

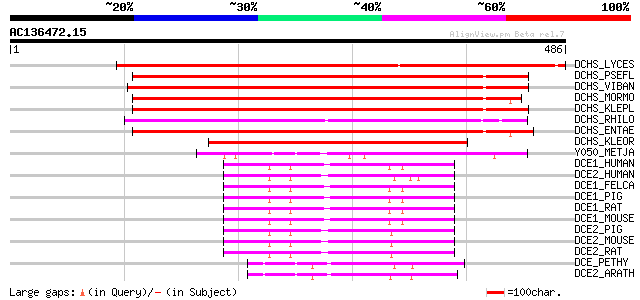
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136472.15 + phase: 0
(486 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
DCHS_LYCES (P54772) Histidine decarboxylase (EC 4.1.1.22) (HDC) ... 482 e-135
DCHS_PSEFL (P95477) Histidine decarboxylase (EC 4.1.1.22) (HDC) 283 7e-76
DCHS_VIBAN (Q56581) Histidine decarboxylase (EC 4.1.1.22) (HDC) 283 9e-76
DCHS_MORMO (P05034) Histidine decarboxylase (EC 4.1.1.22) (HDC) 282 2e-75
DCHS_KLEPL (P28578) Histidine decarboxylase (EC 4.1.1.22) (HDC) 276 7e-74
DCHS_RHILO (Q98A07) Histidine decarboxylase (EC 4.1.1.22) (HDC) 270 6e-72
DCHS_ENTAE (P28577) Histidine decarboxylase (EC 4.1.1.22) (HDC) 261 2e-69
DCHS_KLEOR (Q8L0Z4) Histidine decarboxylase (EC 4.1.1.22) (HDC) ... 196 9e-50
Y050_METJA (Q60358) Hypothetical protein MJ0050 85 5e-16
DCE1_HUMAN (Q99259) Glutamate decarboxylase, 67 kDa isoform (EC ... 71 5e-12
DCE2_HUMAN (Q05329) Glutamate decarboxylase, 65 kDa isoform (EC ... 69 2e-11
DCE1_FELCA (P14748) Glutamate decarboxylase, 67 kDa isoform (EC ... 69 3e-11
DCE1_PIG (P48319) Glutamate decarboxylase, 67 kDa isoform (EC 4.... 69 4e-11
DCE1_RAT (P18088) Glutamate decarboxylase, 67 kDa isoform (EC 4.... 68 5e-11
DCE1_MOUSE (P48318) Glutamate decarboxylase, 67 kDa isoform (EC ... 68 6e-11
DCE2_PIG (P48321) Glutamate decarboxylase, 65 kDa isoform (EC 4.... 66 2e-10
DCE2_MOUSE (P48320) Glutamate decarboxylase, 65 kDa isoform (EC ... 66 2e-10
DCE2_RAT (Q05683) Glutamate decarboxylase, 65 kDa isoform (EC 4.... 65 3e-10
DCE_PETHY (Q07346) Glutamate decarboxylase (EC 4.1.1.15) (GAD) 62 3e-09
DCE2_ARATH (Q42472) Glutamate decarboxylase 2 (EC 4.1.1.15) (GAD 2) 59 2e-08
>DCHS_LYCES (P54772) Histidine decarboxylase (EC 4.1.1.22) (HDC)
(TOM92)
Length = 413
Score = 482 bits (1240), Expect = e-135
Identities = 229/396 (57%), Positives = 301/396 (75%), Gaps = 5/396 (1%)
Query: 94 VLARYRKSLTERTKYHLGYPYNLDFDYGA-LSQLQHFSINNLGDPFIESNYGVHSRQFEV 152
+L +Y ++L+ER KYH+GYP N+ +++ A L+ L F +NN GDPF + HS+ FEV
Sbjct: 17 ILTQYLETLSERKKYHIGYPINMCYEHHATLAPLLQFHLNNCGDPFTQHPTDFHSKDFEV 76
Query: 153 GVLDWFARLWELEKNEYWGYITNCGTEGNLHGILVGR-EVLPDGILYASRESHYSIFKAA 211
VLDWFA+LWE+EK+EYWGYIT+ GTEGNLHG +GR E+LP+G LYAS++SHYSIFKAA
Sbjct: 77 AVLDWFAQLWEIEKDEYWGYITSGGTEGNLHGFWLGRRELLPNGYLYASKDSHYSIFKAA 136
Query: 212 RMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLE 271
RMYRME + + TL +GEID +D ++KLL +++KPAIIN+NIGTT KGA+DDLD VIQ LE
Sbjct: 137 RMYRMELQTINTLVNGEIDYEDLQSKLLVNKNKPAIININIGTTFKGAIDDLDFVIQTLE 196
Query: 272 EAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITR 331
G+S D +YIH D AL GL++PF+K A K+TFKKPIGS+S+SGHKF+GCPM CGVQITR
Sbjct: 197 NCGYSNDNYYIHCDRALCGLILPFIKHAKKITFKKPIGSISISGHKFLGCPMSCGVQITR 256
Query: 332 LEHINALSRNVEYLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFK 391
+++ LS+ +EY+ S DATI GSRNG PIFLWY L++KG+ Q++ C+ NA Y K
Sbjct: 257 RSYVSTLSK-IEYINSADATISGSRNGFTPIFLWYCLSKKGHARLQQDSITCIENARYLK 315
Query: 392 DRLIEAGIGAMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLN 451
