

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136451.6 + phase: 0 /pseudo
(210 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
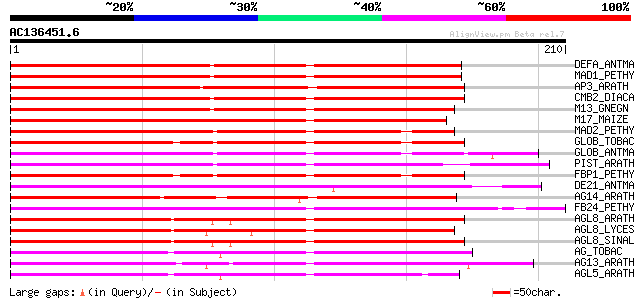
Score E
Sequences producing significant alignments: (bits) Value
DEFA_ANTMA (P23706) Floral homeotic protein DEFICIENS 214 9e-56
MAD1_PETHY (Q07472) Floral homeotic protein PMADS 1 (Green petal... 207 1e-53
AP3_ARATH (P35632) Floral homeotic protein APETALA3 191 1e-48
CMB2_DIACA (Q42498) MADS box protein CMB2 184 2e-46
M13_GNEGN (Q9XGJ4) MADS box protein GGM13 135 5e-32
M17_MAIZE (Q8VWM8) MADS box protein ZMM17 134 2e-31
MAD2_PETHY (Q07474) Floral homeotic protein PMADS 2 133 3e-31
GLOB_TOBAC (Q03416) Floral homeotic protein GLOBOSA 129 5e-30
GLOB_ANTMA (Q03378) Floral homeotic protein GLOBOSA 127 2e-29
PIST_ARATH (P48007) Floral homeotic protein PISTILLATA (Transcri... 125 5e-29
FBP1_PETHY (Q03488) Floral homeotic protein FBP1 (Floral binding... 125 7e-29
DE21_ANTMA (Q8RVL4) MADS box protein defh21 (DEFICIENS homolog 21) 124 2e-28
AG14_ARATH (Q38838) Agamous-like MADS box protein AGL14 122 8e-28
FB24_PETHY (Q9ATE5) MADS box protein FBP24 (Floral binding prote... 118 9e-27
AGL8_ARATH (Q38876) Agamous-like MADS box protein AGL8 (Floral h... 116 3e-26
AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog (... 115 6e-26
AGL8_SINAL (Q41274) Agamous-like MADS box protein AGL8 homolog (... 115 1e-25
AG_TOBAC (Q43585) Floral homeotic protein AGAMOUS (NAG1) 114 1e-25
AG13_ARATH (Q38837) Agamous-like MADS box protein AGL13 114 1e-25
AGL5_ARATH (P29385) Agamous-like MADS box protein AGL5 114 2e-25
>DEFA_ANTMA (P23706) Floral homeotic protein DEFICIENS
Length = 227
Score = 214 bits (546), Expect = 9e-56
Identities = 106/171 (61%), Positives = 138/171 (79%), Gaps = 4/171 (2%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGKI+IK IEN TNRQVTYSKRRNG+FKKAHELSVLCDAKVS+IM S K+HEYI+P
Sbjct: 1 MARGKIQIKRIENQTNRQVTYSKRRNGLFKKAHELSVLCDAKVSIIMISSTQKLHEYISP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+TK++ DQYQK +G +DLW SHYEKM E+LKKL ++N LRR+IR R+GE L+DL
Sbjct: 61 TTATKQLFDQYQKAVG-VDLWSSHYEKMQEHLKKLNEVNRNLRREIRQRMGE---SLNDL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLE 171
++Q+ +L EDM++S+ IRERK+ VI + DT +KKVR++E+++ NL+LE
Sbjct: 117 GYEQIVNLIEDMDNSLKLIRERKYKVISNQIDTSKKKVRNVEEIHRNLVLE 167
>MAD1_PETHY (Q07472) Floral homeotic protein PMADS 1 (Green petal
homeotic protein)
Length = 231
Score = 207 bits (527), Expect = 1e-53
Identities = 101/171 (59%), Positives = 139/171 (81%), Gaps = 4/171 (2%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGKI+IK IEN TNRQVTYSKRRNG+FKKA+EL+VLCDAKVS+IM S K+HE+I+P
Sbjct: 1 MARGKIQIKRIENQTNRQVTYSKRRNGLFKKANELTVLCDAKVSIIMISSTGKLHEFISP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
++TK++ D YQKT+G +DLW SHYEKM E L+KLK++N LR++IR R+GE L+DL
