

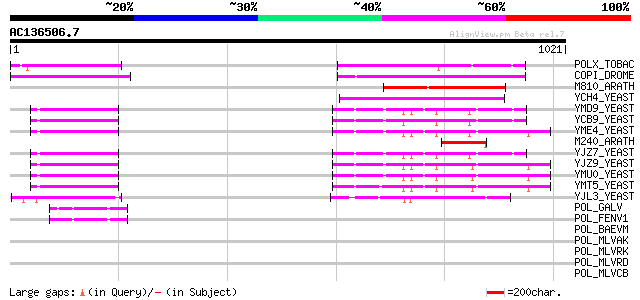
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136506.7 - phase: 0 /pseudo
(1021 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from tran... 205 4e-52
COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contain... 185 6e-46
M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810... 179 3e-44
YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein 129 4e-29
YMD9_YEAST (Q03434) Transposon Ty1 protein B 79 5e-14
YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B) 79 5e-14
YME4_YEAST (Q04711) Transposon Ty1 protein B 78 1e-13
M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240... 78 1e-13
YJZ7_YEAST (P47098) Transposon Ty1 protein B 78 1e-13
YJZ9_YEAST (P47100) Transposon Ty1 protein B 77 2e-13
YMU0_YEAST (Q04670) Transposon Ty1 protein B 76 4e-13
YMT5_YEAST (Q04214) Transposon Ty1 protein B 76 5e-13
YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein 73 4e-12
POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23... 47 2e-04
POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse transcript... 45 8e-04
POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.2... 45 0.001
POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse transcript... 44 0.002
POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC 3.4.2... 44 0.002
POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.2... 44 0.002
POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse transcript... 44 0.002
>POLX_TOBAC (P10978) Retrovirus-related Pol polyprotein from
transposon TNT 1-94 [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase (EC 2.7.7.49); Endonuclease]
Length = 1328
Score = 205 bits (522), Expect = 4e-52
Identities = 127/357 (35%), Positives = 197/357 (54%), Gaps = 13/357 (3%)
Query: 604 LHQLDVNNAFLHGDLHEDVYMTVLPGVTTS-KPNQVCKLSKSLYGLKQASRKWYEKLTGL 662
+ QLDV AFLHGDL E++YM G + K + VCKL+KSLYGLKQA R+WY K
Sbjct: 918 VEQLDVKTAFLHGDLEEEIYMEQPEGFEVAGKKHMVCKLNKSLYGLKQAPRQWYMKFDSF 977
Query: 663 LISNGYQQATSDASLFTKKASV-SLTILLVYVDDIILAGDSLIEIAFIKNVINQAFKIKD 721
+ S Y + SD ++ K+ S + ILL+YVDD+++ G IA +K ++++F +KD
Sbjct: 978 MKSQTYLKTYSDPCVYFKRFSENNFIILLLYVDDMLIVGKDKGLIAKLKGDLSKSFDMKD 1037
Query: 722 LGTLKYFLGLEVAHSQSG--ISLCQRKYCLDLLHDLGLLGSKPVTTPSDPSIKLHNDSSP 779
LG + LG+++ ++ + L Q KY +L + +KPV+TP +KL P
Sbjct: 1038 LGPAQQILGMKIVRERTSRKLWLSQEKYIERVLERFNMKNAKPVSTPLAGHLKLSKKMCP 1097
Query: 780 LFADVSA------YRRLVGRLIY-LNTTRPDITFITQQLSQFLSKPTHTHYNAALRVIKY 832
+ Y VG L+Y + TRPDI +S+FL P H+ A +++Y
Sbjct: 1098 TTVEEKGNMAKVPYSSAVGSLMYAMVCTRPDIAHAVGVVSRFLENPGKEHWEAVKWILRY 1157
Query: 833 LKGSPGRGIFFPRSSSLHIQGYTNADWAGCKDTRRSISCRCFFLGQSLISWRTKK*PTIS 892
L+G+ G + F S + ++GYT+AD AG D R+S + F ISW++K ++
Sbjct: 1158 LRGTTGDCLCFGGSDPI-LKGYTDADMAGDIDNRKSSTGYLFTFSGGAISWQSKLQKCVA 1216
Query: 893 RFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLYCDNQSVMHIAANPDFHSR 949
++EAEY A E+ WL L++L + K V+YCD+QS + ++ N +H+R
Sbjct: 1217 LSTTEAEYIAATETGKEMIWLKRFLQELGLH-QKEYVVYCDSQSAIDLSKNSMYHAR 1272
Score = 122 bits (306), Expect = 5e-27
Identities = 75/213 (35%), Positives = 115/213 (53%), Gaps = 9/213 (4%)
Query: 1 IWHFRLGHLSNQRLSKMHQLYPSISVDNKAT----CDICPFAKQRKLPYNFSHSIAKSKF 56
+WH R+GH+S + L + + S+ K T CD C F KQ ++ + S +
Sbjct: 424 LWHKRMGHMSEKGLQILAK--KSLISYAKGTTVKPCDYCLFGKQHRVSFQTSSERKLNIL 481
Query: 57 ELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHH 116
+L+Y D+ GP+ S+ G+KYF+T +DD SR LWV +LK K +V + F +++
Sbjct: 482 DLVYSDVCGPMEIESMGGNKYFVTFIDDASRKLWVYILKTKDQVFQVFQKFHALVERETG 541
Query: 117 ITPKFIRSDNGPEFLLLEF---YASKGIIHQKSCVETPEQNARVERKHQHILNVARALPF 173
K +RSDNG E+ EF +S GI H+K+ TP+ N ER ++ I+ R++
Sbjct: 542 RKLKRLRSDNGGEYTSREFEEYCSSHGIRHEKTVPGTPQHNGVAERMNRTIVEKVRSMLR 601
Query: 174 QFKLPNIFWSYAVLYSVFLINRVPTPFLHHKSP 206
KLP FW AV + +LINR P+ L + P
Sbjct: 602 MAKLPKSFWGEAVQTACYLINRSPSVPLAFEIP 634
>COPI_DROME (P04146) Copia protein (Gag-int-pol protein) [Contains:
Copia VLP protein; Copia protease (EC 3.4.23.-)]
Length = 1409
Score = 185 bits (469), Expect = 6e-46
