

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136505.11 + phase: 0
(445 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
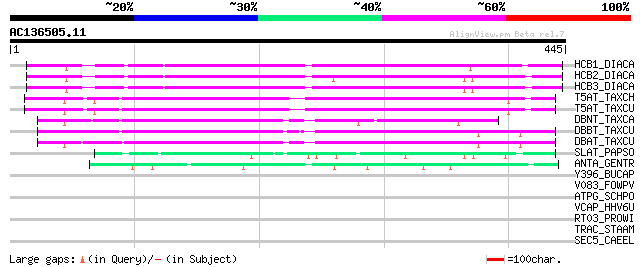
Score E
Sequences producing significant alignments: (bits) Value
HCB1_DIACA (O24645) Anthranilate N-benzoyltransferase protein 1 ... 207 4e-53
HCB2_DIACA (O23917) Anthranilate N-benzoyltransferase protein 2 ... 205 2e-52
HCB3_DIACA (O23918) Anthranilate N-benzoyltransferase protein 3 ... 198 2e-50
T5AT_TAXCH (Q8S9G6) Taxadien-5-alpha-ol O-acetyltransferase (EC ... 172 2e-42
T5AT_TAXCU (Q9M6F0) Taxadien-5-alpha-ol O-acetyltransferase (EC ... 167 4e-41
DBNT_TAXCA (Q8LL69) 3'-N-debenzoyl-2'-deoxytaxol N-benzoyltransf... 149 1e-35
DBBT_TAXCU (Q9FPW3) 2-alpha-hydroxytaxane 2-O-benzoyltransferase... 144 5e-34
DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III 10-O-acetyltransfera... 129 1e-29
SLAT_PAPSO (Q94FT4) Salutaridinol 7-O-acetyltransferase (EC 2.3.... 77 9e-14
ANTA_GENTR (Q9ZWR8) Anthocyanin 5-aromatic acyltransferase (EC 2... 69 3e-11
Y396_BUCAP (Q8K9E3) Hypothetical protein BUsg396 35 0.39
V083_FOWPV (O72903) Protein FPV083 35 0.39
ATPG_SCHPO (O74754) ATP synthase gamma chain, mitochondrial prec... 34 0.66
VCAP_HHV6U (P17887) Major capsid protein (MCP) 33 1.1
RT03_PROWI (P46740) Mitochondrial ribosomal protein S3 33 1.1
TRAC_STAAM (P06698) Transposase for transposon Tn554 31 5.6
SEC5_CAEEL (Q22706) Probable exocyst complex component Sec5 30 9.6
>HCB1_DIACA (O24645) Anthranilate N-benzoyltransferase protein 1 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 1)
Length = 445
Score = 207 bits (527), Expect = 4e-53
Identities = 149/467 (31%), Positives = 226/467 (47%), Gaps = 59/467 (12%)
Query: 14 MKVTIHKTSMIFPSKQTENKSMFLSNIDKVL---------------------NFYVKTVH 52
M + I +++M+ P+++T NKS++LSNID +L N + +
Sbjct: 1 MSIQIKQSTMVRPAEETPNKSLWLSNIDMILRTPYSHTGAVLIYKQPDNNEDNIHPSSSM 60
Query: 53 FFEANKDFPPQIVTEKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASS 112
+F+AN L +AL ALV + +AGRLK+N + R EIDCNAEG FV A S
Sbjct: 61 YFDANI----------LIEALSKALVPFYPMAGRLKINGD--RYEIDCNAEGALFVEAES 108
Query: 113 EYKLNQIGDLAYPNQAFAQFVHNAKDFLK-IGDLPLCVVQLTSFKCGGFAIGFTTSHVTF 171
+ L GD PN + + D+ K I PL +VQLT F+CGG +IGF H
Sbjct: 109 SHVLEDFGDFR-PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHVC 167
Query: 172 DGLSFKNFLDNLASLATNMKLTATPFHDRQL-LAARSPPHVNFPHPEMIK-LDNLPLGIK 229
DG++ F ++ A +A + P HDR L L R+PP + + H + + +LP +
Sbjct: 168 DGMAHFEFNNSWARIAKGLLPALEPVHDRYLHLRPRNPPQIKYSHSQFEPFVPSLPNELL 227
Query: 230 SGVFEASSEEFDFMVFQLTYEDINNLKEKAK-GNNTSRVISFNVVAAHIWRCKALSKSYD 288
G S +F L+ E IN LK+K NNT+R+ ++ VVAAH+WR + ++
Sbjct: 228 DGKTNKSQ-----TLFILSREQINTLKQKLDLSNNTTRLSTYEVVAAHVWRSVSKARGLS 282
Query: 289 PNRSSIILYAVDIRSRL-NPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSR 347
+ ++ VD RSR+ NP LP+ Y GN V A +A +L + V+E +
Sbjct: 283 DHEEIKLIMPVDGRSRINNPSLPKGYCGNVVFLAVCTATVGDLSCNPLTDTAGKVQEALK 342