Sbjct: 61 SITTKQLFDLYQKTVG-VDLWNSHYEKMQEQLRKLKEVNRNLRKEIRQRMGE---SLNDL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLE 171
+++QL L E++++S+ IRERK+ VI + +T +KKVR++E+++ NLLLE
Sbjct: 117 NYEQLEELMENVDNSLKLIRERKYKVIGNQIETFKKKVRNVEEIHRNLLLE 167
>AP3_ARATH (P35632) Floral homeotic protein APETALA3
Length = 232
Score = 191 bits (484), Expect = 1e-48
Identities = 94/172 (54%), Positives = 129/172 (74%), Gaps = 4/172 (2%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGKI+IK IEN TNRQVTYSKRRNG+FKKAHEL+VLCDA+VS+IMFS +NK+HEYI+P
Sbjct: 1 MARGKIQIKRIENQTNRQVTYSKRRNGLFKKAHELTVLCDARVSIIMFSSSNKLHEYISP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+TK+I+D YQ T+ D+D+W + YE+M E +KL + N LR QI+ R+GE LD+L
Sbjct: 61 NTTTKEIVDLYQ-TISDVDVWATQYERMQETKRKLLETNRNLRTQIKQRLGEC---LDEL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
Q+LR LE++M ++ +RERKF + + +T +KK +S + + NL+ EL
Sbjct: 117 DIQELRRLEDEMENTFKLVRERKFKSLGNQIETTKKKNKSQQDIQKNLIHEL 168
>CMB2_DIACA (Q42498) MADS box protein CMB2
Length = 214
Score = 184 bits (466), Expect = 2e-46
Identities = 89/172 (51%), Positives = 131/172 (75%), Gaps = 4/172 (2%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGK+EI+ IEN TNRQVT+SKRRNGI KKA EL+VLCDAKVSL+M S +K+H Y++P
Sbjct: 1 MGRGKLEIRKIENKTNRQVTFSKRRNGIMKKAQELTVLCDAKVSLLMISSTHKLHHYLSP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
G+S KK+ D+YQK G +DLWR +E+M E +K+ ++N LRR+I R+G +L+ L
Sbjct: 61 GVSLKKMYDEYQKIEG-VDLWRKQWERMQEQHRKVLELNSLLRREISRRMGG---DLEGL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
+ +L +L+++M +I +IR +K+H IK +T T RKK+++LE+ + +L++EL
Sbjct: 117 TLVELSALQQEMEEAIIQIRNKKYHTIKNQTGTTRKKIKNLEERHTDLVMEL 168
>M13_GNEGN (Q9XGJ4) MADS box protein GGM13
Length = 237
Score = 135 bits (341), Expect = 5e-32
Identities = 74/168 (44%), Positives = 108/168 (64%), Gaps = 4/168 (2%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN TNRQVT+SKRR G+ KKAHELSVLCDA++ LI+FS + K+ EY +
Sbjct: 1 MGRGKIEIKRIENTTNRQVTFSKRRGGLLKKAHELSVLCDAELGLIIFSSSGKLFEYSSA 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
S KKII++YQK G + + + + ++K+ N KL+ IR +GE +L L
Sbjct: 61 SSSMKKIIERYQKVSG-ARITEYDNQHLYCEMTRMKNENEKLQTNIRRMMGE---DLTSL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+ +L L + + S+ +++R RK ++ + + R+K R LE N +L
Sbjct: 117 TMTELHHLGQQLESASSRVRSRKNQLMLQQLENLRRKERILEDQNSHL 164
>M17_MAIZE (Q8VWM8) MADS box protein ZMM17
Length = 259
Score = 134 bits (336), Expect = 2e-31
Identities = 69/165 (41%), Positives = 104/165 (62%), Gaps = 3/165 (1%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN TNRQVT+SKRR G+ KKA+EL+VLCDA+V +++FS KM EY +P
Sbjct: 1 MGRGKIEIKRIENSTNRQVTFSKRRGGLLKKANELAVLCDARVGVVIFSSTGKMFEYCSP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
S +++I+QYQ +H +++L + ++K+ KL IR G+ +L L
Sbjct: 61 ACSLRELIEQYQHATNSHFEEINHDQQILLEMTRMKNEMEKLETGIRRYTGD---DLSSL 117