DRL+EAGI MLN+ S TVVFERP D +FIR+W L C +AHVV+MP +T E +D F
Sbjct: 316 DRLLEAGISVMLNDFSITVVFERPCDHKFIRRWNLCCLRGMAHVVIMPGITRETIDSFFK 375
Query: 452 ELVQKR-ATWFEDGTFQPYCIASDVGENSCLCAQHK 486
+L+Q+R W++D P C+A D+ N C+C+ K
Sbjct: 376 DLMQERNYKWYQDVKALPPCLADDLALN-CMCSNKK 410
>DCHS_PSEFL (P95477) Histidine decarboxylase (EC 4.1.1.22) (HDC)
Length = 405
Score = 283 bits (724), Expect = 7e-76
Identities = 142/349 (40%), Positives = 216/349 (61%), Gaps = 3/349 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNL-GDPFIESNYGVHSRQFEVGVLDWFARLWELEK 166
+++GYP + DFDY L + FSINNL G SNY ++S FE V+ +FA L+ +
Sbjct: 24 FNIGYPESADFDYSQLHRFLQFSINNLLGTGNEYSNYLLNSFDFEKDVMTYFAELFNIAL 83
Query: 167 NEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS 226
+ WGY+TN GTEGN+ G +GRE+ PDG LY S+++HYS+ K ++ R++C VE+L +
Sbjct: 84 EDSWGYVTNGGTEGNMFGCYLGRELFPDGTLYYSKDTHYSVAKIVKLLRIKCRAVESLPN 143
Query: 227 GEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDG 286
GEID DD AK+ Q++ II NIGTT++GA+D++ + Q+L++AG ++ +Y+H D
Sbjct: 144 GEIDYDDLMAKITADQERHPIIFANIGTTMRGALDNIVTIQQRLQQAGIARHDYYLHADA 203
Query: 287 ALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLA 346
AL G+++PFV +F I S+ VSGHK +G P+PCG+ + + ++ +S V+Y+
Sbjct: 204 ALSGMILPFVDHPQPFSFADGIDSICVSGHKMIGSPIPCGIVVAKRNNVARISVEVDYIR 263
Query: 347 SRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNEL 406
+ D TI GSRNGH P+ +W L + ++ ++ L A Y DR +GI A NE
Sbjct: 264 AHDKTISGSRNGHTPLMMWAALRSYSWAEWRHRIKHSLDTAQYAVDRFQASGIDAWRNEN 323
Query: 407 SSTVVFERPHDEEFIRKWQLACKGNIAHVVVMP-NVTIEKLDDFLNELV 454
S TVVF P E K+ LA GN AH++ P + +D ++E+V
Sbjct: 324 SITVVFPCP-SERIATKYCLATSGNSAHLITTPHHHDCSMIDALIDEVV 371
>DCHS_VIBAN (Q56581) Histidine decarboxylase (EC 4.1.1.22) (HDC)
Length = 386
Score = 283 bits (723), Expect = 9e-76
Identities = 140/352 (39%), Positives = 219/352 (61%), Gaps = 2/352 (0%)
Query: 104 ERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWE 163
E +++GYP + FDY L + FSINN GD ESNY ++S +FE V+ +F++L++
Sbjct: 20 ENQYFNVGYPESAAFDYSILEKFMKFSINNCGDWREESNYKLNSFEFEKEVMRFFSQLFK 79
Query: 164 LEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVET 223
+ N+ WGYI+N GTEGN+ + RE+ P +Y S E+HYS+ K R+ + K+ +
Sbjct: 80 IPYNDSWGYISNGGTEGNMFSCYLAREIFPTAYIYYSEETHYSVDKIVRLLNIPARKIRS 139
Query: 224 LNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIH 283
L SGEID + ++ + + K II NIGTT++GA D++ + Q L G ++ +YIH
Sbjct: 140 LPSGEIDYQNLVDQIQKDKQKNPIIFANIGTTMRGATDNIQRIQQDLASIGLERNDYYIH 199
Query: 284 VDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVE 343
D AL G++MPFV++ +F+ I S+SVSGHK +G P+PCG+ + + ++ +S V+
Sbjct: 200 ADAALSGMIMPFVEQPHPYSFEDGIDSISVSGHKMIGSPIPCGIVLAKRHMVDQISVEVD 259
Query: 344 YLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAML 403
Y++SRD TI GSRNGH+ +F+W + + +Q +V +CL A Y R E GI A
Sbjct: 260 YISSRDQTISGSRNGHSALFMWTAIKSHSFVDWQGKVNQCLNMAEYTVQRFQEVGINAWR 319
Query: 404 NELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVT-IEKLDDFLNELV 454
N+ S+TVVF P E RK LA G++AH++ MP++ +KLD + +++
Sbjct: 320 NKNSNTVVFPCP-SEPVWRKHSLANSGSVAHIITMPHLDGPDKLDPLIEDVI 370