Query: 348 RMSDEYARSIIDWGELYNGFP-----------NGDVLVSSWWRLGFEEVEYPWGKPKYCC 396
+ D+Y RS ID E G P +VLV+SW R+ ++ +++ WG P +
Sbjct: 343 GLDDDYLRSAIDHTESKPGLPVPYMGSPEKTLYPNVLVNSWGRIPYQAMDFGWGSPTFFG 402
Query: 397 PVVYHRKDIILLFPSFNGGEGGGDGVNIIVALPRDEMEKFKSLFYTF 443
L PS G + + + L + +FK FY F
Sbjct: 403 ISNIFYDGQCFLIPS----RDGDGSMTLAINLFSSHLSRFKKYFYDF 445
>HCB2_DIACA (O23917) Anthranilate N-benzoyltransferase protein 2 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 2)
Length = 446
Score = 205 bits (521), Expect = 2e-52
Identities = 152/469 (32%), Positives = 227/469 (47%), Gaps = 62/469 (13%)
Query: 14 MKVTIHKTSMIFPSKQTENKSMFLSNIDKVL---------------------NFYVKTVH 52
M + I +++M+ P+++T NKS++LS ID +L N + +
Sbjct: 1 MSIQIKQSTMVRPAEETPNKSLWLSKIDMILRTPYSHTGAVLIYKQPDNNEDNIHPSSSM 60
Query: 53 FFEANKDFPPQIVTEKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASS 112
+F+AN L +AL ALV Y +AGRLK+N + R EIDCNAEG FV A S
Sbjct: 61 YFDANI----------LIEALSKALVPYYPMAGRLKINGD--RYEIDCNAEGALFVEAES 108
Query: 113 EYKLNQIGDLAYPNQAFAQFVHNAKDFLK-IGDLPLCVVQLTSFKCGGFAIGFTTSHVTF 171
+ L GD PN + + D+ K I PL +VQLT F+CGG +IGF H
Sbjct: 109 SHVLEDFGDFR-PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHAC 167
Query: 172 DGLSFKNFLDNLASLATNMKLTATPFHDRQL-LAARSPPHVNFPHPEMIK-LDNLPLGIK 229
DG+S F ++ A +A + P HDR L L R+PP + + H + + +LP +
Sbjct: 168 DGMSHFEFNNSWARIAKGLLPALEPVHDRYLHLRLRNPPQIKYTHSQFEPFVPSLPNELL 227
Query: 230 SGVFEASSEEFDFMVFQLTYEDINNLKEK--AKGNNTSRVISFNVVAAHIWRCKALSKSY 287
G S +F+L+ E IN LK+K N T+R+ ++ VVA H+WR + ++
Sbjct: 228 DGKTNKSQ-----TLFKLSREQINTLKQKLDLSSNTTTRLSTYEVVAGHVWRSVSKARGL 282
Query: 288 DPNRSSIILYAVDIRSRL-NPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGS 346
+ ++ VD RSR+ NP LP+ Y GN V A +A +L + V+E
Sbjct: 283 SDHEEIKLIMPVDGRSRINNPSLPKGYCGNVVFLAVCTATVGDLSCNPLTDTAGKVQEAL 342
Query: 347 RRMSDEYARSIIDWGEL-------YNGFPN----GDVLVSSWWRLGFEEVEYPWGKPKYC 395
+ + D+Y RS ID E Y G P +VLV+SW R+ ++ +++ WG P +
Sbjct: 343 KGLDDDYLRSAIDHTESKPDLPVPYMGSPEKTLYPNVLVNSWGRIPYQAMDFGWGSPTFF 402
Query: 396 CPVVYHRKDIILLFPSFNGGEGGGDG-VNIIVALPRDEMEKFKSLFYTF 443
L PS N GDG + + + L + FK FY F
Sbjct: 403 GISNIFYDGQCFLIPSQN-----GDGSMTLAINLFSSHLSLFKKYFYDF 446
>HCB3_DIACA (O23918) Anthranilate N-benzoyltransferase protein 3 (EC
2.3.1.144) (Anthranilate
N-hydroxycinnamoyl/benzoyltransferase 3)
Length = 445
Score = 198 bits (504), Expect = 2e-50
Identities = 150/468 (32%), Positives = 224/468 (47%), Gaps = 61/468 (13%)
Query: 14 MKVTIHKTSMIFPSKQTENKSMFLSNIDKVL---------------------NFYVKTVH 52
M + I +++M+ P+++T NKS++LS ID +L N +
Sbjct: 1 MSIHIKQSTMVRPAEETPNKSLWLSKIDMILRTPYSHTGAVLIYKQPDNNEDNIQPSSSM 60
Query: 53 FFEANKDFPPQIVTEKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASS 112
+F+AN L +AL ALV Y +AGRLK+N + R EIDCN EG FV A S
Sbjct: 61 YFDANI----------LIEALSKALVPYYPMAGRLKINGD--RYEIDCNGEGALFVEAES 108
Query: 113 EYKLNQIGDLAYPNQAFAQFVHNAKDFLK-IGDLPLCVVQLTSFKCGGFAIGFTTSHVTF 171
