

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136840.7 - phase: 0 /pseudo
(638 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
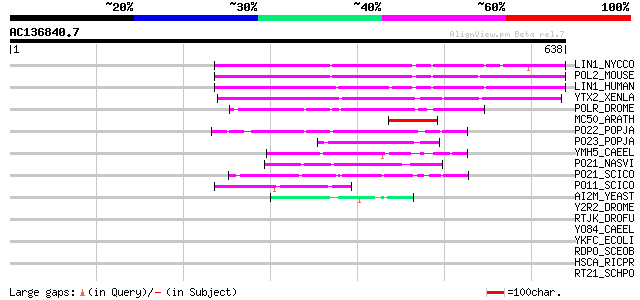
Score E
Sequences producing significant alignments: (bits) Value
LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog 149 3e-35
POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains... 144 9e-34
LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog 139 3e-32
YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein ... 119 2e-26
POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type... 89 3e-17
MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250... 74 2e-12
PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type... 71 8e-12
PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type... 69 3e-11
YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III 66 3e-10
PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type... 62 6e-09
PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type... 57 1e-07
PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type... 55 7e-07
AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein 47 2e-04
Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I retro... 42 0.006
RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile elem... 37 0.16
YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III 36 0.35
YKFC_ECOLI (Q47688) Hypothetical protein ykfC 34 1.3
RDPO_SCEOB (P19593) Probable reverse transcriptase (EC 2.7.7.49) 33 1.8
HSCA_RICPR (Q9ZDW5) Chaperone protein hscA homolog 32 5.1
RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protei... 32 6.7
>LIN1_NYCCO (P08548) LINE-1 reverse transcriptase homolog
Length = 1260
Score = 149 bits (375), Expect = 3e-35
Identities = 119/410 (29%), Positives = 193/410 (47%), Gaps = 16/410 (3%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
++ ++ + + + K+PGPDGF F++ + + + NL + + +
Sbjct: 452 ISSSEIASTIQNLPKKKSPGPDGFTSEFYQTFKEELVPILLNLFQNIEKEGILPNTFYEA 511
Query: 296 LIALIPKVDPPLTYKD-FRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRST 354
I LIPK T K+ +RPISL NI KI+ K+L +R++ + II Q F+PG
Sbjct: 512 NITLIPKPGKDPTRKENYRPISLMNIDAKILNKILTNRIQQHIKKIIHHDQVGFIPGSQG 571
Query: 355 TDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
N VI ++ K K ++ ID EKAFDN+ F+ L G T +KLI
Sbjct: 572 WFNIRKSINVIQHIN-KLKNKDHMILSIDAEKAFDNIQHPFMIRTLKKIGIEGTFLKLIE 630
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
+ + +I+ NG +L F G RQG PLSP LF + ME L+ AI ++ + + I
Sbjct: 631 AIYSKPTANIILNGVKLKSFPLRSGTRQGCPLSPLLFNIVMEVLAIAIR---EEKAIKGI 687
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGV 534
H+ + KLS LFADD++++ + + ++ +S SG KIN HKS AF +
Sbjct: 688 HIGSEEIKLS--LFADDMIVYLENTRDSTTKLLEVIKEYSNVSGYKINTHKSVAFIYTNN 745
Query: 535 TQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQKRLASWQNKLLNKP 594
Q + SI KYLG + K K + + ++K +A NK N P
Sbjct: 746 NQAEKTVKDSIP-FTVVPKKMKYLGVYLTK--DVKDLYKENYETLRKEIAEDVNKWKNIP 802
Query: 595 ----GRLALASFVLSSISTYYMQINWL--PQSVCDNIDQVTRNFIWRDSR 638
GR+ + + + Y + P S +++++ +FIW +
Sbjct: 803 CSWLGRINIVKMSILPKAIYNFNAIPIKAPLSYFKDLEKIILHFIWNQKK 852
>POL2_MOUSE (P11369) Retrovirus-related Pol polyprotein [Contains:
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1300
Score = 144 bits (362), Expect = 9e-34
Identities = 113/409 (27%), Positives = 187/409 (45%), Gaps = 14/409 (3%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
++ K++ A ++S K+PGPDGF F++ + + + L + S +
Sbjct: 479 ISPKEIEAVINSLPTKKSPGPDGFSAEFYQTFKEDLIPILHKLFHKIEVEGTLPNSFYEA 538
Query: 296 LIALIPKVDP-PLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRST 354
I LIPK P ++FRPISL NI KI+ K+L +R++ + II P Q F+PG
Sbjct: 539 TITLIPKPQKDPTKIENFRPISLMNIDAKILNKILANRIQEHIKAIIHPDQVGFIPGMQG 598
Query: 355 TDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
N VIHY+ K K ++ +D EKAFD + F+ L G + +I
Sbjct: 599 WFNIRKSINVIHYIN-KLKDKNHMIISLDAEKAFDKIQHPFMIKVLERSGIQGPYLNMIK 657
Query: 415 HCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPI 474
+ +I NG +L G RQG PLSPYLF + +E L+ AI QQ + I
Sbjct: 658 AIYSKPVANIKVNGEKLEAIPLKSGTRQGCPLSPYLFNIVLEVLARAIR---QQKEIKGI 714
Query: 475 HVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAF-YSSG 533
+ K+S L ADD++++ R + +L + F + G KIN +KS AF Y+
Sbjct: 715 QIGKEEVKIS--LLADDMIVYISDPKNSTRELLNLINSFGEVVGYKINSNKSMAFLYTKN 772
Query: 534 VTQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPKKSD--FLFIIDKMQKRLASWQNKLL 591
+K + + T+ KYLG + K D F + ++++ L W++
Sbjct: 773 KQAEK--EIRETTPFSIVTNNIKYLGVTLTKEVKDLYDKNFKSLKKEIKEDLRRWKDLPC 830
Query: 592 NKPGRLALASFVLSSISTYYMQI--NWLPQSVCDNIDQVTRNFIWRDSR 638
+ GR+ + + + Y +P + ++ F+W + +
Sbjct: 831 SWIGRINIVKMAILPKAIYRFNAIPIKIPTQFFNELEGAICKFVWNNKK 879
>LIN1_HUMAN (P08547) LINE-1 reverse transcriptase homolog
Length = 1259
Score = 139 bits (349), Expect = 3e-32
Identities = 112/410 (27%), Positives = 191/410 (46%), Gaps = 16/410 (3%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
+ ++ A ++S K+PGP+GF F+++Y + + L S S +
Sbjct: 452 ITSSEIEAIINSLPNKKSPGPEGFTAEFYQRYKEELVPFLLKLFQSIEKEGILPNSFYEA 511
Query: 296 LIALIPKVDPPLTYKD-FRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRST 354
I LIPK T K+ FRPISL NI KI+ K+L ++++ + +I Q F+P
Sbjct: 512 SIILIPKPGRDTTKKENFRPISLMNIDAKILNKILANQIQQHIKKLIHHDQVGFIPAMQG 571
Query: 355 TDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIM 414
N +I ++ +K ++ ID EKAFD + F+ L G T +K+I
Sbjct: 572 WFNIRKSINIIQHINRTKDTN-HMIISIDAEKAFDKIQQPFMLKPLNKLGIDGTYLKIIR 630
Query: 415 HCVTSSSFSILWNGNRL--PPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWE 472
+ +I+ NG +L PP + G RQG PLSP L + +E L+ AI Q+ +
Sbjct: 631 AIYDKPTANIILNGQKLEAPPLKT--GTRQGCPLSPLLPNIVLEVLARAIR---QEKEIK 685