>DCHS_MORMO (P05034) Histidine decarboxylase (EC 4.1.1.22) (HDC)
Length = 377
Score = 282 bits (721), Expect = 2e-75
Identities = 140/345 (40%), Positives = 218/345 (62%), Gaps = 5/345 (1%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++FA L+++
Sbjct: 23 FNIGYPESADFDYTNLERFLRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFADLFKIPFE 82
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
+ WGY+TN GTEGN+ G +GRE+ PDG LY S+++HYS+ K ++ R++ + VE+ +G
Sbjct: 83 QSWGYVTNGGTEGNMFGCYLGREIFPDGTLYYSKDTHYSVAKIVKLLRIKSQVVESQPNG 142
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
EID DD K+ ++ II NIGTTV+GA+DD+ + ++L+ AG ++ +Y+H D A
Sbjct: 143 EIDYDDLMKKIADDKEAHPIIFANIGTTVRGAIDDIAEIQKRLKAAGIKREDYYLHADAA 202
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV A TF I S+ VSGHK +G P+PCG+ + + E+++ +S ++Y+++
Sbjct: 203 LSGMILPFVDDAQPFTFADGIDSIGVSGHKMIGSPIPCGIVVAKKENVDRISVEIDYISA 262
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ LW + +++ + + L A Y DR+ +AGI A N+ S
Sbjct: 263 HDKTITGSRNGHTPLMLWEAIRSHSTEEWKRRITRSLDMAQYAVDRMQKAGINAWRNKNS 322
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVV----MPNVTIEKLDD 448
TVVF P E R+ LA G++AH++ + V I+KL D
Sbjct: 323 ITVVFPCP-SERVWREHCLATSGDVAHLITTAHHLDTVQIDKLID 366
>DCHS_KLEPL (P28578) Histidine decarboxylase (EC 4.1.1.22) (HDC)
Length = 377
Score = 276 bits (707), Expect = 7e-74
Identities = 133/348 (38%), Positives = 217/348 (62%), Gaps = 2/348 (0%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++FA+L+++
Sbjct: 23 FNIGYPESADFDYTILERFMRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFAQLFKIPFE 82
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
E WGY+TN GTEGN+ G +GRE+ P+G LY S+++HYS+ K ++ R++ VE+ +G
Sbjct: 83 ESWGYVTNGGTEGNMFGCYLGREIFPNGTLYYSKDTHYSVAKIVKLLRIKSTLVESQPNG 142
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
E+D D K+ R +K II NIGTTV+GA+D++ ++ Q + E G + +Y+H D A
Sbjct: 143 EMDYADLIKKIKRDNEKHPIIFANIGTTVRGAIDNIAIIQQSISELGIERKDYYLHADAA 202
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV F I S+ VSGHK +G P+PCG+ + + ++++ +S ++Y+++
Sbjct: 203 LSGMILPFVDNPQPFNFADGIDSIGVSGHKMIGSPIPCGIVVAKKKNVDRISVEIDYISA 262
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ +W + + +++ +++ L A Y DR AGI A N+ S
Sbjct: 263 HDKTISGSRNGHTPLMMWEAIRSHSWEEWRRRIERSLNMAQYAVDRFQSAGIDAWRNKNS 322
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVVMP-NVTIEKLDDFLNELV 454
TVVF P E +K LA G+IAH++ ++ K+D +++++
Sbjct: 323 ITVVFPCP-SEAVWKKHCLATSGDIAHLIATAHHLDSSKIDALIDDVI 369
>DCHS_RHILO (Q98A07) Histidine decarboxylase (EC 4.1.1.22) (HDC)
Length = 369
Score = 270 bits (690), Expect = 6e-72
Identities = 138/353 (39%), Positives = 204/353 (57%), Gaps = 4/353 (1%)
Query: 101 SLTERTKYHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFAR 160
S+ E LGYP+ DFDY L + + NNLGDPF Y V+S FE V+D+FAR
Sbjct: 16 SMQEANGCFLGYPFAKDFDYEPLWRFMSLTGNNLGDPFEPGTYRVNSHAFECDVVDFFAR 75
Query: 161 LWELEKNEYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEK 220
L+ E WGY+TN GTEGN++G+ + RE+ P+ + Y S+++HYS+ K R+ R+E
Sbjct: 76 LFRACSCEVWGYVTNGGTEGNIYGLYLARELYPNAVAYFSQDTHYSVSKGVRLLRLEHSV 135