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMN 165
+ + LE+ + S++K+R RK ++ + D R+K + LE N
Sbjct: 118 TLDDVSDLEQQLEYSVSKVRARKHQLLNQQLDNLRRKEQILEDQN 162
>MAD2_PETHY (Q07474) Floral homeotic protein PMADS 2
Length = 212
Score = 133 bits (335), Expect = 3e-31
Identities = 72/168 (42%), Positives = 109/168 (64%), Gaps = 8/168 (4%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN +NRQVTYSKRRNGI KKA E++VLCDAKVSLI+F + KMHEY +P
Sbjct: 1 MGRGKIEIKRIENSSNRQVTYSKRRNGIIKKAKEITVLCDAKVSLIIFGNSGKMHEYCSP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+ ++D YQKT G LW + +E + + ++K N ++ ++RH GE +++ L
Sbjct: 61 STTLPDMLDGYQKTSGR-RLWDAKHENLSNEIDRIKKENDNMQVKLRHLKGE---DINSL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
+ ++L LEE + + ++ I ++ +++ RK + LE+ + L
Sbjct: 117 NHKELMVLEEGLTNGLSSISAKQSEILR----MVRKNDQILEEEHKQL 160
>GLOB_TOBAC (Q03416) Floral homeotic protein GLOBOSA
Length = 209
Score = 129 bits (324), Expect = 5e-30
Identities = 73/172 (42%), Positives = 111/172 (64%), Gaps = 10/172 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN +NRQVTYSKRRNGI KKA E+SVLCDA+VS+I+F+ + KMHE+ +
Sbjct: 1 MGRGKIEIKRIENSSNRQVTYSKRRNGILKKAKEISVLCDARVSVIIFASSGKMHEFSST 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
L I+DQY K G LW + +E + + K+K N ++ ++RH GE ++ L
Sbjct: 61 SL--VDILDQYHKLTGR-RLWDAKHENLDNEINKVKKDNDNMQIELRHLKGE---DITSL 114
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
+ ++L LE+ +++ + IR ++ +++ RKK +S+E+ L +L
Sbjct: 115 NHRELMMLEDALDNGLTSIRNKQNDLLR----MMRKKTQSMEEEQDQLNWQL 162
>GLOB_ANTMA (Q03378) Floral homeotic protein GLOBOSA
Length = 215
Score = 127 bits (319), Expect = 2e-29
Identities = 74/206 (35%), Positives = 118/206 (56%), Gaps = 15/206 (7%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN +NRQVTYSKRRNGI KKA E+SVLCDA VS+I+F+ + KMHE+ +P
Sbjct: 1 MGRGKIEIKRIENSSNRQVTYSKRRNGIMKKAKEISVLCDAHVSVIIFASSGKMHEFCSP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+ ++D Y K G LW +E + + ++K N ++ ++RH GE ++ L
Sbjct: 61 STTLVDMLDHYHKLSGK-RLWDPKHEHLDNEINRVKKENDSMQIELRHLKGE---DITTL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTLFLSS 180
++++L LE+ + + + ++ ++ ++ RK +E+ N +L +L R + L
Sbjct: 117 NYKELMVLEDALENGTSALKNKQMEFVR----MMRKHNEMVEEENQSLQFKL-RQMHLDP 171
Query: 181 Y------LYAFCQHHSHLNLPHHHGE 200
A HH H N+ + +
Sbjct: 172 MNDNVMESQAVYDHHHHQNIADYEAQ 197
>PIST_ARATH (P48007) Floral homeotic protein PISTILLATA
(Transcription factor PI)
Length = 208
Score = 125 bits (315), Expect = 5e-29
Identities = 70/204 (34%), Positives = 112/204 (54%), Gaps = 14/204 (6%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN NR VT+SKRRNG+ KKA E++VLCDAKV+LI+F+ N KM +Y P
Sbjct: 1 MGRGKIEIKRIENANNRVVTFSKRRNGLVKKAKEITVLCDAKVALIIFASNGKMIDYCCP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