Query: 473 PIHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSS 532
I + KLS LFADD++++ + + + L FSK SG KIN+ KS+AF +
Sbjct: 686 GIQLGKEEVKLS--LFADDMIVYLENPIVSAQNLLKLISNFSKVSGYKINVQKSQAFLYT 743
Query: 533 GVTQDKINTLTSISGIRCTTSLDKYLGFPIIKGRPK--KSDFLFIIDKMQKRLASWQNKL 590
Q + ++ + + KYLG + + K ++ +++++++ W+N
Sbjct: 744 NNRQTESQIMSELP-FTIASKRIKYLGIQLTRDVKDLFKENYKPLLNEIKEDTNKWKNIP 802
Query: 591 LNKPGRLALASFVLSSISTYYMQI--NWLPQSVCDNIDQVTRNFIWRDSR 638
+ GR+ + + Y LP + +++ T FIW R
Sbjct: 803 CSWVGRINIVKMAILPKVIYRFNAIPIKLPMTFFTELEKTTLKFIWNQKR 852
>YTX2_XENLA (P14381) Transposon TX1 hypothetical 149 kDa protein
(ORF 2)
Length = 1308
Score = 119 bits (298), Expect = 2e-26
Identities = 96/398 (24%), Positives = 182/398 (45%), Gaps = 13/398 (3%)
Query: 240 KVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIAL 299
+++ AL K+PG DG FF+ + +G D + T F S +++L
Sbjct: 453 ELSQALRLMPHNKSPGLDGLTIEFFQFFWDTLGPDFHRVLTEAFKKGELPLSCRRAVLSL 512
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
+PK K++RP+SL + YKI+ K + RL+ +L +I P QS +PGR+ DN
Sbjct: 513 LPKKGDLRLIKNWRPVSLLSTDYKIVAKAISLRLKSVLAEVIHPDQSYTVPGRTIFDNVF 572
Query: 360 VLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTS 419
++++++H+ R + +++ +D EKAFD V+ +L L + F + + S
Sbjct: 573 LIRDLLHFARRTGLSLAFLS--LDQEKAFDRVDHQYLIGTLQAYSFGPQFVGYLKTMYAS 630
Query: 420 SSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNP 479
+ + N + P G+RQG PLS L+ L +E + + + + P
Sbjct: 631 AECLVKINWSLTAPLAFGRGVRQGCPLSGQLYSLAIEPFLCLLRKRL-----TGLVLKEP 685
Query: 480 GPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKI 539
++ +ADDV+L + + L ++ + ++ S +IN KS + D +
Sbjct: 686 DMRVVLSAYADDVILVAQ-DLVDLERAQECQEVYAAASSARINWSKSSGLLEGSLKVDFL 744
Query: 540 NTLTSISGIRCTTSLDKYLG-FPIIKGRPKKSDFLFIIDKMQKRLASWQN--KLLNKPGR 596
+ I + + KYLG + + P +F+ + + + RL W+ K+L+ GR
Sbjct: 745 P--PAFRDISWESKIIKYLGVYLSAEEYPVSQNFIELEECVLTRLGKWKGFAKVLSMRGR 802
Query: 597 LALASFVLSSISTYYMQINWLPQSVCDNIDQVTRNFIW 634
+ + +++S Y + Q I + +F+W
Sbjct: 803 ALVINQLVASQIWYRLICLSPTQEFIAKIQRRLLDFLW 840
>POLR_DROME (P16423) Retrovirus-related Pol polyprotein from type II
retrotransposable element R2DM [Contains: Protease (EC
3.4.23.-); Reverse transcriptase (EC 2.7.7.49);
Endonuclease]
Length = 1057
Score = 89.4 bits (220), Expect = 3e-17
Identities = 78/294 (26%), Positives = 128/294 (43%), Gaps = 16/294 (5%)
Query: 253 APGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKDF 312
+PGPDG I K + + + + SI IPK +DF
Sbjct: 359 SPGPDG---ITPKSAREVPSGIMLRIMNLILWCGNLPHSIRLARTVFIPKTVTAKRPQDF 415
Query: 313 RPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRISK 372
RPIS+ +++ + + +L RL +N P Q FLP DNA ++ V+ +
Sbjct: 416 RPISVPSVLVRQLNAILATRLNSSIN--WDPRQRGFLPTDGCADNATIVDLVLRHSH-KH 472
Query: 373 KKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNRLP 432