Query: 221 VETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRF 280
V + ++GEI+ DD K R++ +PA++ NIGTT+K DD + L + G S
Sbjct: 136 VRSQSNGEINYDDLAQKATRYRTRPAVVVANIGTTMKEGKDDTLKIRAVLHDVGIS--AI 193
Query: 281 YIHVDGALFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSR 340
Y+H D AL G P + P F S+++SGHKF+G PMPCGV ++ H+ + R
Sbjct: 194 YVHSDAALCGPYAPLLNPKPAFDFADGADSITLSGHKFLGAPMPCGVVLSHKLHVQRVMR 253
Query: 341 NVEYLASRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIG 400
N++Y+ S D T+ GSRN PI LWY + G G ++ Q+C R A Y D L G+
Sbjct: 254 NIDYIGSSDTTLSGSRNAFTPIILWYAIRSLGIEGIKQTFQQCERLAAYTADELNVRGVS 313
Query: 401 AMLNELSSTVVFERPHDEEFIRKWQLACKGNIAHVVVMPNVTIEKLDDFLNEL 453
A N + TVV P ++ KWQ+A + +++H+VV P T ++ D + +
Sbjct: 314 AWRNPNALTVVLP-PVEDSIKTKWQIATQ-DVSHLVVTPGTTKQQADALIETI 364
>DCHS_ENTAE (P28577) Histidine decarboxylase (EC 4.1.1.22) (HDC)
Length = 377
Score = 261 bits (668), Expect = 2e-69
Identities = 133/356 (37%), Positives = 217/356 (60%), Gaps = 6/356 (1%)
Query: 108 YHLGYPYNLDFDYGALSQLQHFSINNLGDPFIESNYGVHSRQFEVGVLDWFARLWELEKN 167
+++GYP + DFDY L + FSINN GD NY ++S FE V+++F+ ++++
Sbjct: 23 FNIGYPESADFDYTMLERFLRFSINNCGDWGEYCNYLLNSFDFEKEVMEYFSGIFKIPFA 82
Query: 168 EYWGYITNCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSG 227
E WGY+TN GTE N+ G +GRE+ P+G LY S+++HYS+ K ++ R++ + VE+ G
Sbjct: 83 ESWGYVTNGGTESNMFGCYLGRELFPEGTLYYSKDTHYSVAKIVKLLRIKSQLVESQPDG 142
Query: 228 EIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGA 287
E+D DD K+ ++ II NIGTTV+GAVD++ + +++ G ++ +Y+H D A
Sbjct: 143 EMDYDDLINKIRTSGERHPIIFANIGTTVRGAVDNIAEIQKRIAALGIPREDYYLHADAA 202
Query: 288 LFGLMMPFVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLAS 347
L G+++PFV+ TF I S+ VSGHK +G P+PCG+ + + +++ +S ++Y+++
Sbjct: 203 LSGMILPFVEDPQPFTFADGIDSIGVSGHKMIGSPIPCGIVVAKKANVDRISVEIDYISA 262
Query: 348 RDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGAMLNELS 407
D TI GSRNGH P+ +W + + + + L A Y DR AGI A+ ++ S
Sbjct: 263 HDKTISGSRNGHTPLMMWAAVRSHTDAEWHRRIGHSLNMAKYAVDRFKAAGIDALCHKNS 322
Query: 408 STVVFERPHDEEFIRKWQLACKGNIAHVVV----MPNVTIEKL-DDFLNELVQKRA 458
TVVF +P E +K LA GN+AH++ + + I+ L DD + +L Q+ A
Sbjct: 323 ITVVFPKP-SEWVWKKHCLATSGNVAHLITTAHHLDSSRIDALIDDVIADLAQRAA 377
>DCHS_KLEOR (Q8L0Z4) Histidine decarboxylase (EC 4.1.1.22) (HDC)
(Fragment)
Length = 228
Score = 196 bits (499), Expect = 9e-50
Identities = 87/227 (38%), Positives = 143/227 (62%)
Query: 175 NCGTEGNLHGILVGREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNSGEIDCDDF 234
N GTEGN+ G +GRE+ P+G LY S+++HYS+ K ++ R++ VE+ +GE+D D
Sbjct: 1 NGGTEGNMFGCYLGREIFPNGTLYYSKDTHYSVAKIVKLLRIKSTLVESQPNGEMDYADL 60
Query: 235 KAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMP 294
K+ R +K II NIGTTV+GA+D++ ++ Q + E G + +Y+H D AL G+++P
Sbjct: 61 IKKIKRDNEKHPIIFANIGTTVRGAIDNIAIIQQSISELGIERKDYYLHADAALSGMILP 120
Query: 295 FVKRAPKVTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHINALSRNVEYLASRDATIMG 354
FV F I S+ VSGHK +G P+PCG+ + + ++++ +S ++Y+++ D TI G
Sbjct: 121 FVDNPQPFNFADGIDSIGVSGHKMIGSPIPCGIVVAKKKNVDRISVEIDYISAHDKTISG 180