+ L GD PN + + D+ K I PL +VQLT F+CGG +IGF H
Sbjct: 109 SHVLEDFGDFR-PNDELHRVMVPTCDYSKGISSFPLLMVQLTRFRCGGVSIGFAQHHHVC 167
Query: 172 DGLSFKNFLDNLASLATNMKLTATPFHDRQL-LAARSPPHVNFPHPEMIK-LDNLPLGIK 229
D +S F ++ A +A + P HDR L L R+PP + + H + + +LP +
Sbjct: 168 DRMSHFEFNNSWARIAKGLLPALEPVHDRYLHLCPRNPPQIKYTHSQFEPFVPSLPKELL 227
Query: 230 SGVFEASSEEFDFMVFQLTYEDINNLKEKAK-GNNTSRVISFNVVAAHIWRCKALSKSYD 288
G S +F+L+ E IN LK+K N T+R+ ++ VVA H+WR + ++
Sbjct: 228 DGKTSKSQ-----TLFKLSREQINTLKQKLDWSNTTTRLSTYEVVAGHVWRSVSKARGLS 282
Query: 289 PNRSSIILYAVDIRSRL-NPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSR 347
+ ++ VD RSR+ NP LP+ Y GN V A +A +L + V+E +
Sbjct: 283 DHEEIKLIMPVDGRSRINNPSLPKGYCGNVVFLAVCTATVGDLACNPLTDTAGKVQEALK 342
Query: 348 RMSDEYARSIIDWGEL-------YNGFPN----GDVLVSSWWRLGFEEVEYPWGKPKYCC 396
+ D+Y RS ID E Y G P +VLV+SW R+ ++ +++ WG P +
Sbjct: 343 GLDDDYLRSAIDHTESKPDLPVPYMGSPEKTLYPNVLVNSWGRIPYQAMDFGWGNPTFFG 402
Query: 397 PVVYHRKDIILLFPSFNGGEGGGDG-VNIIVALPRDEMEKFKSLFYTF 443
L PS N GDG + + + L + FK FY F
Sbjct: 403 ISNIFYDGQCFLIPSQN-----GDGSMTLAINLFSSHLSLFKKHFYDF 445
>T5AT_TAXCH (Q8S9G6) Taxadien-5-alpha-ol O-acetyltransferase (EC
2.3.1.162)
(Taxa-4(20),11(12)-dien-5alpha-ol-O-acetyltransferase)
(Taxadienol acetyltransferase)
Length = 439
Score = 172 bits (435), Expect = 2e-42
Identities = 125/440 (28%), Positives = 213/440 (48%), Gaps = 34/440 (7%)
Query: 13 DMKVTIHKTSMIFPSKQTENKSMFLSNIDK---VLNFYVKTVHFFEANKDFPPQIVT--- 66
D+ V +++ M+ PS ++ LS+ID V + + A+ P +V+
Sbjct: 5 DLHVNLNEKVMVGPSLPLPKTTLQLSSIDNLPGVRGSIFNALLIYNASPS--PTMVSADP 62
Query: 67 -EKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYP 125
+ +++AL LV Y AGRL+ TE LE++C EG F+ A ++ +L+ +GD
Sbjct: 63 AKLIREALAKILVYYPPFAGRLR-ETENGDLEVECTGEGAMFLEAMADNELSVLGDFDDS 121
Query: 126 NQAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLAS 185
N +F Q + + DLPL VVQ+T F CGGF +G + H DG FL LA
Sbjct: 122 NPSFQQLLFSLPLDTNFKDLPLLVVQVTRFTCGGFVVGVSFHHGVCDGRGAAQFLKGLAE 181
Query: 186 LAT-NMKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMV 244
+A +KL+ P +R+L+ P ++ F H E ++ ++ E+
Sbjct: 182 MARGEVKLSLEPIWNRELVKLDDPKYLQFFHFEFLRAPSI------------VEKIVQTY 229
Query: 245 FQLTYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSR 304
F + +E IN +K+ SF V +A W + + + IL+ +D+R+
Sbjct: 230 FIIDFETINYIKQSVMEECKEFCSSFEVASAMTWIARTRAFQIPESEYVKILFGMDMRNS 289
Query: 305 LNPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEY-ARSIIDWGEL 363
NPPLP Y GN++ TA A ++L G R + ++++ ++D + +R+++ EL
Sbjct: 290 FNPPLPSGYYGNSIGTACAVDNVQDLLSGSLLRAIMIIKKSKVSLNDNFKSRAVVKPSEL 349
Query: 364 YNGFPNGDVLV-SSWWRLGFEEVEYPWGKPKYCCPV-----VYHRKDIILLFPSFNGGEG 417
+ +V+ + W RLGF+EV++ WG PV + + + L PS N
Sbjct: 350 DVNMNHENVVAFADWSRLGFDEVDFGWGNAVSVSPVQQQCELAMQNYFLFLKPSKN---- 405
Query: 418 GGDGVNIIVALPRDEMEKFK 437
DG+ I++ LP +M+ FK
Sbjct: 406 KPDGIKILMFLPLSKMKSFK 425
>T5AT_TAXCU (Q9M6F0) Taxadien-5-alpha-ol O-acetyltransferase (EC