+ Y+A +D+ KAFD+++ + L +G P + + + S+ +G
Sbjct: 473 FRSCYIA-NLDVSKAFDSLSHASIYDTLRAYGAPKGFVDYVQNTYEGGGTSLNGDGWSSE 531
Query: 433 PFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFADDV 492
F P G++QGDPLSP LF L M++L + + + G+ ++N FADD+
Sbjct: 532 EFVPARGVKQGDPLSPILFNLVMDRLLRTLPSEI--GAKVGNAITNAA------AFADDL 583
Query: 493 LLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKINTLTSIS 546
+LF + L+ + D F GLK+N K G + K L + S
Sbjct: 584 VLFAETR-MGLQVLLDKTLDFLSIVGLKLNADKCFTVGIKGQPKQKCTVLEAQS 636
>MC50_ARATH (P92555) Hypothetical mitochondrial protein AtMg01250
(ORF102)
Length = 122
Score = 73.6 bits (179), Expect = 2e-12
Identities = 35/56 (62%), Positives = 40/56 (70%)
Query: 436 PTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFADD 491
P+ GLRQGDPLSPYLFILC E LS A +QG I VSN P+++HLLFADD
Sbjct: 24 PSRGLRQGDPLSPYLFILCTEVLSGLCRRAQEQGRLPGIRVSNNSPRINHLLFADD 79
>PO22_POPJA (Q03274) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 711
Score = 71.2 bits (173), Expect = 8e-12
Identities = 72/297 (24%), Positives = 128/297 (42%), Gaps = 22/297 (7%)
Query: 233 Y*ALNKKKVTAALHSTKPYKAPGPDGF--HCIFFKQYSHIVGDDVFNLATSTFLTSHFDP 290
Y + ++++ A+ KP APG DG I + N L H
Sbjct: 3 YRPIAREEIQCAIKGWKP-SAPGSDGLTVQAITRTRLPR-------NFVQLHLLRGHVPT 54
Query: 291 SISDTLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLP 350
+ LIPK ++RPI++ + + +++ ++L RL + + P Q +
Sbjct: 55 PWTAMRTTLIPKDGDLENPSNWRPITIASALQRLLHRILAKRLEAAVE--LHPAQKGYAR 112
Query: 351 GRSTTDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTI 410
T N+++L I R +++K Y +D+ KAFD V+ + L G + T
Sbjct: 113 IDGTLVNSLLLDTYISSRR--EQRKTYNVVSLDVRKAFDTVSHSSICRALQRLGIDEGTS 170
Query: 411 KLIMHCVTSSSFSI-LWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQG 469
I ++ S+ +I + G++ G++QGDPLSP+LF +++L ++ + G
Sbjct: 171 NYITGSLSDSTTTIRVGPGSQTRKICIRRGVKQGDPLSPFLFNAVLDELLCSLQSTPGIG 230
Query: 470 SWEPIHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKS 526
+ K+ L FADD+LL + N L F + G+ +N KS
Sbjct: 231 G------TIGEEKIPVLAFADDLLLL-EDNDVLLPTTLATVANFFRLRGMSLNAKKS 280
>PO23_POPJA (Q05118) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 606
Score = 69.3 bits (168), Expect = 3e-11
Identities = 45/142 (31%), Positives = 74/142 (51%), Gaps = 10/142 (7%)
Query: 354 TTDNAIVLQEVIHYMRISKKK-KGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKL 412
T N I+L+ HY+++ + K K Y +D+ KAFD V+ + + FG D
Sbjct: 2 TLANIIMLE---HYIKLRRLKGKTYNVVSLDIRKAFDTVSHPAILRAMRAFGIDDGMQDF 58
Query: 413 IMHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWE 472
IM +T + +I+ G +G++QGDPLSP LF + +++L + +N+ +
Sbjct: 59 IMSTITDAYTNIVVGGRTTNKIYIRNGVKQGDPLSPVLFNIVLDELVTRLNDE------Q 112
Query: 473 PIHVSNPGPKLSHLLFADDVLL 494
P P K++ L FADD+LL
Sbjct: 113 PGASMTPACKIASLAFADDLLL 134
>YMH5_CAEEL (P34472) Hypothetical protein F58A4.5 in chromosome III