Query: 355 SRNGHAPIFLWYTLNRKGYRGFQKEVQKCLRNAHYFKDRLIEAGIGA 401
SRNGH P+ +W + + +++ +++ L A Y DR AGI A
Sbjct: 181 SRNGHTPLMMWEAIRSHSWEEWRRRIERSLNMAQYAVDRFQSAGIDA 227
>Y050_METJA (Q60358) Hypothetical protein MJ0050
Length = 396
Score = 84.7 bits (208), Expect = 5e-16
Identities = 81/317 (25%), Positives = 137/317 (42%), Gaps = 34/317 (10%)
Query: 164 LEKNEYWGYITNCGTEGNLHGIL----VGREVLPDGI-------LYASRESHYSIFKAAR 212
L + +G+I + GTE NL + + RE G+ + +H+S K
Sbjct: 81 LNNKDAYGHIVSGGTEANLMALRCIKNIWREKRRKGLSKNEHPKIIVPITAHFSFEKGRE 140
Query: 213 MYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDLDLVIQKLEE 272
M +E ID + F + D II + GTT G +D+++ + + +E
Sbjct: 141 MMDLEYIYAPIKEDYTID-EKFVKDAVEDYDVDGIIGI-AGTTELGTIDNIEELSKIAKE 198
Query: 273 AGFSQDRFYIHVDGALFGLMMPFV-----KRAPKVTFKKPIG--SVSVSGHKFVGCPMPC 325
+ YIHVD A GL++PF+ K+ F +G S+++ HK CP+P
Sbjct: 199 -----NNIYIHVDAAFGGLVIPFLDDKYKKKGVNYKFDFSLGVDSITIDPHKMGHCPIPS 253
Query: 326 G-VQITRLEHINALSRNVEYLA-SRDATIMGSRNGHAPIFLWYTLNRKGYRGFQKEVQKC 383
G + + + L + YL +R ATI+G+R G + L G G +K V +C
Sbjct: 254 GGILFKDIGYKRYLDVDAPYLTETRQATILGTRVGFGGACTYAVLRYLGREGQRKIVNEC 313
Query: 384 LRNAHYFKDRLIEAGIGAMLNELSSTVVFERPHDEEFIRK------WQLACK-GNIAHVV 436
+ N Y +L E ++ + + V E +E +K + C +V
Sbjct: 314 MENTLYLYKKLKENNFKPVIEPILNIVAIEDEDYKEVCKKLRDRGIYVSVCNCVKALRIV 373
Query: 437 VMPNVTIEKLDDFLNEL 453
VMP++ E +D+F+ L
Sbjct: 374 VMPHIKREHIDNFIEIL 390
>DCE1_HUMAN (Q99259) Glutamate decarboxylase, 67 kDa isoform (EC
4.1.1.15) (GAD-67) (67 kDa glutamic acid decarboxylase)
Length = 594
Score = 71.2 bits (173), Expect = 5e-12
Identities = 62/220 (28%), Positives = 95/220 (43%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P +L+ S +SHYSI KA + V + G+I DF+AK+L + K
Sbjct: 275 GMAAVPKLVLFTSEQSHYSIKKAGAALGFGTDNVILIKCNERGKIIPADFEAKILEAKQK 334
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +N GTTV GA D + + E+ ++HVD A G ++ K K
Sbjct: 335 GYVPFYVNATAGTTVYGAFDPIQEIADICEKYNL-----WLHVDAAWGGGLLMSRKHRHK 389
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITR----LEHINALSRNV------EYLASRDAT 351
+ + SV+ + HK +G + C + + L+ N + +Y S D
Sbjct: 390 LNGIERANSVTWNPHKMMGVLLQCSAILVKEKGILQGCNQMCAGYLFQPDKQYDVSYDTG 449
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H IF W KG GF+ ++ KCL A Y
Sbjct: 450 DKAIQCGRHVDIFKFWLMWKAKGTVGFENQINKCLELAEY 489
>DCE2_HUMAN (Q05329) Glutamate decarboxylase, 65 kDa isoform (EC
4.1.1.15) (GAD-65) (65 kDa glutamic acid decarboxylase)
Length = 585
Score = 69.3 bits (168), Expect = 2e-11
Identities = 60/220 (27%), Positives = 97/220 (43%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G LP I + S SH+S+ K A + + V + G++ D + ++L + K
Sbjct: 266 GMAALPRLIAFTSEHSHFSLKKGAAALGIGTDSVILIKCDERGKMIPSDLERRILEAKQK 325
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +++ GTTV GA D L V ++ + ++HVD A G ++ K K
Sbjct: 326 GFVPFLVSATAGTTVYGAFDPLLAVADICKKY-----KIWMHVDAAWGGGLLMSRKHKWK 380
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITRLEHI--NALSRNVEYLASRD-----ATIMG 354
++ + SV+ + HK +G P+ C + R E + N + YL +D + G