Identities = 114/352 (32%), Positives = 192/352 (54%), Gaps = 8/352 (2%)
Query: 604 LHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASRKWYEKLTGLL 663
+HQ+DV AFL+G L E++YM + G++ + N VCKL+K++YGLKQA+R W+E L
Sbjct: 998 VHQMDVKTAFLNGTLKEEIYMRLPQGISCNSDN-VCKLNKAIYGLKQAARCWFEVFEQAL 1056
Query: 664 ISNGYQQATSDASLFT-KKASVSLTI-LLVYVDDIILAGDSLIEIAFIKNVINQAFKIKD 721
+ ++ D ++ K +++ I +L+YVDD+++A + + K + + F++ D
Sbjct: 1057 KECEFVNSSVDRCIYILDKGNINENIYVLLYVDDVVIATGDMTRMNNFKRYLMEKFRMTD 1116
Query: 722 LGTLKYFLGLEVAHSQSGISLCQRKYCLDLLHDLGLLGSKPVTTPSDPSIKLHNDSSPLF 781
L +K+F+G+ + + I L Q Y +L + V+TP I +S
Sbjct: 1117 LNEIKHFIGIRIEMQEDKIYLSQSAYVKKILSKFNMENCNAVSTPLPSKINYELLNSDED 1176
Query: 782 ADVSAYRRLVGRLIYLNT-TRPDITFITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRG 840
+ R L+G L+Y+ TRPD+T LS++ SK + RV++YLKG+
Sbjct: 1177 CNTPC-RSLIGCLMYIMLCTRPDLTTAVNILSRYSSKNNSELWQNLKRVLRYLKGTIDMK 1235
Query: 841 IFFPRSSSLH--IQGYTNADWAGCKDTRRSISCRCFFLGQ-SLISWRTKK*PTISRFSSE 897
+ F ++ + I GY ++DWAG + R+S + F + +LI W TK+ +++ S+E
Sbjct: 1236 LIFKKNLAFENKIIGYVDSDWAGSEIDRKSTTGYLFKMFDFNLICWNTKRQNSVAASSTE 1295
Query: 898 AEYRAMASATCELQWLLYLLRDLHIRCVKLLVLYCDNQSVMHIAANPDFHSR 949
AEY A+ A E WL +LL ++I+ + +Y DNQ + IA NP H R
Sbjct: 1296 AEYMALFEAVREALWLKFLLTSINIKLENPIKIYEDNQGCISIANNPSCHKR 1347
Score = 100 bits (248), Expect = 3e-20
Identities = 67/236 (28%), Positives = 119/236 (50%), Gaps = 14/236 (5%)
Query: 1 IWHFRLGHLSNQRLS--KMHQLYPSISVDN--KATCDICP---FAKQRKLPYNF--SHSI 51
+WH R GH+S+ +L K ++ S+ N + +C+IC KQ +LP+ +
Sbjct: 417 LWHERFGHISDGKLLEIKRKNMFSDQSLLNNLELSCEICEPCLNGKQARLPFKQLKDKTH 476
Query: 52 AKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQII 111
K +++ D+ GP+ ++ YF+ VD F+ + L+K K++V + FV
Sbjct: 477 IKRPLFVVHSDVCGPITPVTLDDKNYFVIFVDQFTHYCVTYLIKYKSDVFSMFQDFVAKS 536
Query: 112 DTHHHITPKFIRSDNGPEFL---LLEFYASKGIIHQKSCVETPEQNARVERKHQHILNVA 168
+ H ++ ++ DNG E+L + +F KGI + + TP+ N ER + I A
Sbjct: 537 EAHFNLKVVYLYIDNGREYLSNEMRQFCVKKGISYHLTVPHTPQLNGVSERMIRTITEKA 596
Query: 169 RALPFQFKLPNIFWSYAVLYSVFLINRVPTPFL--HHKSPY*VLYDSLPDIQFFKI 222
R + KL FW AVL + +LINR+P+ L K+PY + ++ P ++ ++
Sbjct: 597 RTMVSGAKLDKSFWGEAVLTATYLINRIPSRALVDSSKTPYEMWHNKKPYLKHLRV 652
>M810_ARATH (P92519) Hypothetical mitochondrial protein AtMg00810
(ORF240b)
Length = 240
Score = 179 bits (454), Expect = 3e-44
Identities = 91/225 (40%), Positives = 143/225 (63%), Gaps = 3/225 (1%)
Query: 689 LLVYVDDIILAGDSLIEIAFIKNVINQAFKIKDLGTLKYFLGLEVAHSQSGISLCQRKYC 748
LL+YVDDI+L G S + + ++ F +KDLG + YFLG+++ SG+ L Q KY
Sbjct: 3 LLLYVDDILLTGSSNTLLNMLIFQLSSTFSMKDLGPVHYFLGIQIKTHPSGLFLSQTKYA 62
Query: 749 LDLLHDLGLLGSKPVTTPSDPSIKLHND-SSPLFADVSAYRRLVGRLIYLNTTRPDITFI 807
+L++ G+L KP++TP +KL++ S+ + D S +R +VG L YL TRPDI++
Sbjct: 63 EQILNNAGMLDCKPMSTPLP--LKLNSSVSTAKYPDPSDFRSIVGALQYLTLTRPDISYA 120
Query: 808 TQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSLHIQGYTNADWAGCKDTRR 867
+ Q + +PT ++ RV++Y+KG+ G++ ++S L++Q + ++DWAGC TRR
Sbjct: 121 VNIVCQRMHEPTLADFDLLKRVLRYVKGTIFHGLYIHKNSKLNVQAFCDSDWAGCTSTRR 180
Query: 868 SISCRCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQW 912
S + C FLG ++ISW K+ PT+SR S+E EYRA+A EL W
Sbjct: 181 STTGFCTFLGCNIISWSAKRQPTVSRSSTETEYRALALTAAELTW 225
>YCH4_YEAST (P25600) Transposon Ty5-1 34.5 kDa hypothetical protein
Length = 308
Score = 129 bits (324), Expect = 4e-29
Identities = 91/308 (29%), Positives = 150/308 (48%), Gaps = 4/308 (1%)
Query: 607 LDVNNAFLHGDLHEDVYMTVLPG-VTTSKPNQVCKLSKSLYGLKQASRKWYEKLTGLLIS 665
+DV+ AFL+ + E +Y+ PG V P+ V +L +YGLKQA W E + L
Sbjct: 1 MDVDTAFLNSTMDEPIYVKQPPGFVNERNPDYVWELYGGMYGLKQAPLLWNEHINNTLKK 60
Query: 666 NGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVINQAFKIKDLGTL 725
G+ + + L+ + S + VYVDD+++A S +K + + + +KDLG +
Sbjct: 61 IGFCRHEGEHGLYFRSTSDGPIYIGVYVDDLLVAAPSPKIYDRVKQELTKLYSMKDLGKV 120
Query: 726 KYFLGLEVAHSQSG-ISLCQRKYCLDLLHDLGLLGSKPVTTPSDPSIKLHNDSSPLFADV 784
FLGL + S +G I+L + Y + + K TP S L +SP D+
Sbjct: 121 DKFLGLNIHQSTNGDITLSLQDYIAKAASESEINTFKLTQTPLCNSKPLFETTSPHLKDI 180