+ ++DQYQK G LW + +E + + ++K N L+ ++RH GE ++ L
Sbjct: 61 SMDLGAMLDQYQKLSGK-KLWDAKHENLSNEIDRIKKENDSLQLELRHLKGE---DIQSL 116
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTLFLSS 180
+ + L ++E + + K+R+ + ++ ++ + Q+ T L
Sbjct: 117 NLKNLMAVEHAIEHGLDKVRDHQMEILISKRRNEKMMAEEQRQL----------TFQLQQ 166
Query: 181 YLYAFCQHHSHLNLPHHHGEEGYK 204
A + + + H G+ GY+
Sbjct: 167 QEMAIASNARGMMMRDHDGQFGYR 190
>FBP1_PETHY (Q03488) Floral homeotic protein FBP1 (Floral binding
protein 1)
Length = 210
Score = 125 bits (314), Expect = 7e-29
Identities = 73/172 (42%), Positives = 109/172 (62%), Gaps = 10/172 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIEIK IEN +NRQVTYSKRRNGI KKA E+SVLCDA+VS+I+F+ + KMHE+ +
Sbjct: 1 MGRGKIEIKRIENSSNRQVTYSKRRNGILKKAKEISVLCDARVSVIIFASSGKMHEFSST 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
L I+DQY K G L + +E + + K+K N ++ ++RH GE ++ L
Sbjct: 61 SL--VDILDQYHKLTGR-RLLDAKHENLDNEINKVKKDNDNMQIELRHLKGE---DITSL 114
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
+ ++L LE+ + + + IR ++ V++ RKK +S+E+ L +L
Sbjct: 115 NHRELMILEDALENGLTSIRNKQNEVLR----MMRKKTQSMEEEQDQLNCQL 162
>DE21_ANTMA (Q8RVL4) MADS box protein defh21 (DEFICIENS homolog 21)
Length = 247
Score = 124 bits (311), Expect = 2e-28
Identities = 72/204 (35%), Positives = 113/204 (55%), Gaps = 14/204 (6%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIE+K IEN T+RQVT+SKRR+G+ KK HELSVLCDA++ LI+FS K+ EY TP
Sbjct: 1 MGRGKIEVKRIENNTSRQVTFSKRRSGLMKKTHELSVLCDAQIGLIVFSTKGKLTEYCTP 60
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
S K+IID+Y K G + + +N + +K++ + ++ + DDL
Sbjct: 61 PFSMKQIIDRYVKAKGILPEMENRAGPHADNDQVIKELTRMKEETLNLQLNLQRYKGDDL 120
Query: 121 S---FQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTLF 177
S F++L LE+ ++ S+ K+R RK ++ + + ++ LE+ N + L+
Sbjct: 121 STVRFEELTELEKLLDQSLNKVRARKLELLHEQMENLKRTEFMLEKENQEMYHWLMSN-- 178
Query: 178 LSSYLYAFCQHHSHLNLPHHHGEE 201
Q + HHH ++
Sbjct: 179 ---------QIQRQAEVEHHHQQQ 193
>AG14_ARATH (Q38838) Agamous-like MADS box protein AGL14
Length = 221
Score = 122 bits (305), Expect = 8e-28
Identities = 72/176 (40%), Positives = 110/176 (61%), Gaps = 15/176 (8%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
M RGK E+K IEN T+RQVT+SKRRNG+ KKA ELSVLCDA+V+LI+FS K++E+ +
Sbjct: 1 MVRGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEF-SS 59
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRH-------RIGEG 113
S K +++YQK + D+ K +N ++ KD + L R+I H +GEG
Sbjct: 60 SSSIPKTVERYQKRIQDL----GSNHKRNDNSQQSKDETYGLARKIEHLEISTRKMMGEG 115
Query: 114 GMELDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLL 169
LD S ++L+ LE ++ S+ KIR +K+ +++ T+ ++K R+L N L+
Sbjct: 116 ---LDASSIEELQQLENQLDRSLMKIRAKKYQLLREETEKLKEKERNLIAENKMLM 168
>FB24_PETHY (Q9ATE5) MADS box protein FBP24 (Floral binding protein
24)
Length = 268
Score = 118 bits (296), Expect = 9e-27