Sbjct: 381 LSGVERANSVTWNPHKMMGVPLQCSALLVREEGLMQNCNQMHASYLFQQDKHYDLSYDTG 440
Query: 355 SR----NGHAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ H +F LW KG GF+ V KCL A Y
Sbjct: 441 DKALQCGRHVDVFKLWLMWRAKGTTGFEAHVDKCLELAEY 480
>DCE1_FELCA (P14748) Glutamate decarboxylase, 67 kDa isoform (EC
4.1.1.15) (GAD-67) (67 kDa glutamic acid decarboxylase)
Length = 594
Score = 68.9 bits (167), Expect = 3e-11
Identities = 61/220 (27%), Positives = 94/220 (42%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P +L+ S SHYSI KA + V + G+I D +AK+L + K
Sbjct: 275 GMAAVPKLVLFTSEHSHYSIKKAGAALGFGTDNVILIKCNERGKIIPADLEAKILEAKQK 334
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +N GTTV GA D + + E+ ++HVD A G ++ K K
Sbjct: 335 GYVPLYVNATAGTTVYGAFDPIQEIADICEKYNL-----WLHVDAAWGGGLLMSRKHRHK 389
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITR----LEHINALSRNV------EYLASRDAT 351
++ + SV+ + HK +G + C + + L+ N + +Y S D
Sbjct: 390 LSGIERANSVTWNPHKMMGVLLQCSAILVKEKGILQGCNQMCAGYLFQPDKQYDVSYDTG 449
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H IF W KG GF+ ++ KCL A Y
Sbjct: 450 DKAIQCGRHVDIFKFWLMWKAKGTVGFENQINKCLELAEY 489
>DCE1_PIG (P48319) Glutamate decarboxylase, 67 kDa isoform (EC
4.1.1.15) (GAD-67) (67 kDa glutamic acid decarboxylase)
Length = 594
Score = 68.6 bits (166), Expect = 4e-11
Identities = 61/220 (27%), Positives = 94/220 (42%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P +L+ S SHYSI KA + V + G+I D +AK+L + K
Sbjct: 275 GMAAVPKLVLFTSEHSHYSIKKAGAALGFGTDNVILIKCNERGKIIPADLEAKILEAKQK 334
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +N GTTV GA D + + E+ ++HVD A G ++ K K
Sbjct: 335 GYIPLYVNATAGTTVYGAFDPIQEIADICEKYNL-----WLHVDAAWGGGLLMSRKHRHK 389
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITR----LEHINALSRNV------EYLASRDAT 351
++ + SV+ + HK +G + C + + L+ N + +Y S D
Sbjct: 390 LSGIERADSVTWNPHKMMGVLLQCSAILVKEKGILQGCNQMCAGYLFQPDKQYDVSYDTG 449
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H IF W KG GF+ ++ KCL A Y
Sbjct: 450 DKAIQCGRHVDIFKFWLMWKAKGTVGFENQINKCLELAEY 489
>DCE1_RAT (P18088) Glutamate decarboxylase, 67 kDa isoform (EC
4.1.1.15) (GAD-67) (67 kDa glutamic acid decarboxylase)
Length = 593
Score = 68.2 bits (165), Expect = 5e-11
Identities = 61/220 (27%), Positives = 94/220 (42%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P +L+ S SHYSI KA + V + G+I D +AK+L + K
Sbjct: 274 GMAAVPKLVLFTSEHSHYSIKKAGAALGFGTDNVILIKCNERGKIIPADLEAKILDAKQK 333
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +N GTTV GA D + + E+ ++HVD A G ++ K K
Sbjct: 334 GFVPLYVNATAGTTVYGAFDPIQEIADICEKYNL-----WLHVDAAWGGGLLMSRKHRHK 388
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITR----LEHINALSRNV------EYLASRDAT 351
++ + SV+ + HK +G + C + + L+ N + +Y S D
Sbjct: 389 LSGIERANSVTWNPHKMMGVLLQCSAILVKEKGILQGCNQMCAGYLFQPDKQYDVSYDTG 448
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H IF W KG GF+ ++ KCL A Y
Sbjct: 449 DKAIQCGRHVDIFKFWLMWKAKGTVGFENQINKCLELAEY 488
>DCE1_MOUSE (P48318) Glutamate decarboxylase, 67 kDa isoform (EC
4.1.1.15) (GAD-67) (67 kDa glutamic acid decarboxylase)