Length = 1222
Score = 65.9 bits (159), Expect = 3e-10
Identities = 68/236 (28%), Positives = 104/236 (43%), Gaps = 29/236 (12%)
Query: 296 LIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTT 355
+I IPK P + ++RPISL + +I+ +++ R+R ++++ P+Q FL RS
Sbjct: 669 VIIPIPKKGNPSSPSNYRPISLTDPFARIMERIICSRIRSEYSHLLSPHQHGFLNFRSCP 728
Query: 356 DNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMH 415
+ +++ + Y I K +K D KAFD V+ L L FG T
Sbjct: 729 SS--LVRSISLYHSILKNEKSLDILFFDFAKAFDKVSHPILLKKLALFGLDKLTCSWFKE 786
Query: 416 CVTSSSFSILWN----GNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSW 471
+ +FS+ N N P + G+ QG P LFIL + L +
Sbjct: 787 FLHLRTFSVKINKFVSSNAYP---ISSGVPQGSVSGPLLFILFINDLLIDL--------- 834
Query: 472 EP-IHVSNPGPKLSHLLFADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKS 526
EP IHVS FADD+ +F ST I D ++SK + L + KS
Sbjct: 835 EPNIHVS---------CFADDIKIFHHNPSTLQNSI-DTIVKWSKKNKLPLAPAKS 880
>PO21_NASVI (Q03278) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1025
Score = 61.6 bits (148), Expect = 6e-09
Identities = 53/204 (25%), Positives = 91/204 (43%), Gaps = 12/204 (5%)
Query: 294 DTLIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRS 353
D+ LIPK + FRP+S+ ++ + ++L +R+ + ++ Q +F+
Sbjct: 371 DSRTVLIPKEPGTMDPACFRPLSIASVALRHFHRILANRIGE--HGLLDTRQRAFIVADG 428
Query: 354 TTDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLI 413
+N +L +I R+ K KG +D++KAFD+V + L P I
Sbjct: 429 VAENTSLLSAMIKEARM--KIKGLYIAILDVKKAFDSVEHRSILDALRRKKLPLEMRNYI 486
Query: 414 MHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEP 473
M +S + + P G+RQGDPLSP LF + + +AV + E
Sbjct: 487 MWVYRNSKTRLEVVKTKGRWIRPARGVRQGDPLSPLLF--------NCVMDAVLRRLPEN 538
Query: 474 IHVSNPGPKLSHLLFADDVLLFTK 497
K+ L+FADD++L +
Sbjct: 539 TGFLMGAEKIGALVFADDLVLLAE 562
>PO21_SCICO (Q03279) Retrovirus-related Pol polyprotein from type I
retrotransposable element R2 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 869
Score = 57.4 bits (137), Expect = 1e-07
Identities = 71/277 (25%), Positives = 119/277 (42%), Gaps = 19/277 (6%)
Query: 252 KAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPS-ISDTLIALIPKVDPPLTYK 310
K GPDG + + + D L + F+ PS I + PK++
Sbjct: 173 KGAGPDGVT----PRSWNALDDRYKRLLYNIFVFYGRVPSPIKGSRTVFTPKIEGGPDPG 228
Query: 311 DFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRI 370
FRP+S+C++I + K+L R + Q+++LP N +L +I +
Sbjct: 229 VFRPLSICSVILREFNKILARRF--VSCYTYDERQTAYLPIDGVCINVSMLTAIIAEAKR 286
Query: 371 SKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNR 430
+K+ ++A +DL KAF++V L + + G P + I + + + G +
Sbjct: 287 LRKEL-HIAI-LDLVKAFNSVYHSALIDAITEAGCPPGVVDYIADMYNNVITEMQFEG-K 343
Query: 431 LPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFAD 490
G+ QGDPLS LF L EK A+NN +G ++ V +++ ++D
Sbjct: 344 CELASILAGVYQGDPLSGPLFTLAYEKALRALNN---EGRFDIADV-----RVNASAYSD 395