Query: 785 SAYRRLVGRLIYL-NTTRPDITFITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFF 843
+ Y+ +VG+L++ NT RPDI++ LS+FL +P H +A RV++YL + + +
Sbjct: 181 TPYQSIVGQLLFCANTGRPDISYPVSLLSRFLREPRAIHLESARRVLRYLYTTRSMCLKY 240
Query: 844 PRSSSLHIQGYTNADWAGCKDTRRSISCRCFFLGQSLISWRTKK*P-TISRFSSEAEYRA 902
S + + Y +A D S L + ++W +KK I S+EAEY
Sbjct: 241 RSGSQVALTVYCDASHGAIHDLPHSTGGYVTLLAGAPVTWSSKKLKGVIPVPSTEAEYIT 300
Query: 903 MASATCEL 910
+ E+
Sbjct: 301 ASETVMEI 308
>YMD9_YEAST (Q03434) Transposon Ty1 protein B
Length = 1328
Score = 79.3 bits (194), Expect = 5e-14
Identities = 87/382 (22%), Positives = 178/382 (45%), Gaps = 33/382 (8%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ LA N++++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KSLYGLKQ+
Sbjct: 905 LSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGA 962
Query: 654 KWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVI 713
WYE + LI + S K + V++ + +VDD+IL L I +
Sbjct: 963 NWYETIKSYLIKQCGMEEVRGWSCVFKNSQVTICL---FVDDMILFSKDLNANKKIITTL 1019
Query: 714 NQAF--KIKDLG----TLKY-FLGLEVAHSQS-----GISLCQRKYCLDLLHDLGLLGSK 761
+ + KI +LG ++Y LGLE+ + + G+ + L L G K
Sbjct: 1020 KKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRK 1079
Query: 762 PVTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQFL 815
++ P P + + D + D V ++L+G Y+ R D+ + L+Q +
Sbjct: 1080 -LSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHI 1138
Query: 816 SKPTHTHYNAALRVIKYLKGSPGRGIFF----PRSSSLHIQGYTNADWAGCKDTRRSISC 871
P+ + +I+++ + + + + P + ++A + G + +S
Sbjct: 1139 LFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPYYKSQIG 1197
Query: 872 RCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLY 931
+ L +I ++ K ++EAE A++ + L L YL+++L+ + + + L
Sbjct: 1198 NIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPI-IKGLL 1256
Query: 932 CDNQSVMHIAANPD---FHSRF 950
D++S + I + + F +RF
Sbjct: 1257 TDSRSTISIIKSTNEEKFRNRF 1278
Score = 50.1 bits (118), Expect = 3e-05
Identities = 42/167 (25%), Positives = 73/167 (43%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ ++F WV L ++
Sbjct: 225 KGSRLKYQNSYE----PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRR 280
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F GI +
Sbjct: 281 EDSI-LDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSR 339
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +L+ R LPN W A+ +S + N + +P
Sbjct: 340 AHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
>YCB9_YEAST (P25384) Transposon Ty2 protein B (Ty1-17 protein B)
Length = 1770
Score = 79.3 bits (194), Expect = 5e-14
Identities = 89/381 (23%), Positives = 178/381 (46%), Gaps = 31/381 (8%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ +A N +++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KSLYGLKQ+
Sbjct: 1347 LSIALDNDYYITQLDISSAYLYADIKEELYIRPPPHLGLN--DKLLRLRKSLYGLKQSGA 1404
Query: 654 KWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVI 713
WYE + LI+ Q S K + V++ + +VDD+IL L I +
Sbjct: 1405 NWYETIKSYLINCCDMQEVRGWSCVFKNSQVTICL---FVDDMILFSKDLNANKKIITTL 1461
Query: 714 NQAF--KIKDLG----TLKY-FLGLEVAHSQSG-ISLCQRKYCLDLLHDLGL---LGSKP 762
+ + KI +LG ++Y LGLE+ + +S + L K + L L + K
Sbjct: 1462 KKQYDTKIINLGESDNEIQYDILGLEIKYQRSKYMKLGMEKSLTEKLPKLNVPLNPKGKK 1521
Query: 763 VTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQFLS 816
+ P P + D + D V ++L+G Y+ R D+ + L+Q +
Sbjct: 1522 LRAPGQPGHYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHIL 1581
Query: 817 KPTHTHYNAALRVIKYLKGSPGRGIFF----PRSSSLHIQGYTNADWAGCKDTRRSISCR 872
P+ + +I+++ + + + + P + ++A + G + +S
Sbjct: 1582 FPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTKPDNKLVAISDASY-GNQPYYKSQIGN 1640
Query: 873 CFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLYC 932
F L +I ++ K ++EAE A++ A L L +L+++L+ + + + L
Sbjct: 1641 IFLLNGKVIGGKSTKASLTCTSTTEAEIHAVSEAIPLLNNLSHLVQELNKKPI-IKGLLT 1699
Query: 933 DNQSVMHIAANPD---FHSRF 950
D++S + I + + F +RF
Sbjct: 1700 DSRSTISIIKSTNEEKFRNRF 1720
Score = 60.1 bits (144), Expect = 3e-08