2.3.1.162)
(Taxa-4(20),11(12)-dien-5alpha-ol-O-acetyltransferase)
(Taxadienol acetyltransferase)
Length = 439
Score = 167 bits (424), Expect = 4e-41
Identities = 123/440 (27%), Positives = 211/440 (47%), Gaps = 34/440 (7%)
Query: 13 DMKVTIHKTSMIFPSKQTENKSMFLSNIDK---VLNFYVKTVHFFEANKDFPPQIVT--- 66
D+ V + + M+ PS ++ LS+ID V + + A+ P +++
Sbjct: 5 DLHVNLIEKVMVGPSPPLPKTTLQLSSIDNLPGVRGSIFNALLIYNASPS--PTMISADP 62
Query: 67 -EKLKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYP 125
+ +++AL LV Y AGRL+ TE LE++C EG F+ A ++ +L+ +GD
Sbjct: 63 AKPIREALAKILVYYPPFAGRLR-ETENGDLEVECTGEGAMFLEAMADNELSVLGDFDDS 121
Query: 126 NQAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLAS 185
N +F Q + + DL L VVQ+T F CGGF +G + H DG FL LA
Sbjct: 122 NPSFQQLLFSLPLDTNFKDLSLLVVQVTRFTCGGFVVGVSFHHGVCDGRGAAQFLKGLAE 181
Query: 186 LAT-NMKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMV 244
+A +KL+ P +R+L+ P ++ F H E ++ ++ E+
Sbjct: 182 MARGEVKLSLEPIWNRELVKLDDPKYLQFFHFEFLRAPSI------------VEKIVQTY 229
Query: 245 FQLTYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSR 304
F + +E IN +K+ SF V +A W + + + IL+ +D+R+
Sbjct: 230 FIIDFETINYIKQSVMEECKEFCSSFEVASAMTWIARTRAFQIPESEYVKILFGMDMRNS 289
Query: 305 LNPPLPRAYTGNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEY-ARSIIDWGEL 363
NPPLP Y GN++ TA A ++L G R + ++++ ++D + +R+++ EL
Sbjct: 290 FNPPLPSGYYGNSIGTACAVDNVQDLLSGSLLRAIMIIKKSKVSLNDNFKSRAVVKPSEL 349
Query: 364 YNGFPNGDVLV-SSWWRLGFEEVEYPWGKPKYCCPV-----VYHRKDIILLFPSFNGGEG 417
+ +V+ + W RLGF+EV++ WG PV + + + L PS N
Sbjct: 350 DVNMNHENVVAFADWSRLGFDEVDFGWGNAVSVSPVQQQSALAMQNYFLFLKPSKN---- 405
Query: 418 GGDGVNIIVALPRDEMEKFK 437
DG+ I++ LP +M+ FK
Sbjct: 406 KPDGIKILMFLPLSKMKSFK 425
>DBNT_TAXCA (Q8LL69) 3'-N-debenzoyl-2'-deoxytaxol
N-benzoyltransferase (EC 2.3.1.-) (DBTNBT)
Length = 441
Score = 149 bits (377), Expect = 1e-35
Identities = 117/378 (30%), Positives = 179/378 (46%), Gaps = 24/378 (6%)
Query: 23 MIFPSKQTENKSMFLSNIDK--VLNFYVKTVHFFEANKDFPPQIVTEKLKKALEDALVAY 80
M+ PS + ++ LS +D + T+ F A + V + +++AL LV Y
Sbjct: 18 MVAPSLPSPKATVQLSVVDSLTICRGIFNTLLVFNAPDNISADPV-KIIREALSKVLVYY 76
Query: 81 DFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAFAQFVHNAKDFL 140
LAGRL+ + E LE++C +G FV A E ++ + DL N +F Q V
Sbjct: 77 FPLAGRLR-SKEIGELEVECTGDGALFVEAMVEDTISVLRDLDDLNPSFQQLVFWHPLDT 135
Query: 141 KIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLATN-MKLTATPFHD 199
I DL L +VQ+T F CGG A+G T H DG F+ LA +A +K + P +
Sbjct: 136 AIEDLHLVIVQVTRFTCGGIAVGVTLPHSVCDGRGAAQFVTALAEMARGEVKPSLEPIWN 195
Query: 200 RQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQLTYEDINNLKEKA 259
R+LL P H+ + I P + + +AS F + + I +K+
Sbjct: 196 RELLNPEDPLHLQLNQFDSI----CPPPMLEELGQAS--------FVINVDTIEYMKQCV 243
Query: 260 KGNNTSRVISFNVVAAHIW--RCKALSKSYDPNRSSIILYAVDIRSRLNPPLPRAYTGNA 317
SF VVAA +W R KAL + N +L+A+D+R NPPLP Y GNA
Sbjct: 244 MEECNEFCSSFEVVAALVWIARTKALQIPHTENVK--LLFAMDLRKLFNPPLPNGYYGNA 301