Query: 491 DVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSR 527
D LL L+ D F GL+IN KS+
Sbjct: 396 DGLLLA-MTVIGLQHNLDKFGETLAKIGLRINSRKSK 431
>PO11_SCICO (Q03277) Retrovirus-related Pol polyprotein from type I
retrotransposable element R1 [Contains: Reverse
transcriptase (EC 2.7.7.49); Endonuclease] (Fragment)
Length = 1004
Score = 54.7 bits (130), Expect = 7e-07
Identities = 43/165 (26%), Positives = 75/165 (45%), Gaps = 15/165 (9%)
Query: 236 LNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDT 295
L +V+ ++ K K+PGPDG + + + +F L L S+F
Sbjct: 405 LEMDEVSDSVRRCKVRKSPGPDGIVGEMVRAVWGAIPEYMFCLYKQCLLESYFPQKWKIA 464
Query: 296 LIALIPKV------DPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNI-IGPYQSSF 348
+ ++ K+ DP +RPI L + + K++ ++V RL L ++ + PYQ +F
Sbjct: 465 SLVILLKLLDRIRSDPG----SYRPICLLDNLGKVLEGIMVKRLDQKLMDVEVSPYQFAF 520
Query: 349 LPGRSTTDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNW 393
G+ST D +Q + + K + ID + AFDN+ W
Sbjct: 521 TYGKSTEDAWRCVQRHVE----CSEMKYVIGLNIDFQGAFDNLGW 561
>AI2M_YEAST (P03876) Putative COX1/OXI3 intron 2 protein
Length = 789
Score = 47.0 bits (110), Expect = 2e-04
Identities = 46/169 (27%), Positives = 67/169 (39%), Gaps = 21/169 (12%)
Query: 300 IPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAI 359
+ +V+ P T FRP+S+ N KI+ + + L I NN Y F P S I
Sbjct: 278 VRRVEIPKTSGGFRPLSVGNPREKIVQESMRMMLEIIYNNSFSYYSHGFRPNLSCLTAII 337
Query: 360 VLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCL----CDFGFPDTTIKLIMH 415
+ + Y K+DL K FD + + L + L D GF D KL+
Sbjct: 338 QCKNYMQYCN--------WFIKVDLNKCFDTIPHNMLINVLNERIKDKGFMDLLYKLLRA 389
Query: 416 CVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINN 464
N N T G+ QG +SP L + ++KL + N
Sbjct: 390 GYVDK------NNNY---HNTTLGIPQGSVVSPILCNIFLDKLDKYLEN 429
>Y2R2_DROME (P16425) Hypothetical 115 kDa protein in type I
retrotransposable element R1DM (ORF 2)
Length = 1021
Score = 41.6 bits (96), Expect = 0.006
Identities = 56/268 (20%), Positives = 103/268 (37%), Gaps = 27/268 (10%)
Query: 235 ALNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSIS- 293
AL +V + K ++PG DG + K + + + +L + +F
Sbjct: 433 ALEVFEVDTCVARLKSRRSPGLDGINGTICKAVWRAIPEHLASLFSRCIRLGYFPAEWKC 492
Query: 294 DTLIALIPKVDPPLTY-KDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGR 352
+++L+ D +R I L + K++ ++V+R+R +L +Q F GR
Sbjct: 493 PRVVSLLKGPDKDKCEPSSYRGICLLPVFGKVLEAIMVNRVREVLPEGCR-WQFGFRQGR 551
Query: 353 STTDNAIVLQEVIHYMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKL 412
D + V + S + F +D + AFDNV W S L D G +
Sbjct: 552 CVED---AWRHVKSSVGASAAQYVLGTF-VDFKGAFDNVEWSAALSRLADLGCREMG--- 604
Query: 413 IMHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWE 472
+ S +++ + + T G QG P+++ + M+ L + Q +
Sbjct: 605 LWQSFFSGRRAVIRSSSGTVEVPVTRGCPQGSISGPFIWDILMDVLLQRLQPYCQLSA-- 662