Identities = 46/167 (27%), Positives = 77/167 (45%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ +RF WV L ++
Sbjct: 648 KGSRLKYQESYE----PFQYLHTDIFGPVHHLPKSAPSYFISFTDEKTRFQWVYPLHDRR 703
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F+ ++GI +
Sbjct: 704 EESI-LNVFTSILAFIKNQFNARVLVIQMDRGSEYTNKTLHKFFTNRGITACYTTTADSR 762
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +LN R L LPN W AV +S + N + +P
Sbjct: 763 AHGVAERLNRTLLNDCRTLLHCSGLPNHLWFSAVEFSTIIRNSLVSP 809
>YME4_YEAST (Q04711) Transposon Ty1 protein B
Length = 1328
Score = 78.2 bits (191), Expect = 1e-13
Identities = 96/432 (22%), Positives = 193/432 (44%), Gaps = 38/432 (8%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ LA N++++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KSLYGLKQ+
Sbjct: 905 LSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGA 962
Query: 654 KWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVI 713
WYE + LI + S K + V++ + +VDD+IL L I +
Sbjct: 963 NWYETIKSYLIKQCGMEEVRGWSCVFKNSQVTICL---FVDDMILFSKDLNANKKIITTL 1019
Query: 714 NQAF--KIKDLG----TLKY-FLGLEVAHSQS-----GISLCQRKYCLDLLHDLGLLGSK 761
+ + KI +LG ++Y LGLE+ + + G+ + L L G K
Sbjct: 1020 KKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRK 1079
Query: 762 PVTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQFL 815
++ P P + + D + D V ++L+G Y+ R D+ + L+Q +
Sbjct: 1080 -LSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHI 1138
Query: 816 SKPTHTHYNAALRVIKYLKGSPGRGIFF----PRSSSLHIQGYTNADWAGCKDTRRSISC 871
P+ + +I+++ + + + + P + ++A + G + +S
Sbjct: 1139 LFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPYYKSQIG 1197
Query: 872 RCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLY 931
+ L +I ++ K ++EAE A++ + L L +L+++L+ + + +L
Sbjct: 1198 NIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPITKGLLT 1257
Query: 932 CDNQSVMHIAANPD--FHSRFS*T------HKAP*N*LPHCEGKASRWFVQIVT*SPLMI 983
++ I +N + F +RF T + N L C + + ++T PL I
Sbjct: 1258 DSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMT-KPLPI 1316
Query: 984 NLQIFLQNPYFH 995
L N + H
Sbjct: 1317 KTFKLLTNKWIH 1328
Score = 50.1 bits (118), Expect = 3e-05
Identities = 42/167 (25%), Positives = 73/167 (43%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ ++F WV L ++
Sbjct: 225 KGSRLKYQNSYE----PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRR 280
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F GI +
Sbjct: 281 EDSI-LDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSR 339
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +L+ R LPN W A+ +S + N + +P
Sbjct: 340 AHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
>M240_ARATH (P93290) Hypothetical mitochondrial protein AtMg00240
(ORF111a)
Length = 111
Score = 78.2 bits (191), Expect = 1e-13
Identities = 36/88 (40%), Positives = 55/88 (61%), Gaps = 5/88 (5%)
Query: 795 IYLNTTRPDITFITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSLHIQGY 854
+YL TRPD+TF +LSQF S A +V+ Y+KG+ G+G+F+ +S L ++ +
Sbjct: 1 MYLTITRPDLTFAVNRLSQFSSASRTAQMQAVYKVLHYVKGTVGQGLFYSATSDLQLKAF 60
Query: 855 TNADWAGCKDTRRSISCRC-----FFLG 877
++DWA C DTRRS++ C +FLG
Sbjct: 61 ADSDWASCPDTRRSVTGFCSLVPLWFLG 88
>YJZ7_YEAST (P47098) Transposon Ty1 protein B
Length = 1755
Score = 77.8 bits (190), Expect = 1e-13
Identities = 87/382 (22%), Positives = 177/382 (45%), Gaps = 33/382 (8%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ LA N++++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KS YGLKQ+
Sbjct: 1332 LSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSHYGLKQSGA 1389
Query: 654 KWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVI 713
WYE + LI + S K + V++ + +VDD+IL L I +
Sbjct: 1390 NWYETIKSYLIKQCGMEEVRGWSCVFKNSQVTICL---FVDDMILFSKDLNANKKIITTL 1446
Query: 714 NQAF--KIKDLG----TLKY-FLGLEVAHSQS-----GISLCQRKYCLDLLHDLGLLGSK 761
+ + KI +LG ++Y LGLE+ + + G+ + L L G K
Sbjct: 1447 KKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRK 1506
Query: 762 PVTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQFL 815
++ P P + + D + D V ++L+G Y+ R D+ + L+Q +