Query: 318 VLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSII---DWGELYNGFPNGDVLV 374
+ TAYA ++L G R + ++++ + D Y+RS + + N + + +
Sbjct: 302 IGTAYAMDNVQDLLNGSLLRAIMIIKKAKADLKDNYSRSRVVTNPYSLDVNKKSDNILAL 361
Query: 375 SSWWRLGFEEVEYPWGKP 392
S W RLGF E ++ WG P
Sbjct: 362 SDWRRLGFYEADFGWGGP 379
>DBBT_TAXCU (Q9FPW3) 2-alpha-hydroxytaxane 2-O-benzoyltransferase
(EC 2.3.1.166) (TBT) (2-debenzoyl-7,13-diacetylbaccatin
III-2-O-benzoyl transferase) (DBBT)
Length = 440
Score = 144 bits (363), Expect = 5e-34
Identities = 107/422 (25%), Positives = 189/422 (44%), Gaps = 20/422 (4%)
Query: 23 MIFPSKQTENKSMFLSNIDKVLNFYVKTVHFFEANKDFPPQIVTEK-LKKALEDALVAYD 81
++ P Q+ + LS ID + + A++ K +++AL LV Y
Sbjct: 13 IVAPCLQSPKNILHLSPIDNKTRGLTNILSVYNASQRVSVSADPAKTIREALSKVLVYYP 72
Query: 82 FLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAFAQFVHNAKDFLK 141
AGRL+ NTE LE++C EG FV A ++ L+ + D + +F Q V N ++ +
Sbjct: 73 PFAGRLR-NTENGDLEVECTGEGAVFVEAMADNDLSVLQDFNEYDPSFQQLVFNLREDVN 131
Query: 142 IGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLAT-NMKLTATPFHDR 200
I DL L VQ+T F CGGF +G H DG L + +A K + P +R
Sbjct: 132 IEDLHLLTVQVTRFTCGGFVVGTRFHHSVSDGKGIGQLLKGMGEMARGEFKPSLEPIWNR 191
Query: 201 QLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQLTYEDINNLKEKAK 260
+++ ++ F H + I + PL ++ + +AS +++E IN +K
Sbjct: 192 EMVKPEDIMYLQFDHFDFI---HPPLNLEKSI-QAS--------MVISFERINYIKRCMM 239
Query: 261 GNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPPLPRAYTGNAVLT 320
+F VV A IW + S PN I++ +D+R+ + PLP+ Y GNA+
Sbjct: 240 EECKEFFSAFEVVVALIWLARTKSFRIPPNEYVKIIFPIDMRNSFDSPLPKGYYGNAIGN 299
Query: 321 AYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYNGFPNGDVLV--SSWW 378
A A ++L G + ++++ +++ + I+ + + +V W
Sbjct: 300 ACAMDNVKDLLNGSLLYALMLIKKSKFALNENFKSRILTKPSTLDANMKHENVVGCGDWR 359
Query: 379 RLGFEEVEYPWGKPKYCCPVVYHRKDIILL---FPSFNGGEGGGDGVNIIVALPRDEMEK 435
LGF E ++ WG P+ R+ + + F + DG+ I++ +P ++
Sbjct: 360 NLGFYEADFGWGNAVNVSPMQQQREHELAMQNYFLFLRSAKNMIDGIKILMFMPASMVKP 419
Query: 436 FK 437
FK
Sbjct: 420 FK 421
>DBAT_TAXCU (Q9M6E2) 10-deacetylbaccatin III 10-O-acetyltransferase
(EC 2.3.1.167) (DBAT)
Length = 440
Score = 129 bits (325), Expect = 1e-29
Identities = 110/425 (25%), Positives = 194/425 (44%), Gaps = 24/425 (5%)
Query: 23 MIFPSKQTENKSMFLSNIDK---VLNFYVKTVHFFEANKDFPPQIVTEKLKKALEDALVA 79
M+ PS+ + + LS +D V T+ + A+ D + +++AL LV
Sbjct: 16 MVAPSQPSPKAFLQLSTLDNLPGVRENIFNTLLVYNAS-DRVSVDPAKVIRQALSKVLVY 74
Query: 80 YDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQAFAQFVHNAKDF 139
Y AGRL+ E LE++C EG FV A ++ L+ +GDL + + Q +
Sbjct: 75 YSPFAGRLRKK-ENGDLEVECTGEGALFVEAMADTDLSVLGDLDDYSPSLEQLLFCLPPD 133
Query: 140 LKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLAT-NMKLTATPFH 198
I D+ VVQ+T F CGGF +G + H DGL FL + +A +K ++ P
Sbjct: 134 TDIEDIHPLVVQVTRFTCGGFVVGVSFCHGICDGLGAGQFLIAMGEMARGEIKPSSEPIW 193
Query: 199 DRQLLAARSPPH-VNFPHPEMIKLDNLPLGIKSGVFEASSEEFDFMVFQLTYEDINNLKE 257