Length = 593
Score = 67.8 bits (164), Expect = 6e-11
Identities = 61/220 (27%), Positives = 94/220 (42%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P +L+ S SHYSI KA + V + G+I D +AK+L + K
Sbjct: 274 GMAAVPKLVLFTSEHSHYSIKKAGAALGFGTDNVILIKCNERGKIIPADLEAKILDAKQK 333
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +N GTTV GA D + + E+ ++HVD A G ++ K K
Sbjct: 334 GYVPLYVNATAGTTVYGAFDPIQEIADICEKYNL-----WLHVDAAWGGGLLMSRKHRHK 388
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITR----LEHINALSRNV------EYLASRDAT 351
++ + SV+ + HK +G + C + + L+ N + +Y S D
Sbjct: 389 LSGIERANSVTWNPHKMMGVLLQCSAILVKEKGILQGCNQMCAGYLFQPDKQYDVSYDTG 448
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H IF W KG GF+ ++ KCL A Y
Sbjct: 449 DKAIQCGRHVDIFKFWLMWKAKGTVGFENQINKCLELADY 488
>DCE2_PIG (P48321) Glutamate decarboxylase, 65 kDa isoform (EC
4.1.1.15) (GAD-65) (65 kDa glutamic acid decarboxylase)
Length = 585
Score = 66.2 bits (160), Expect = 2e-10
Identities = 59/220 (26%), Positives = 97/220 (43%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P I + S SH+S+ K A + + V + G++ D + ++L + K
Sbjct: 266 GMAAVPRLIAFTSEHSHFSLKKGAAALGIGTDSVILIKCDERGKMIPSDLERRILEAKQK 325
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +++ GTTV GA D L V ++ + ++HVD A G ++ K K
Sbjct: 326 GFVPFLVSATAGTTVYGAFDPLLAVADICKKY-----KIWMHVDAAWGGGLLMSRKHKWK 380
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITRLE---------HINAL-SRNVEYLASRDAT 351
++ + SV+ + HK +G P+ C + R E H + L ++ Y S D
Sbjct: 381 LSGVERANSVTWNPHKMMGVPLQCSALLVREEGLMQSCNQMHASYLFQQDKHYDLSYDTG 440
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H +F LW KG GF+ + KCL A Y
Sbjct: 441 DKALQCGRHVDVFKLWLMWRAKGTTGFEAHIDKCLELAEY 480
>DCE2_MOUSE (P48320) Glutamate decarboxylase, 65 kDa isoform (EC
4.1.1.15) (GAD-65) (65 kDa glutamic acid decarboxylase)
Length = 585
Score = 65.9 bits (159), Expect = 2e-10
Identities = 59/220 (26%), Positives = 97/220 (43%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P I + S SH+S+ K A + + V + G++ D + ++L + K
Sbjct: 266 GMAAVPRLIAFTSEHSHFSLKKGAAALGIGTDSVILIKCDERGKMIPSDLERRILEVKQK 325
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +++ GTTV GA D L V ++ + ++HVD A G ++ K K
Sbjct: 326 GFVPFLVSATAGTTVYGAFDPLLAVADICKKY-----KIWMHVDAAWGGGLLMSRKHKWK 380
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITRLE---------HINAL-SRNVEYLASRDAT 351
++ + SV+ + HK +G P+ C + R E H + L ++ Y S D
Sbjct: 381 LSGVERANSVTWNPHKMMGVPLQCSALLVREEGLMQSCNQMHASYLFQQDKHYDLSYDTG 440
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H +F LW KG GF+ + KCL A Y
Sbjct: 441 DKALQCGRHVDVFKLWLMWRAKGTTGFEAHIDKCLELAEY 480
>DCE2_RAT (Q05683) Glutamate decarboxylase, 65 kDa isoform (EC
4.1.1.15) (GAD-65) (65 kDa glutamic acid decarboxylase)
Length = 585
Score = 65.5 bits (158), Expect = 3e-10
Identities = 59/220 (26%), Positives = 96/220 (42%), Gaps = 23/220 (10%)
Query: 188 GREVLPDGILYASRESHYSIFKAARMYRMECEKVETLNS---GEIDCDDFKAKLLRHQDK 244
G +P I + S SH+S+ K A + + V + G++ D + ++L + K
Sbjct: 266 GMAAVPRLIAFTSEHSHFSLKKGAAALGIGTDSVILIKCDERGKMIPSDLERRILEVKQK 325
