 5/210 (2%)
Query: 3 TFEIVAVKRLKRKFCFWE-EYTNLREIKALRKMNHQNIIKLREVVRENNELFFIFEYMDC 61
T EIVA+K+ K E + T LRE+K LR + +NI++L+E R +L+ +FEY++
Sbjct: 35 THEIVAIKKFKDSEENEEVKETTLRELKMLRTLKQENIVELKEAFRRRGKLYLVFEYVEK 94
Query: 62 NLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVLKIADF 120
N+ +L++E E+++ ++ Q+++ + HK HRD+KPENLL++ NDVLK+ DF
Sbjct: 95 NMLELLEEMPNGVPPEKVKSYIYQLIKAIHWCHKNDIVHRDIKPENLLISHNDVLKLCDF 154
Query: 121 GLAREVS--SMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIFPG 178
G AR +S + YT+YV+TRWYR+PE+LL +P Y +VDMW++G IL EL P+FPG
Sbjct: 155 GFARNLSEGNNANYTEYVATRWYRSPELLLGAP-YGKSVDMWSVGCILGELSDGQPLFPG 213
Query: 179 ESEIDQMYKIYCILGMPDSTCFTIGANNSR 208
ESEIDQ++ I +LG S + +N R
Sbjct: 214 ESEIDQLFTIQKVLGPLPSEQMKLFYSNPR 243
>IME2_YEAST (P32581) Meiosis induction protein kinase IME2/SME1 (EC
2.7.1.-)
Length = 645
Score = 169 bits (427), Expect = 1e-41
Identities = 90/216 (41%), Positives = 134/216 (61%), Gaps = 22/216 (10%)
Query: 2 RTFEIVAVKRLKRKFCFWEEYTNLREIKALRKMN-HQNIIKLREVV--RENNELFFIFEY 58
+T +VA+K + K ++YT +REIK + + + ++I++ EV EN +L + E
Sbjct: 89 KTQGVVAIKTMMTKLHTLQDYTRVREIKFILAIPANDHLIQIFEVFIDSENYQLHIVMEC 148
Query: 59 MDCNLYQLIKEREKP-FSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT------ 111
M+ NLYQ++K R + FS ++ + Q+L GL H+H+ FFHRDLKPEN+L+T
Sbjct: 149 MEQNLYQMMKHRRRRVFSIPSLKSILSQILAGLKHIHEHNFFHRDLKPENILITPSTQYF 208
Query: 112 ------------NDVLKIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDM 159
N V+K+ADFGLAR V + PYT YVSTRWYR+PE+LL+S Y+ +D+
Sbjct: 209 EKEYMNQIGYQDNYVIKLADFGLARHVENKNPYTAYVSTRWYRSPEILLRSGYYSKPLDI 268
Query: 160 WAIGAILAELFTLTPIFPGESEIDQMYKIYCILGMP 195
WA G + E+ +FPG +EIDQ++KI +LG P
Sbjct: 269 WAFGCVAVEVTVFRALFPGANEIDQIWKILEVLGTP 304
>CDL2_HUMAN (Q9UQ88) PITSLRE serine/threonine-protein kinase CDC2L2
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 2) (CDK11)
Length = 780
Score = 165 bits (417), Expect = 2e-40
Identities = 89/199 (44%), Positives = 130/199 (64%), Gaps = 5/199 (2%)
Query: 2 RTFEIVAVKRLK-RKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFEY 58
+T EIVA+KRLK K T+LREI + K H NI+ +RE+V +N +++ + Y
Sbjct: 444 KTDEIVALKRLKMEKEKEGFPITSLREINTILKAQHPNIVTVREIVVGSNMDKIYIVMNY 503
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKI 117
++ +L L++ ++PF E++ M Q+L+G+ H+H HRDLK NLL+++ +LK+
Sbjct: 504 VEHDLKSLMETMKQPFLPGEVKTLMIQLLRGVKHLHDNWILHRDLKTSNLLLSHAGILKV 563
Query: 118 ADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIF 176
DFGLARE S + YT V T+WYRAPE+LL + Y+ AVDMW++G I EL T P+F
Sbjct: 564 GDFGLAREYGSPLKAYTPVVVTQWYRAPELLLGAKEYSTAVDMWSVGCIFGELLTQKPLF 623
Query: 177 PGESEIDQMYKIYCILGMP 195
PG SEIDQ+ K++ LG P
Sbjct: 624 PGNSEIDQINKVFKELGTP 642
>CDL1_HUMAN (P21127) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1) (CLK-1)
(CDK11) (p58 CLK-1)
Length = 795