Sbjct: 1507 -LSAPGQPGLYIDQDELEIDEDEYKEKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHI 1565
Query: 816 SKPTHTHYNAALRVIKYLKGSPGRGIFF----PRSSSLHIQGYTNADWAGCKDTRRSISC 871
P+ + +I+++ + + + + P + ++A + G + +S
Sbjct: 1566 LFPSRQVLDMTYELIQFMWDTRDKQLIWHKNKPTEPDNKLVAISDASY-GNQPYYKSQIG 1624
Query: 872 RCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLY 931
F L +I ++ K ++EAE A++ + L L YL+++L+ + + + L
Sbjct: 1625 NIFLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELNKKPI-IKGLL 1683
Query: 932 CDNQSVMHIAANPD---FHSRF 950
D++S + I + + F +RF
Sbjct: 1684 TDSRSTISIIKSTNEEKFRNRF 1705
Score = 50.1 bits (118), Expect = 3e-05
Identities = 42/167 (25%), Positives = 73/167 (43%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ ++F WV L ++
Sbjct: 652 KGSRLKYQNSYE----PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRR 707
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F GI +
Sbjct: 708 EDSI-LDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSR 766
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +L+ R LPN W A+ +S + N + +P
Sbjct: 767 AHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 813
>YJZ9_YEAST (P47100) Transposon Ty1 protein B
Length = 1755
Score = 77.0 bits (188), Expect = 2e-13
Identities = 96/432 (22%), Positives = 193/432 (44%), Gaps = 38/432 (8%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ LA N++++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KSLYGLKQ+
Sbjct: 1332 LSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGA 1389
Query: 654 KWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVI 713
WYE + LI + S K + V++ + +VDD++L +L I +
Sbjct: 1390 NWYETIKSYLIQQCGMEEVRGWSCVFKNSQVTICL---FVDDMVLFSKNLNSNKRIIEKL 1446
Query: 714 NQAF--KIKDLG----TLKY-FLGLEVAHSQS-----GISLCQRKYCLDLLHDLGLLGSK 761
+ KI +LG ++Y LGLE+ + + G+ + L L G K
Sbjct: 1447 KMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRK 1506
Query: 762 PVTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQFL 815
++ P P + + L D V ++L+G Y+ R D+ + L+Q +
Sbjct: 1507 -LSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHI 1565
Query: 816 SKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSL----HIQGYTNADWAGCKDTRRSISC 871
P+ + +I+++ + + + + +S + + ++A + G + +S
Sbjct: 1566 LFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIG 1624
Query: 872 RCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLY 931
+ L +I ++ K ++EAE A++ + L L YL+++L + + +L
Sbjct: 1625 NIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITKGLLT 1684
Query: 932 CDNQSVMHIAANPD--FHSRFS*T------HKAP*N*LPHCEGKASRWFVQIVT*SPLMI 983
++ I +N + F +RF T + N L C + + ++T PL I
Sbjct: 1685 DSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMT-KPLPI 1743
Query: 984 NLQIFLQNPYFH 995
L N + H
Sbjct: 1744 KTFKLLTNKWIH 1755
Score = 50.1 bits (118), Expect = 3e-05
Identities = 42/167 (25%), Positives = 73/167 (43%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ ++F WV L ++
Sbjct: 652 KGSRLKYQNSYE----PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRR 707
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F GI +
Sbjct: 708 EDSI-LDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSR 766
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +L+ R LPN W A+ +S + N + +P
Sbjct: 767 AHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 813
>YMU0_YEAST (Q04670) Transposon Ty1 protein B
Length = 1328
Score = 76.3 bits (186), Expect = 4e-13
Identities = 96/432 (22%), Positives = 194/432 (44%), Gaps = 38/432 (8%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ LA N++++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KSLYGLKQ+
Sbjct: 905 LSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGA 962
Query: 654 KWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNVI 713
WYE + LI + S K + V++ + +VDD+IL L I +
Sbjct: 963 NWYETIKSYLIKQCGMEEVRGWSCVFKNSQVTICL---FVDDMILFSKDLNANKKIITTL 1019
Query: 714 NQAF--KIKDLG----TLKY-FLGLEVAHSQS-----GISLCQRKYCLDLLHDLGLLGSK 761
+ + KI +LG ++Y LGLE+ + + G+ + L L G K
Sbjct: 1020 KKQYDTKIINLGESDNEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGRK 1079