Query: 245 ---PAIINVNIGTTVKGAVDDLDLVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPK 301
P +++ GTTV GA D L V ++ + ++HVD A G ++ K K
Sbjct: 326 GFVPFLVSATAGTTVYGAFDPLLAVADICKKY-----KIWMHVDAAWGGGLLMSRKHKWK 380
Query: 302 VTFKKPIGSVSVSGHKFVGCPMPCGVQITRLE---------HINAL-SRNVEYLASRDAT 351
+ + SV+ + HK +G P+ C + R E H + L ++ Y S D
Sbjct: 381 LNGVERANSVTWNPHKMMGVPLQCSALLVREEGLMQSCNQMHASYLFQQDKHYDLSYDTG 440
Query: 352 IMGSRNG-HAPIF-LWYTLNRKGYRGFQKEVQKCLRNAHY 389
+ G H +F LW KG GF+ + KCL A Y
Sbjct: 441 DKALQCGRHVDVFKLWLMWRAKGTTGFEAHIDKCLELAEY 480
>DCE_PETHY (Q07346) Glutamate decarboxylase (EC 4.1.1.15) (GAD)
Length = 500
Score = 62.0 bits (149), Expect = 3e-09
Identities = 54/199 (27%), Positives = 99/199 (49%), Gaps = 14/199 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V+ L+ G D KA + ++ + + +G+T+ G +D+ D
Sbjct: 169 KFARYFEVELKEVK-LSEGYYVMDPEKAVEMVDENTICVAAI-LGSTLNGEFEDVKRLND 226
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKFVGCPM 323
L+++K +E G+ IHVD A G + PF+ + F+ P + S++VSGHK+
Sbjct: 227 LLVEKNKETGWDTP---IHVDAASGGFIAPFIYPELEWDFRLPLVKSINVSGHKYGLVYA 283
Query: 324 PCGVQITRLEHI--NALSRNVEYLASRDAT--IMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
G + R + + L ++ YL + T + S+ I +Y L R GY G++
Sbjct: 284 GIGWVVWRNKDDLPDELIFHINYLGADQPTFTLNFSKGSSQVIAQYYQLIRLGYEGYKNV 343
Query: 380 VQKCLRNAHYFKDRLIEAG 398
++ C NA ++ L + G
Sbjct: 344 MENCQENASVLREGLEKTG 362
>DCE2_ARATH (Q42472) Glutamate decarboxylase 2 (EC 4.1.1.15) (GAD 2)
Length = 494
Score = 59.3 bits (142), Expect = 2e-08
Identities = 52/193 (26%), Positives = 95/193 (48%), Gaps = 14/193 (7%)
Query: 209 KAARMYRMECEKVETLNSGEIDCDDFKAKLLRHQDKPAIINVNIGTTVKGAVDDL----D 264
K AR + +E ++V L+ G D KA + ++ + + +G+T+ G +D+ D
Sbjct: 168 KFARYFEVELKEVN-LSEGYYVMDPDKAAEMVDENTICVAAI-LGSTLNGEFEDVKRLND 225
Query: 265 LVIQKLEEAGFSQDRFYIHVDGALFGLMMPFVKRAPKVTFKKP-IGSVSVSGHKFVGCPM 323
L+++K EE G++ IHVD A G + PF+ + F+ P + S++VSGHK+
Sbjct: 226 LLVKKNEETGWNTP---IHVDAASGGFIAPFIYPELEWDFRLPLVKSINVSGHKYGLVYA 282
Query: 324 PCGVQITRL--EHINALSRNVEYLASRDA--TIMGSRNGHAPIFLWYTLNRKGYRGFQKE 379
G + R + L ++ YL + T+ S+ I +Y L R G+ G++
Sbjct: 283 GIGWVVWRAAEDLPEELIFHINYLGADQPTFTLNFSKGSSQIIAQYYQLIRLGFEGYKNV 342
Query: 380 VQKCLRNAHYFKD 392
++ C+ N K+
Sbjct: 343 MENCIENMVVLKE 355
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.139 0.421
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 59,411,940
Number of Sequences: 164201
Number of extensions: 2586844
Number of successful extensions: 5850
Number of sequences better than 10.0: 84
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 64
Number of HSP's that attempted gapping in prelim test: 5786
Number of HSP's gapped (non-prelim): 89
length of query: 486
length of database: 59,974,054
effective HSP length: 114
effective length of query: 372
effective length of database: 41,255,140
effective search space: 15346912080
effective search space used: 15346912080
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 68 (30.8 bits)
Medicago: description of AC136472.15