R+LL P + + H ++I P + + S +T E IN +K+
Sbjct: 194 KRELLKPEDPLYRFQYYHFQLI----CPPSTFGKIVQGS--------LVITSETINCIKQ 241
Query: 258 KAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAVDIRSRLNPPLPRAYTGNA 317
+ + +F VV+A W + + + + +++A+D+R NPPL + Y GN
Sbjct: 242 CLREESKEFCSAFEVVSALAWIARTRALQIPHSENVKLIFAMDMRKLFNPPLSKGYYGNF 301
Query: 318 VLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYNGFPNGDVLV--S 375
V T A ++L G R+V ++++ +++ + +I+ + N + +V
Sbjct: 302 VGTVCAMDNVKDLLSGSLLRVVRIIKKAKVSLNEHFTSTIVTPRSGSDESINYENIVGFG 361
Query: 376 SWWRLGFEEVEYPWGKPKYCCPVVYHRKDIILL---FPSFNGGEGGGDGVNIIVALPRDE 432
RLGF+EV++ WG V + KD+ ++ F + DG+ I+ +P
Sbjct: 362 DRRRLGFDEVDFGWGHADNVSLVQHGLKDVSVVQSYFLFIRPPKNNPDGIKILSFMPPSI 421
Query: 433 MEKFK 437
++ FK
Sbjct: 422 VKSFK 426
>SLAT_PAPSO (Q94FT4) Salutaridinol 7-O-acetyltransferase (EC
2.3.1.150) (salAT)
Length = 474
Score = 77.0 bits (188), Expect = 9e-14
Identities = 93/412 (22%), Positives = 167/412 (39%), Gaps = 61/412 (14%)
Query: 69 LKKALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGFVVASSEYKLNQIGDLAYPNQA 128
LK +L LV + +AGR+ N + +DC+ +G+ F K+ + ++ P+
Sbjct: 71 LKSSLSKTLVHFYPMAGRMIDN-----ILVDCHDQGINFYKVKIRGKMCEF--MSQPDVP 123
Query: 129 FAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSFKNFLDNLASLAT 188
+Q + + + L +VQ+ F CGG AI + SH D + F+ + AS
Sbjct: 124 LSQLLPSEVVSASVPKEALVIVQVNMFDCGGTAICSSVSHKIADAATMSTFIRSWASTTK 183
Query: 189 NMKL--TATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLGIKSGVFEASSE--EFDFMV 244
+ + D++L+ + + FP E + P G+ F ++ E E D V
Sbjct: 184 TSRSGGSTAAVTDQKLIPSFDSASL-FPPSERL---TSPSGMSEIPFSSTPEDTEDDKTV 239
Query: 245 ---FQLTYEDINNLKEKAK-----GNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIIL 296
F + I +++EK + + R VV + IW K++ KS ++
Sbjct: 240 SKRFVFDFAKITSVREKLQVLMHDNYKSRRQTRVEVVTSLIW--KSVMKSTPAGFLPVVH 297
Query: 297 YAVDIRSRLNPPLPRAYTGN------------AVLTAYASAKCEELKEGEFSRLVEMVEE 344
+AV++R +++PPL GN T A K E ++ + +
Sbjct: 298 HAVNLRKKMDPPLQDVSFGNLSVTVSAFLPATTTTTTNAVNKTINSTSSESQVVLHELHD 357
Query: 345 GSRRMSDEYARSIIDWGEL---YNGFPNG--------------DVLVSSWWRLGFEEVEY 387
+M E + D G L F +G + +SSW R+G E+++
Sbjct: 358 FIAQMRSEIDKVKGDKGSLEKVIQNFASGHDASIKKINDVEVINFWISSWCRMGLYEIDF 417
Query: 388 PWGKPKYCC--PVVYHRKDIILLFPSFNGGEGGGDGVNIIVALPRDEMEKFK 437
WGKP + P + K+ F G+G+ + + D+M KF+
Sbjct: 418 GWGKPIWVTVDPNIKPNKNCF-----FMNDTKCGEGIEVWASFLEDDMAKFE 464
>ANTA_GENTR (Q9ZWR8) Anthocyanin 5-aromatic acyltransferase (EC
2.3.1.153) (5AT)
Length = 469
Score = 68.6 bits (166), Expect = 3e-11
Identities = 97/412 (23%), Positives = 156/412 (37%), Gaps = 48/412 (11%)
Query: 65 VTEKLKKALEDALVAYDFLAGRLKMNTETNRLE----IDCNAEGVGFVVASSE------- 113
V LK +L L Y L+G L M ++ + + + +VA S+
Sbjct: 62 VIPNLKASLSLTLKHYVPLSGNLLMPIKSGEMPKFQYSRDEGDSITLIVAESDQDFDYLK 121
Query: 114 -YKLNQIGDLAYPNQAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFD 172
++L DL + + +D+ I PL VQ+T F G A+ T H D