Query: 762 PVTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQFL 815
++ P P + + L D V ++L+G Y+ R D+ + L+Q +
Sbjct: 1080 -LSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQHI 1138
Query: 816 SKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSL----HIQGYTNADWAGCKDTRRSISC 871
P+ + +I+++ + + + + +S + + ++A + G + +S
Sbjct: 1139 LFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQIG 1197
Query: 872 RCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVLY 931
+ L +I ++ K ++EAE A++ + L L +L+++L+ + + +L
Sbjct: 1198 NIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSHLVQELNKKPITKGLLT 1257
Query: 932 CDNQSVMHIAANPD--FHSRFS*T------HKAP*N*LPHCEGKASRWFVQIVT*SPLMI 983
++ I +N + F +RF T + N L C + + ++T PL I
Sbjct: 1258 DSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMT-KPLPI 1316
Query: 984 NLQIFLQNPYFH 995
L N + H
Sbjct: 1317 KTFKLLTNKWIH 1328
Score = 50.1 bits (118), Expect = 3e-05
Identities = 42/167 (25%), Positives = 73/167 (43%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ ++F WV L ++
Sbjct: 225 KGSRLKYQNSYE----PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRR 280
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F GI +
Sbjct: 281 EDSI-LDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSR 339
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +L+ R LPN W A+ +S + N + +P
Sbjct: 340 AHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
>YMT5_YEAST (Q04214) Transposon Ty1 protein B
Length = 1328
Score = 75.9 bits (185), Expect = 5e-13
Identities = 96/433 (22%), Positives = 195/433 (44%), Gaps = 40/433 (9%)
Query: 594 IGLASINHWFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQVCKLSKSLYGLKQASR 653
+ LA N++++ QLD+++A+L+ D+ E++Y+ P + + +++ +L KSLYGLKQ+
Sbjct: 905 LSLALDNNYYITQLDISSAYLYADIKEELYIRPPPHLGMN--DKLIRLKKSLYGLKQSGA 962
Query: 654 KWYEKLTGLLISN-GYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLIEIAFIKNV 712
WYE + LI G ++ + +F S + ++VDD++L +L I +
Sbjct: 963 NWYETIKSYLIKQCGMEEVRGWSCVFEN----SQVTICLFVDDMVLFSKNLNSNKRIIDK 1018
Query: 713 INQAF--KIKDLG----TLKY-FLGLEVAHSQS-----GISLCQRKYCLDLLHDLGLLGS 760
+ + KI +LG ++Y LGLE+ + + G+ + L L G
Sbjct: 1019 LKMQYDTKIINLGESDEEIQYDILGLEIKYQRGKYMKLGMENSLTEKIPKLNVPLNPKGR 1078
Query: 761 KPVTTPSDPSIKLHNDSSPLFAD-----VSAYRRLVGRLIYLN-TTRPDITFITQQLSQF 814
K ++ P P + + L D V ++L+G Y+ R D+ + L+Q
Sbjct: 1079 K-LSAPGQPGLYIDQQELELEEDDYKMKVHEMQKLIGLASYVGYKFRFDLLYYINTLAQH 1137
Query: 815 LSKPTHTHYNAALRVIKYLKGSPGRGIFFPRSSSL----HIQGYTNADWAGCKDTRRSIS 870
+ P+ + +I+++ + + + + +S + + ++A + G + +S
Sbjct: 1138 ILFPSKQVLDMTYELIQFIWNTRDKQLIWHKSKPVKPTNKLVVISDASY-GNQPYYKSQI 1196
Query: 871 CRCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDLHIRCVKLLVL 930
+ L +I ++ K ++EAE A++ + L L YL+++L + + +L
Sbjct: 1197 GNIYLLNGKVIGGKSTKASLTCTSTTEAEIHAISESVPLLNNLSYLIQELDKKPITKGLL 1256
Query: 931 YCDNQSVMHIAANPD--FHSRFS*T------HKAP*N*LPHCEGKASRWFVQIVT*SPLM 982
++ I +N + F +RF T + N L C + + ++T PL
Sbjct: 1257 TDSKSTISIIISNNEEKFRNRFFGTKAMRLRDEVSGNHLHVCYIETKKNIADVMT-KPLP 1315
Query: 983 INLQIFLQNPYFH 995
I L N + H
Sbjct: 1316 IKTFKLLTNKWIH 1328
Score = 50.1 bits (118), Expect = 3e-05
Identities = 42/167 (25%), Positives = 73/167 (43%), Gaps = 11/167 (6%)
Query: 39 KQRKLPYNFSHSIAKSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVILLKNKA 98
K +L Y S+ F+ L+ DI+GP+ YF++ D+ ++F WV L ++
Sbjct: 225 KGSRLKYQNSYE----PFQYLHTDIFGPVHNLPKSAPSYFISFTDETTKFRWVYPLHDRR 280
Query: 99 EVSLPVKIFVQI---IDTHHHITPKFIRSDNGPEF---LLLEFYASKGIIHQKSCVETPE 152
E S+ + +F I I + I+ D G E+ L +F GI +
Sbjct: 281 EDSI-LDVFTTILAFIKNQFQASVLVIQMDRGSEYTNRTLHKFLEKNGITPCYTTTADSR 339
Query: 153 QNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSVFLINRVPTP 199
+ ER ++ +L+ R LPN W A+ +S + N + +P
Sbjct: 340 AHGVAERLNRTLLDDCRTQLQCSGLPNHLWFSAIEFSTIVRNSLASP 386
>YJL3_YEAST (P47024) Transposon Ty4 207.7 kDa hypothetical protein
Length = 1803
Score = 72.8 bits (177), Expect = 4e-12
Identities = 85/356 (23%), Positives = 157/356 (43%), Gaps = 36/356 (10%)