Score = 164 bits (415), Expect = 3e-40
Identities = 89/199 (44%), Positives = 130/199 (64%), Gaps = 5/199 (2%)
Query: 2 RTFEIVAVKRLK-RKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFEY 58
+T EIVA+KRLK K T+LREI + K H NI+ +RE+V +N +++ + Y
Sbjct: 459 KTDEIVALKRLKMEKEKEGFPITSLREINTILKAQHPNIVTVREIVVGSNMDKIYIVMNY 518
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKI 117
++ +L L++ ++PF E++ M Q+L+G+ H+H HRDLK NLL+++ +LK+
Sbjct: 519 VEHDLKSLMETMKQPFLPGEVKTLMIQLLRGVKHLHDNWILHRDLKTSNLLLSHAGILKV 578
Query: 118 ADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIF 176
DFGLARE S + YT V T WYRAPE+LL + Y+ AVDMW++G I EL T P+F
Sbjct: 579 GDFGLAREYGSPLKAYTPVVVTLWYRAPELLLGAKEYSTAVDMWSVGCIFGELLTQKPLF 638
Query: 177 PGESEIDQMYKIYCILGMP 195
PG+SEIDQ+ K++ LG P
Sbjct: 639 PGKSEIDQINKVFKDLGTP 657
>KP58_DROME (Q9VPC0) Serine/threonine-protein kinase PITSLRE (EC
2.7.1.37) (Cell division cycle 2-like)
Length = 952
Score = 163 bits (413), Expect = 6e-40
Identities = 89/203 (43%), Positives = 133/203 (64%), Gaps = 8/203 (3%)
Query: 2 RTFEIVAVKRLK-RKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFEY 58
RT EIVA+KRLK K T+LREI L K H NI+ +RE+V +N ++F + +Y
Sbjct: 579 RTNEIVALKRLKMEKEKEGFPITSLREINTLLKGQHPNIVTVREIVVGSNMDKIFIVMDY 638
Query: 59 MDCNLYQLI---KEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTND-V 114
++ +L L+ K R++ F E++C +Q+L+ ++H+H HRDLK NLL+++ +
Sbjct: 639 VEHDLKSLMETMKNRKQSFFPGEVKCLTQQLLRAVAHLHDNWILHRDLKTSNLLLSHKGI 698
Query: 115 LKIADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLT 173
LK+ DFGLARE S + YT V T WYRAPE+LL SP Y+ +D+W++G I AE +
Sbjct: 699 LKVGDFGLAREYGSPIKKYTSLVVTLWYRAPELLLCSPVYSTPIDVWSVGCIFAEFLQML 758
Query: 174 PIFPGESEIDQMYKIYCILGMPD 196
P+FPG+SEID++ +I+ LG P+
Sbjct: 759 PLFPGKSEIDELNRIFKELGTPN 781
>MPK4_ARATH (Q39024) Mitogen-activated protein kinase homolog 4 (EC
2.7.1.37) (MAP kinase 4) (AtMPK4)
Length = 376
Score = 163 bits (412), Expect = 8e-40
Identities = 96/245 (39%), Positives = 150/245 (61%), Gaps = 12/245 (4%)
Query: 3 TFEIVAVKRLKRKFC-FWEEYTNLREIKALRKMNHQNIIKLREVV----REN-NELFFIF 56
T E VA+K++ F + LREIK L+ M+H+N+I +++++ REN N+++ ++
Sbjct: 65 TGEEVAIKKIGNAFDNIIDAKRTLREIKLLKHMDHENVIAVKDIIKPPQRENFNDVYIVY 124
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + P +++ R F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 125 ELMDTDLHQIIRSNQ-PLTDDHCRFFLYQLLRGLKYVHSANVLHRDLKPSNLLLNANCDL 183
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
K+ DFGLAR S T+YV TRWYRAPE+LL YT A+D+W++G IL E T P+
Sbjct: 184 KLGDFGLARTKSETDFMTEYVVTRWYRAPELLLNCSEYTAAIDIWSVGCILGETMTREPL 243
Query: 176 FPGESEIDQMYKIYCILGMP-DSTCFTIGANNSRLLDFVGHEVVAP-VKLSDIIPNASME 233
FPG+ + Q+ I ++G P DS+ + ++N+R +V P + PN S
Sbjct: 244 FPGKDYVHQLRLITELIGSPDDSSLGFLRSDNAR--RYVRQLPQYPRQNFAARFPNMSAG 301
Query: 234 AIDLI 238
A+DL+
Sbjct: 302 AVDLL 306