Sbjct: 122 GHQLVDSNDLHGLFYVMPRVIRTMQDYKVI---PLVAVQVTVFPNRGIAVALTAHHSIAD 178
Query: 173 GLSFKNFLDNLASL------ATNMKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLP- 225
SF F++ A + A + P DR ++ F + L+
Sbjct: 179 AKSFVMFINAWAYINKFGKDADLLSANLLPSFDRSIIKDLYGLEETFWNEMQDVLEMFSR 238
Query: 226 LGIKSGVFEASSEEFDFMVFQLTYEDINNLKEKA---KGNN-TSRVISFNVVAAHIWRCK 281
G K F + L+ +I LK K +G+ T RV +F + ++W C
Sbjct: 239 FGSKPPRFNKVR-----ATYVLSLAEIQKLKNKVLNLRGSEPTIRVTTFTMTCGYVWTCM 293
Query: 282 ALSK--------SYDPNRSSIILYAVDIRSRLNPPLPRAYTGNAVLTAYASAKCEEL--K 331
SK S D N + D R L PP P Y GN + + A A +EL
Sbjct: 294 VKSKDDVVSEESSNDENELEYFSFTADCRGLLTPPCPPNYFGNCLASCVAKATHKELVGD 353
Query: 332 EGEFSRLVEMVEEGSRRMSDE--YARSIIDWGELYNGFPNGDVL-VSSWWRLGFEEVEYP 388
+G + + E +R+ +E W NG P+ L ++ + V++
Sbjct: 354 KGLLVAVAAIGEAIEKRLHNEKGVLADAKTWLSESNGIPSKRFLGITGSPKFDSYGVDFG 413
Query: 389 WGKPKYCCPVVYHRKDIILLFPSFNGGEGGGDGVNIIVALPRDEMEKFKSLF 440
WGKP ++I + S + + GV I V+LP+ M+ F +F
Sbjct: 414 WGKPAKFDITSVDYAELIYVIQSRDFEK----GVEIGVSLPKIHMDAFAKIF 461
>Y396_BUCAP (Q8K9E3) Hypothetical protein BUsg396
Length = 199
Score = 35.0 bits (79), Expect = 0.39
Identities = 37/144 (25%), Positives = 57/144 (38%), Gaps = 11/144 (7%)
Query: 27 SKQTENKSMFLSNIDKVLNFYVKTVHFFEANKDFPPQIVTEKLKKALEDALVAYDFLAGR 86
+K+++N+ S I V N T + E+NK + A A FL+
Sbjct: 3 TKRSQNRKEIRSYIRIVRNSVTLTKKYDESNKIVRTAFNCNIIYNAKNIAC----FLSFD 58
Query: 87 LKMNTETNRLEIDCNAEGVGFVVASSEYKLNQI-------GDLAYPNQAFAQFVHNAKDF 139
++NT L++ N + V + SS Y L Y Q +N KD
Sbjct: 59 GEINTYPLILKLWLNNKNVFLPIVSSSYSRKLFFVPFTCKSILYYNQYNILQPFYNMKDI 118
Query: 140 LKIGDLPLCVVQLTSFKCGGFAIG 163
+ DL L +V L +F C G +G
Sbjct: 119 ILESDLDLIIVPLVAFDCRGVRLG 142
>V083_FOWPV (O72903) Protein FPV083
Length = 421
Score = 35.0 bits (79), Expect = 0.39
Identities = 23/83 (27%), Positives = 34/83 (40%), Gaps = 13/83 (15%)
Query: 224 LPLGIKSGVFEASSEEFDFMVFQLTYE---------DINNLKEKAKGNNTSRVISFNVVA 274
+P+ + V F+FM YE ++ LK++ + N +SR I F
Sbjct: 180 IPMCYGTEVIYIGQFNFNFMNRHAIYEKSSVFNKNTEVFKLKDRIRDNRSSRFIMFGFCY 239
Query: 275 AHIWRCKALSKSYDPNRSSIILY 297
H W+C YD NR I Y
Sbjct: 240 LHHWKCAI----YDKNRDFICFY 258
>ATPG_SCHPO (O74754) ATP synthase gamma chain, mitochondrial
precursor (EC 3.6.3.14)
Length = 301
Score = 34.3 bits (77), Expect = 0.66
Identities = 25/94 (26%), Positives = 44/94 (46%), Gaps = 15/94 (15%)
Query: 245 FQLTYEDINNLKEKAKG--------NNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIIL 296
F+ + +N+ E AK N + +SF V +++ KA+++S PN S
Sbjct: 167 FEEALQISSNILEHAKDYDRIVLVYNKFASAVSFETVMKNLYTTKAINES--PNLS---- 220
Query: 297 YAVDIRSRLNPPLPRAYTGNAVLTAYASAKCEEL 330
A ++ ++ PL NA+ +A A A C E+
Sbjct: 221 -AYEVSDEVHQPLMEFAFANAIFSAMAEAHCSEM 253
>VCAP_HHV6U (P17887) Major capsid protein (MCP)
Length = 1345
Score = 33.5 bits (75), Expect = 1.1
Identities = 22/78 (28%), Positives = 35/78 (44%), Gaps = 3/78 (3%)
Query: 117 NQIGDLAYPNQAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTSHVTFDGLSF 176