Query: 590 NNCKIGLASINH--WFLHQLDVNNAFLHGDLHEDVYMTVLPGVTTSKPNQ---VCKLSKS 644
N+ KI L N+ F+ LD+N+AFL+ L E++Y+ P+ V KL+K+
Sbjct: 1364 NHIKIFLMIANNRNMFMKTLDINHAFLYAKLEEEIYIP--------HPHDRRCVVKLNKA 1415
Query: 645 LYGLKQASRKWYEKLTGLLISNGYQQATSDASLFTKKASVSLTILLVYVDDIILAGDSLI 704
LYGLKQ+ ++W + L L G + + L+ + ++ VYVDD ++A +
Sbjct: 1416 LYGLKQSPKEWNDHLRQYLNGIGLKDNSYTPGLY--QTEDKNLMIAVYVDDCVIAASNEQ 1473
Query: 705 EIAFIKNVINQAFKIKDLGTL------KYFLGLEVAHS----------QSGISLCQRKYC 748
+ N + F++K GTL LG+++ ++ +S I+ +KY
Sbjct: 1474 RLDEFINKLKSNFELKITGTLIDDVLDTDILGMDLVYNKRLGTIDLTLKSFINRMDKKYN 1533
Query: 749 LDLLHDLGLLGSKPVTTPSDPSIKLHNDSSPLFAD-VSAYRRLVGRLIYL-NTTRPDITF 806
+L T DP + S F V ++L+G L Y+ + R DI F
Sbjct: 1534 EELKKIRKSSIPHMSTYKIDPKKDVLQMSEEEFRQGVLKLQQLLGELNYVRHKCRYDIEF 1593
Query: 807 ITQQLSQFLSKPTHTHYNAALRVIKYLKGSPGRGIFFPR--SSSLHIQGYTNADWAGCKD 864
+++++ ++ P + ++I+YL GI + R + + T+A D
Sbjct: 1594 AVKKVARLVNYPHERVFYMIYKIIQYLVRYKDIGIHYDRDCNKDKKVIAITDASVGSEYD 1653
Query: 865 TRRSISCRCFFLGQSLISWRTKK*PTISRFSSEAEYRAMASATCELQWLLYLLRDL 920
+ I ++ G ++ + + K S+EAE A+ + + L L++L
Sbjct: 1654 AQSRIGVILWY-GMNIFNVYSNKSTNRCVSSTEAELHAIYEGYADSETLKVTLKEL 1708
Score = 68.6 bits (166), Expect = 8e-11
Identities = 61/218 (27%), Positives = 94/218 (42%), Gaps = 20/218 (9%)
Query: 3 HFRLGHLSNQRL--SKMHQLYPS----ISVDNKATCDICPFAKQRKLPYNF----SHSIA 52
H R+GH Q++ S H Y I N+ C C +K K + +HS
Sbjct: 562 HKRMGHTGIQQIENSIKHNHYEESLDLIKEPNEFWCQTCKISKATKRNHYTGSMNNHSTD 621
Query: 53 KSKFELLYFDIWGPLAQTSIHGHKYFLTIVDDFSRFLWVI--LLKNKAEVSLPVKIFVQI 110
DI+GP++ ++ +Y L +VD+ +R+ KN + V+ +Q
Sbjct: 622 HEPGSSWCMDIFGPVSSSNADTKRYMLIMVDNNTRYCMTSTHFNKNAETILAQVRKNIQY 681
Query: 111 IDTHHHITPKFIRSDNGPEFL---LLEFYASKGIIHQKSCVETPEQNARVERKHQHILNV 167
++T + I SD G EF + E++ SKGI H + + N R ER + I+
Sbjct: 682 VETQFDRKVREINSDRGTEFTNDQIEEYFISKGIHHILTSTQDHAANGRAERYIRTIITD 741
Query: 168 ARALPFQFKLPNIFWSYAVLYSVFLINRVPTPFLHHKS 205
A L Q L FW YAV + + N +L HKS
Sbjct: 742 ATTLLRQSNLRVKFWEYAVTSATNIRN-----YLEHKS 774
>POL_GALV (P21414) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1165
Score = 47.4 bits (111), Expect = 2e-04
Identities = 44/150 (29%), Positives = 69/150 (45%), Gaps = 13/150 (8%)
Query: 73 HGHKYFLTIVDDFSRFLWVILLKNKAEVSLPV--KIFVQIIDTHHHITPKFIRSDNGPEF 130
+G+KY L +D FS WV K E +L V KI +I+ PK + SDNGP F
Sbjct: 892 YGNKYLLVFIDTFSG--WVEAFPTKTETALIVCKKILEEILPRFG--IPKVLGSDNGPAF 947
Query: 131 L--LLEFYASK-GIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVL 187
+ + + A++ GI + C P+ + +VER ++ I L + + + L
Sbjct: 948 VAQVSQGLATQLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLALETGGKD----WVTL 1003
Query: 188 YSVFLINRVPTPFLHHKSPY*VLYDSLPDI 217
+ L+ TP +PY +LY P I
Sbjct: 1004 LPLALLRARNTPGRFGLTPYEILYGGPPPI 1033
>POL_FENV1 (P31792) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 1046
Score = 45.4 bits (106), Expect = 8e-04
Identities = 40/147 (27%), Positives = 63/147 (42%), Gaps = 9/147 (6%)
Query: 74 GHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHHITPKFIRSDNGPEF--- 130
G+KY L VD FS ++ + + + KI +I PK I SDNGP F
Sbjct: 778 GYKYLLVFVDTFSGWVEAYPTRQETAHMVAKKILEEIFPRFG--LPKVIGSDNGPAFVSQ 835
Query: 131 LLLEFYASKGIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSV 190
+ + GI + C P+ + +VER ++ I L + L + + L S+
Sbjct: 836 VSQGLARTLGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKD----WRRLLSL 891
Query: 191 FLINRVPTPFLHHKSPY*VLYDSLPDI 217
L+ TP +PY +LY P +
Sbjct: 892 ALLRARNTPNRFGLTPYEILYGGPPPL 918
>POL_BAEVM (P10272) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1189
Score = 44.7 bits (104), Expect = 0.001
Identities = 40/147 (27%), Positives = 62/147 (41%), Gaps = 9/147 (6%)
Query: 74 GHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHHITPKFIRSDNGPEFLLL 133
G+KY L VD FS ++ + + + KI +I PK I SDNGP F+
Sbjct: 921 GYKYLLVFVDTFSGWVEAFPTRQETAHIVAKKILEEIFPRFG--LPKVIGSDNGPAFVSQ 978
Query: 134 EFYASK---GIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVLYSV 190
GI + C P+ + +VER ++ I L + L + + L S+
Sbjct: 979 VSQGLARILGINWKLHCAYRPQSSGQVERMNRTIKETLTKLTLETGLKD----WRRLLSL 1034