>CDL1_MOUSE (P24788) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1)
Length = 434
Score = 162 bits (411), Expect = 1e-39
Identities = 89/199 (44%), Positives = 128/199 (63%), Gaps = 5/199 (2%)
Query: 2 RTFEIVAVKRLK-RKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFEY 58
+T EIVA+KRLK K T+LREI + K H NI+ +RE+V +N +++ + Y
Sbjct: 98 KTDEIVALKRLKMEKEKEGFPITSLREINTILKAQHPNIVTVREIVVGSNMDKIYIVMNY 157
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKI 117
++ +L L++ ++PF E++ M Q+L G+ H+H HRDLK NLL+T+ +LK+
Sbjct: 158 VEHDLKSLMETMKQPFLPGEVKTLMIQLLSGVKHLHDNWILHRDLKTSNLLLTHAGILKV 217
Query: 118 ADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIF 176
DFGLARE S + YT V T WYRAPE+LL + Y+ A DMW++G I EL T P+F
Sbjct: 218 GDFGLAREYGSPLKAYTPVVVTLWYRAPELLLGAKEYSTACDMWSVGCIFGELLTQKPLF 277
Query: 177 PGESEIDQMYKIYCILGMP 195
PG+S+IDQ+ KI+ LG P
Sbjct: 278 PGKSDIDQINKIFKDLGSP 296
>NTF6_TOBAC (Q40531) Mitogen-activated protein kinase homolog NTF6
(EC 2.7.1.37) (P43)
Length = 371
Score = 162 bits (410), Expect = 1e-39
Identities = 99/244 (40%), Positives = 145/244 (58%), Gaps = 12/244 (4%)
Query: 3 TFEIVAVKRLKRKF-CFWEEYTNLREIKALRKMNHQNIIKLREVVR-----ENNELFFIF 56
T E VA+K++ F + LREIK L M+H+NIIK++++VR E N+++ ++
Sbjct: 60 TKEEVAIKKIGNAFENRIDAKRTLREIKLLSHMDHENIIKIKDIVRPPDREEFNDVYIVY 119
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + +++ + F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 120 ELMDTDLHQIIRSSQA-LTDDHCQYFLYQLLRGLKYVHSANVLHRDLKPSNLLLNANCDL 178
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL YT A+D+W++G IL EL P+
Sbjct: 179 KICDFGLARTTSEADFMTEYVVTRWYRAPELLLNCTEYTAAIDIWSVGCILMELIKREPL 238
Query: 176 FPGESEIDQMYKIYCILGMP-DSTCFTIGANNSRLLDFVGHEVVAP-VKLSDIIPNASME 233
FPG Q+ I +LG P DS + ++N+R +V H P S P+ S
Sbjct: 239 FPGRDYAQQLGLIIALLGSPEDSDLGFLRSDNAR--KYVKHLPRVPRHPFSQKFPDVSPL 296
Query: 234 AIDL 237
A+DL
Sbjct: 297 ALDL 300
>MMK2_MEDSA (Q40353) Mitogen-activated protein kinase homolog MMK2
(EC 2.7.1.37)
Length = 371
Score = 161 bits (408), Expect = 2e-39
Identities = 96/245 (39%), Positives = 146/245 (59%), Gaps = 12/245 (4%)
Query: 3 TFEIVAVKRLKRKFCFW-EEYTNLREIKALRKMNHQNIIKLREVVR----EN-NELFFIF 56
T E VA+K++ F + LREIK LR M+H+N++ +++++R EN N ++ +
Sbjct: 59 TREEVAIKKIGNAFDNRIDAKRTLREIKLLRHMDHENVMSIKDIIRPPQKENFNHVYIVS 118
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + P +++ R F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 119 ELMDTDLHQIIRSNQ-PMTDDHCRYFVYQLLRGLKYVHSANVLHRDLKPSNLLLNANCDL 177
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL YT A+D+W++G IL E+ T P+
Sbjct: 178 KIGDFGLARTTSETDFMTEYVVTRWYRAPELLLNCSDYTAAIDIWSVGCILGEIVTRQPL 237
Query: 176 FPGESEIDQMYKIYCILGMPDSTCF-TIGANNSRLLDFVGHEVVAPVK-LSDIIPNASME 233
FPG + Q+ + ++G PD + + N+R +V P + S PN S
Sbjct: 238 FPGRDYVHQLRLVTELIGSPDDASLGFLRSENAR--RYVRQLPQYPKQNFSARFPNMSPG 295