N +GD P AFA + N +++ + + L CG + F ++ FDG F
Sbjct: 658 NHMGDELIPGAAFAHY-RNLVSLIRLVKRTISISNLNEQLCGEPLVNF--ANALFDGRLF 714
Query: 177 KNFLDNLASLATNMKLTA 194
F+ + TN K+TA
Sbjct: 715 CPFVHTMPRNDTNAKITA 732
>RT03_PROWI (P46740) Mitochondrial ribosomal protein S3
Length = 500
Score = 33.5 bits (75), Expect = 1.1
Identities = 54/297 (18%), Positives = 115/297 (38%), Gaps = 58/297 (19%)
Query: 4 TNKETKPLLDM------KVTIHKTSMIFPSKQTENKSMFLSNIDKVLNFYVKTVHFFEAN 57
++ +K LL M K + +K+S+++ +K ++ I++V N K ++ +
Sbjct: 219 SDNSSKSLLKMLLREYYKSSDNKSSILYENKSPKSLENIFGKINEVSNINKKNLYLLNSE 278
Query: 58 KDFPPQIVTEKLK----------KALEDALVAYDFLAGRLKMNTETNRLEIDCNAEGVGF 107
D P + K K K +ED + F+ ++N+++ D
Sbjct: 279 TDNIPNLYKRKSKPTKFLRNEHNKVIEDKKTSLYFMP-------DSNQIKNDIY------ 325
Query: 108 VVASSEYKLNQIGDLAYPNQAFAQFVHNAKDFLKIGDLPLCVVQLTSFKCGGFAIGFTTS 167
+ K+++I ++ +N+KD L +L L L + +G + S
Sbjct: 326 -----QGKISKISSIS----------NNSKDSLTDYNLHLYQAYLHNI------LGNSKS 364
Query: 168 HVTFDGLSFKNFLDNLASLATNMKLTATPFHDRQLLAARSPPHVNFPHPEMIKLDNLPLG 227
V+ + +KN+++N S N+ PF +Q + F E++ +
Sbjct: 365 LVSRNQFKYKNYIENFLSSQYNIDSQLFPFISKQ-----NWQSAGFIADEIVYFIERRVS 419
Query: 228 ---IKSGVFEASSEEFDFMVFQLTYEDINNLKEKAKGNNTSRVISFNVVAAHIWRCK 281
IK+ + +S + ++T K K T + + + H++ CK
Sbjct: 420 FSRIKNRILRQASMQSYVRGIRITCSGRVGGKSKKAQRATQECVKYGETSLHVFECK 476
>TRAC_STAAM (P06698) Transposase for transposon Tn554
Length = 125
Score = 31.2 bits (69), Expect = 5.6
Identities = 29/111 (26%), Positives = 50/111 (44%), Gaps = 16/111 (14%)
Query: 256 KEKAKGNNTSRVISFNVVAAHIWRCKALSKSYDPNRSSIILYAV-DIRSRLNPPLPRAYT 314
K +K + +VI+FN +A + N S LY DIR R+ R T
Sbjct: 28 KAISKFSIEGKVINFNSIAK------------EANVSKSWLYKEHDIRQRIESLRERQIT 75
Query: 315 GNAVLTAYASAKCEELKEGEFSRLVEMVEEGSRRMSDEYARSIIDWGELYN 365
N V S++ EE+ R V +E+ ++++ ++ + +G+LYN
Sbjct: 76 ANVVSKPKKSSRSEEILIKTLKRRVMELEKENKKLQNQIQKL---YGDLYN 123
>SEC5_CAEEL (Q22706) Probable exocyst complex component Sec5
Length = 884
Score = 30.4 bits (67), Expect = 9.6
Identities = 18/43 (41%), Positives = 26/43 (59%), Gaps = 2/43 (4%)
Query: 51 VHFFEANKDFPPQIVTEKL--KKALEDALVAYDFLAGRLKMNT 91
+HF N+D P IVT+ + LE+AL Y LA R+++NT
Sbjct: 793 LHFSRLNQDTSPLIVTQIVIDLTGLEEALSTYATLAIRVQINT 835
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.138 0.411
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 53,220,163
Number of Sequences: 164201
Number of extensions: 2330394
Number of successful extensions: 5241
Number of sequences better than 10.0: 17
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 5194
Number of HSP's gapped (non-prelim): 19
length of query: 445
length of database: 59,974,054
effective HSP length: 113
effective length of query: 332
effective length of database: 41,419,341
effective search space: 13751221212
effective search space used: 13751221212
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 67 (30.4 bits)
Medicago: description of AC136505.11