Query: 191 FLINRVPTPFLHHKSPY*VLYDSLPDI 217
L+ TP +PY +LY P +
Sbjct: 1035 ALLRARNTPNRFGLTPYEILYGGPPPL 1061
>POL_MLVAK (P03357) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 843
Score = 44.3 bits (103), Expect = 0.002
Identities = 42/153 (27%), Positives = 66/153 (42%), Gaps = 12/153 (7%)
Query: 72 IHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPV--KIFVQIIDTHHHITPKFIRSDNGPE 129
++G+KY L VD FS WV K E + V K+ +I P+ + SDNGP
Sbjct: 572 LYGYKYLLVFVDTFSG--WVEAFPTKRETARVVSKKLLEEIFPRFG--MPQVLGSDNGPA 627
Query: 130 FL--LLEFYASKGIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVL 187
F + + A I + C P+ + +VER ++ I L + + +L
Sbjct: 628 FTSQVSQSVADLLGIDKLHCAYRPQSSGQVERMNRTIKETLTKLTLAAGTRD----WVLL 683
Query: 188 YSVFLINRVPTPFLHHKSPY*VLYDSLPDIQFF 220
+ L TP H +PY +LY + P + F
Sbjct: 684 LPLALYRARNTPGPHGLTPYEILYGAPPPLVNF 716
>POL_MLVRK (P31795) Pol polyprotein [Contains: Protease (EC
3.4.23.-); Reverse transcriptase/ribonuclease H (EC
2.7.7.49) (EC 3.1.26.4) (RT); Integrase (IN)] (Fragment)
Length = 581
Score = 43.9 bits (102), Expect = 0.002
Identities = 39/152 (25%), Positives = 68/152 (44%), Gaps = 9/152 (5%)
Query: 72 IHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHHITPKFIRSDNGPEFL 131
++G+KY L VD FS ++ K++ + K+ +I P+ + +DNGP F+
Sbjct: 309 LYGYKYLLVFVDTFSGWVEAFPTKHETAKIVTKKLLEEIFPRFG--MPQVLGTDNGPAFV 366
Query: 132 --LLEFYAS-KGIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVLY 188
+ + A GI + C P+ + +VER ++ I L L + +L
Sbjct: 367 SQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKL----TLATGTRDWVLLL 422
Query: 189 SVFLINRVPTPFLHHKSPY*VLYDSLPDIQFF 220
+ L TP H +PY +LY + P + F
Sbjct: 423 PLALYRARNTPGPHGLTPYEILYGAPPPLVNF 454
>POL_MLVRD (P11227) Pol polyprotein [Contains: Protease (EC 3.4.23.-);
Reverse transcriptase/ribonuclease H (EC 2.7.7.49) (EC
3.1.26.4) (RT); Integrase (IN)]
Length = 1196
Score = 43.9 bits (102), Expect = 0.002
Identities = 39/152 (25%), Positives = 68/152 (44%), Gaps = 9/152 (5%)
Query: 72 IHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHHITPKFIRSDNGPEFL 131
++G+KY L VD FS ++ K++ + K+ +I P+ + +DNGP F+
Sbjct: 924 LYGYKYLLVFVDTFSGWVEAFPTKHETAKIVTKKLLEEIFPRFG--MPQVLGTDNGPAFV 981
Query: 132 --LLEFYAS-KGIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVLY 188
+ + A GI + C P+ + +VER ++ I L L + +L
Sbjct: 982 SQVSQSVAKLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKL----TLATGTRDWVLLL 1037
Query: 189 SVFLINRVPTPFLHHKSPY*VLYDSLPDIQFF 220
+ L TP H +PY +LY + P + F
Sbjct: 1038 PLALYRARNTPGPHGLTPYEILYGAPPPLVNF 1069
>POL_MLVCB (P08361) Pol polyprotein [Contains: Reverse
transcriptase/ribonuclease H (EC 2.7.7.49) (EC 3.1.26.4)
(RT); Integrase (IN)] (Fragment)
Length = 282
Score = 43.9 bits (102), Expect = 0.002
Identities = 39/152 (25%), Positives = 67/152 (43%), Gaps = 9/152 (5%)
Query: 72 IHGHKYFLTIVDDFSRFLWVILLKNKAEVSLPVKIFVQIIDTHHHITPKFIRSDNGPEFL 131
++G+KY L VD FS ++ K + + K+ +I P+ + +DNGP F+
Sbjct: 10 LYGYKYLLVFVDTFSGWIEAFPTKKETAKVVTKKLLEEIFPRFG--MPQVLGTDNGPAFV 67
Query: 132 --LLEFYAS-KGIIHQKSCVETPEQNARVERKHQHILNVARALPFQFKLPNIFWSYAVLY 188
+ + A GI + C P+ + +VER ++ I L L + +L
Sbjct: 68 SKVSQTVADLLGIDWKLHCAYRPQSSGQVERMNRTIKETLTKL----TLATGSRDWVLLL 123
Query: 189 SVFLINRVPTPFLHHKSPY*VLYDSLPDIQFF 220
+ L TP H +PY +LY + P + F
Sbjct: 124 PLALYRARNTPGPHGLTPYEILYGAPPPLVNF 155
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.340 0.149 0.489
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 108,642,209
Number of Sequences: 164201
Number of extensions: 4305199
Number of successful extensions: 14924
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 14774
Number of HSP's gapped (non-prelim): 101
length of query: 1021
length of database: 59,974,054
effective HSP length: 120
effective length of query: 901
effective length of database: 40,269,934
effective search space: 36283210534
effective search space used: 36283210534
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.9 bits)
S2: 71 (32.0 bits)
Medicago: description of AC136506.7