Query: 473 PIHVSNPGPKLSHLLFADDVLLFTKANS 500
+ADD+LL + NS
Sbjct: 663 ---------------YADDLLLLVEGNS 675
>RTJK_DROFU (P21329) RNA-directed DNA polymerase from mobile element
jockey (EC 2.7.7.49) (Reverse transcriptase)
Length = 916
Score = 37.0 bits (84), Expect = 0.16
Identities = 59/276 (21%), Positives = 112/276 (40%), Gaps = 27/276 (9%)
Query: 231 LPY*ALNKKKVTAALHSTKPYKAPGPDGFHCIFFKQYSHIVGDDV---FNLATSTFLTSH 287
LP + ++V + KP KAPG D + + ++ D L ++ L
Sbjct: 433 LPADPVTLEEVKELVSKLKPKKAPGED----LLDNRTIRLLPDQALLYLVLIFNSILRVG 488
Query: 288 FDPSISDT--LIALIPKVDPPLTYKDFRPISLCNIIYKIITKVLVHRL--RPILNNIIGP 343
+ P T +I ++ PL +RP SL + K++ +++++R+ + I
Sbjct: 489 YFPKARPTASIIMILKPGKQPLDVDSYRPTSLLPSLGKMLERLILNRILTSEEVTRAIPK 548
Query: 344 YQSSFLPGRSTTDNAIVLQEVIHYMRISKKKKGYV-AFKIDLEKAFDNVNWDFLRSCLCD 402
+Q F T + L V+++ + +KK Y + +D+++AFD V W
Sbjct: 549 FQFGFRLQHGTPEQ---LHRVVNFALEALEKKEYAGSCFLDIQQAFDRV-WHPGLLYKAK 604
Query: 403 FGFPDTTIKLIMHCVTSSSFSILWNGNRLPPFEPTHGLRQGDPLSPYLFILCMEKLSSAI 462
+LI FS+ +G R G+ QG L P L+ + + +
Sbjct: 605 SLLSPQLFQLIKSFWEGRKFSVTADGCRSSVKFIEAGVPQGSVLGPTLYSIFTADMPN-- 662
Query: 463 NNAVQQGSWEPIHVSNPGPKLSHLLFADDVLLFTKA 498
NAV + + ++ +ADD+ + TK+
Sbjct: 663 QNAVTGLAEGEVLIAT---------YADDIAVLTKS 689
>YO84_CAEEL (P34620) Hypothetical protein ZK1236.4 in chromosome III
Length = 364
Score = 35.8 bits (81), Expect = 0.35
Identities = 22/79 (27%), Positives = 37/79 (45%)
Query: 382 IDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNRLPPFEPTHGLR 441
+D KAFD V+ D L L I+ + +T+ SF + P + G+
Sbjct: 23 LDFSKAFDKVSHDILLDKLTSIKINKHLIRWLDVFLTNRSFKVKVGNTLSEPKKTVCGVP 82
Query: 442 QGDPLSPYLFILCMEKLSS 460
QG +SP LF + + ++S+
Sbjct: 83 QGSVISPVLFGIFVNEISA 101
>YKFC_ECOLI (Q47688) Hypothetical protein ykfC
Length = 376
Score = 33.9 bits (76), Expect = 1.3
Identities = 53/286 (18%), Positives = 108/286 (37%), Gaps = 51/286 (17%)
Query: 247 STKPYKAPGPDGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPP 306
S+K PG DG + + + + L+ H+ P + + IPK +
Sbjct: 39 SSKGAHTPGVDGVNKTMLQARLAVE----LQILRDELLSGHYQPLPARRVY--IPKSNGK 92
Query: 307 LTYKDFRPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIH 366
L RP+ + + +I+ + ++ + PI + F P RS ++ V
Sbjct: 93 L-----RPLGIPALRDRIVQRAMLMAMEPIWESDFHTLSYGFRPERSVHH---AIRTVKL 144
Query: 367 YMRISKKKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSF-SIL 425
+ + +G + DL FD V+ L +K + ++ + F ++L
Sbjct: 145 QLTDCGETRGRWVIEGDLSSYFDTVHHRLL-------------MKAVRRRISDARFMTLL 191
Query: 426 WNGNRLPPFE------PTHGLRQGDPLSPYLFILCMEKLSSAI----------------N 463
W + + + G+ QG +SP L + + + + N
Sbjct: 192 WKTIKAGHIDVGLFRAASEGVPQGGVISPLLSNIMLNEFDQYLHERYLSGKARKDRWYWN 251
Query: 464 NAVQQGSWEPIHVS-NPGPKLSHLLFADDVLLFTKANSTQLRFIRD 508
N++Q+G + + P +++ +ADD +L K Q+ IR+
Sbjct: 252 NSIQRGRSTAVRENWQWKPAVAYCRYADDFVLIVKGTKAQVEAIRE 297
>RDPO_SCEOB (P19593) Probable reverse transcriptase (EC 2.7.7.49)