Query: 234 AIDLI 238
A+DL+
Sbjct: 296 AVDLL 300
>MPK3_ARATH (Q39023) Mitogen-activated protein kinase homolog 3 (EC
2.7.1.37) (MAP kinase 3) (AtMPK3)
Length = 370
Score = 161 bits (407), Expect = 3e-39
Identities = 88/200 (44%), Positives = 126/200 (63%), Gaps = 8/200 (4%)
Query: 3 TFEIVAVKRLKRKFC-FWEEYTNLREIKALRKMNHQNIIKLREVV-----RENNELFFIF 56
T E+VA+K++ F + LREIK LR ++H+NII +R+VV R+ ++++
Sbjct: 60 TNELVAMKKIANAFDNHMDAKRTLREIKLLRHLDHENIIAIRDVVPPPLRRQFSDVYIST 119
Query: 57 EYMDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVT-NDVL 115
E MD +L+Q+I+ + SEE + F+ Q+L+GL ++H HRDLKP NLL+ N L
Sbjct: 120 ELMDTDLHQIIRSNQS-LSEEHCQYFLYQLLRGLKYIHSANIIHRDLKPSNLLLNANCDL 178
Query: 116 KIADFGLAREVSSMPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPI 175
KI DFGLAR S T+YV TRWYRAPE+LL S YT A+D+W++G I EL P+
Sbjct: 179 KICDFGLARPTSENDFMTEYVVTRWYRAPELLLNSSDYTAAIDVWSVGCIFMELMNRKPL 238
Query: 176 FPGESEIDQMYKIYCILGMP 195
FPG+ + QM + +LG P
Sbjct: 239 FPGKDHVHQMRLLTELLGTP 258
>CDL1_RAT (P46892) PITSLRE serine/threonine-protein kinase CDC2L1
(EC 2.7.1.37) (Galactosyltransferase associated protein
kinase p58/GTA) (Cell division cycle 2-like 1)
Length = 436
Score = 161 bits (407), Expect = 3e-39
Identities = 87/199 (43%), Positives = 128/199 (63%), Gaps = 5/199 (2%)
Query: 2 RTFEIVAVKRLK-RKFCFWEEYTNLREIKALRKMNHQNIIKLREVVRENN--ELFFIFEY 58
+T EIVA+KRLK K T++REI + K H NI+ +RE+V +N +++ + Y
Sbjct: 100 KTDEIVALKRLKMEKEKEGFPLTSIREINTILKAQHPNIVTVREIVVGSNMDKIYIVMNY 159
Query: 59 MDCNLYQLIKEREKPFSEEEIRCFMKQMLQGLSHMHKKGFFHRDLKPENLLVTN-DVLKI 117
++ +L L++ ++PF E++ M Q+L G+ H+H HRDLK NLL+T+ +LK+
Sbjct: 160 VEHDLKSLMETMKQPFLPGEVKTLMIQLLSGVKHLHDNWILHRDLKTSNLLLTHAGILKV 219
Query: 118 ADFGLAREVSS-MPPYTQYVSTRWYRAPEVLLQSPCYTPAVDMWAIGAILAELFTLTPIF 176
DFGLARE S + YT V T WYRAPE+LL + Y+ A DMW++G I EL T P+F
Sbjct: 220 GDFGLAREYGSPLKAYTPVVVTLWYRAPELLLGAKEYSTACDMWSVGCIFGELLTQKPLF 279
Query: 177 PGESEIDQMYKIYCILGMP 195
PG+S+IDQ+ KI+ +G P
Sbjct: 280 PGKSDIDQINKIFKDIGTP 298
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.326 0.140 0.423
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 42,869,757
Number of Sequences: 164201
Number of extensions: 1768909
Number of successful extensions: 9543
Number of sequences better than 10.0: 1667
Number of HSP's better than 10.0 without gapping: 1216
Number of HSP's successfully gapped in prelim test: 451
Number of HSP's that attempted gapping in prelim test: 5390
Number of HSP's gapped (non-prelim): 2048
length of query: 381
length of database: 59,974,054
effective HSP length: 112
effective length of query: 269
effective length of database: 41,583,542
effective search space: 11185972798
effective search space used: 11185972798
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC136841.7