Length = 608
Score = 33.5 bits (75), Expect = 1.8
Identities = 47/214 (21%), Positives = 87/214 (39%), Gaps = 21/214 (9%)
Query: 313 RPISLCNIIYKIITKVLVHRLRPILNNIIGPYQSSFLPGRSTTDNAIVLQEVIHYMRISK 372
RP+S+ + + + + + P+ + F P R+ + + V++ +
Sbjct: 132 RPLSIPSYTDRCLQALYKLAIEPMAEEVADLSSYGFRPMRNVS---WAVGRVLNGLNNPL 188
Query: 373 KKKGYVAFKIDLEKAFDNVNWDFLRSCLCDFGFPDTTIKLIMHCVTSSSFSILWNGNRLP 432
YV +ID++ DN+N F+ S + F P + + C I N N L
Sbjct: 189 ANYQYVV-EIDIKGCVDNINHQFI-SQVTPF-IPKKILWAWLKC-----GYIERNSNTLQ 240
Query: 433 PFEPTHGLRQGDPLSPYLFILCMEKLSSAINNAVQQGSWEPIHVSNPGPKLSHLLFADDV 492
P T G+ QG +SP + L ++ L I +Q+ S+ ++ +ADD+
Sbjct: 241 P--TTTGVPQGGIISPLIMNLTLDGLEFHIYKKIQKS-------SSQSKGNTYCRYADDM 291
Query: 493 LLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKS 526
++ T T L F GL++ L K+
Sbjct: 292 VILTTTEETAL-IALPAVKEFLAVRGLEVKLAKT 324
>HSCA_RICPR (Q9ZDW5) Chaperone protein hscA homolog
Length = 593
Score = 32.0 bits (71), Expect = 5.1
Identities = 33/134 (24%), Positives = 53/134 (38%), Gaps = 7/134 (5%)
Query: 257 DGFHCIFFKQYSHIVGDDVFNLATSTFLTSHFDPSISDTLIALIPKVDPPLTYKDFRPIS 316
+G + +++G D ++ + +L + FD S + L K LTYK+ S
Sbjct: 214 EGIFQVIATNGDNMLGGDDIDVVITQYLCNKFDLPHSIETLQLAKKAKEILTYKE----S 269
Query: 317 LCNIIYKIITKVLVHRLRPILNNIIGPYQSSF-LPGRSTTDNAIVLQEVIHYMRISKKKK 375
N I I + L + P++ I Q G D I++ I K +
Sbjct: 270 FNNDIISINKQTLEQLISPLVERTINITQECLEQSGNPNIDGVILVGGTTRIPLI--KDE 327
Query: 376 GYVAFKIDLEKAFD 389
Y AFKID+ D
Sbjct: 328 LYKAFKIDILSDID 341
>RT21_SCHPO (Q05654) Retrotransposable element Tf2 155 kDa protein
type 1
Length = 1333
Score = 31.6 bits (70), Expect = 6.7
Identities = 35/123 (28%), Positives = 59/123 (47%), Gaps = 12/123 (9%)
Query: 482 KLSHLL-FADDVLLFTKANSTQLRFIRDLFDRFSKFSGLKINLHKSRAFYSSGVTQDKIN 540
K SH++ + DD+L+ +K+ S ++ ++D+ + K + L IN K F+ S V I
Sbjct: 557 KESHVVCYMDDILIHSKSESEHVKHVKDVLQKL-KNANLIINQAKCE-FHQSQV--KFIG 612
Query: 541 TLTSISGIR-CTTSLDKYLGFPIIKGRPKKSDFLFIIDKMQK------RLASWQNKLLNK 593
S G C ++DK L + K R + FL ++ ++K +L N LL K
Sbjct: 613 YHISEKGFTPCQENIDKVLQWKQPKNRKELRQFLGSVNYLRKFIPKTSQLTHPLNNLLKK 672
Query: 594 PGR 596
R
Sbjct: 673 DVR 675
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.350 0.156 0.550
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 68,686,446
Number of Sequences: 164201
Number of extensions: 2724973
Number of successful extensions: 10619
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 10580
Number of HSP's gapped (non-prelim): 23
length of query: 638
length of database: 59,974,054
effective HSP length: 117
effective length of query: 521
effective length of database: 40,762,537
effective search space: 21237281777
effective search space used: 21237281777
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 69 (31.2 bits)
Medicago: description of AC136840.7