Identities = 71/210 (33%), Positives = 114/210 (53%), Gaps = 8/210 (3%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGKIE+K IEN T+RQVT+SKRR G+ KK HELSVLCDA++ LI+FS K+ EY +
Sbjct: 4 MGRGKIEVKRIENKTSRQVTFSKRRAGLLKKTHELSVLCDAQIGLIIFSSKGKLFEYCSQ 63
Query: 61 GLSTKKIIDQYQKTLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMELDDL 120
S +II +Y +T G + ++ + + K++ L+ ++ G+ +L
Sbjct: 64 PHSMSQIISRYLQTTGASLPVEDNRVQLYDEVAKMRRDTLNLQLSLQRYKGD---DLSLA 120
Query: 121 SFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRTLFLSS 180
+++L LE+ + ++ KIR RK +++ + + +K + LE+ N ++ L+
Sbjct: 121 QYEELNELEKQLEHALNKIRARKLELMQQQMENLKKTEKMLEKENHDMYQWLMNNQMYKQ 180
Query: 181 YLYAFCQHHSHLNLPHHHGEEGYKNDDLRL 210
A H H HHH E +L L
Sbjct: 181 ESAAM-DHEDH----HHHHEHQQAITELNL 205
>AGL8_ARATH (Q38876) Agamous-like MADS box protein AGL8 (Floral
homeotic protein AGL8) (Transcription factor FRUITFULL)
Length = 242
Score = 116 bits (291), Expect = 3e-26
Identities = 71/176 (40%), Positives = 109/176 (61%), Gaps = 8/176 (4%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR+G+ KKAHE+SVLCDA+V+LI+FS K+ EY T
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSSKGKLFEYSTD 60
Query: 61 GLSTKKIIDQYQKTL-GDIDLWR---SHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
++I+++Y + L D L S E + KLK L + R+ +GE +
Sbjct: 61 S-CMERILERYDRYLYSDKQLVGRDVSQSENWVLEHAKLKARVEVLEKNKRNFMGE---D 116
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
LD LS ++L+SLE ++++I IR RK + +KK ++L+ N +LL ++
Sbjct: 117 LDSLSLKELQSLEHQLDAAIKSIRSRKNQAMFESISALQKKDKALQDHNNSLLKKI 172
>AGL8_LYCES (Q40170) Agamous-like MADS box protein AGL8 homolog
(TM4)
Length = 227
Score = 115 bits (289), Expect = 6e-26
Identities = 65/172 (37%), Positives = 108/172 (62%), Gaps = 8/172 (4%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR+G+ KKAHE+SVLCDA+V LI+FS K+ EY
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVGLIVFSTKGKLFEYAND 60
Query: 61 GLSTKKIIDQYQK-TLGDIDLWRSHYEKMLE---NLKKLKDINHKLRRQIRHRIGEGGME 116
++I+++Y++ + + L + + + +KLK L+R +H +GE +
Sbjct: 61 S-CMERILERYERYSFAEKQLVPTDHTSPVSWTLEHRKLKARLEVLQRNQKHYVGE---D 116
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNL 168
L+ LS ++L++LE ++S++ IR RK ++ +KK R+L++ N L
Sbjct: 117 LESLSMKELQNLEHQLDSALKHIRSRKNQLMHESISVLQKKDRALQEQNNQL 168
>AGL8_SINAL (Q41274) Agamous-like MADS box protein AGL8 homolog
(MADS B)
Length = 241
Score = 115 bits (287), Expect = 1e-25
Identities = 71/176 (40%), Positives = 107/176 (60%), Gaps = 8/176 (4%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRG++++K IEN NRQVT+SKRR+G+ KKAHE+SVLCDA+V+L++FS K+ EY T
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALVIFSSKGKLFEYSTD 60
Query: 61 GLSTKKIIDQYQKTL-GDIDLWR---SHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
+KI+++Y + L D L S E + KLK L + R+ +GE +
Sbjct: 61 S-CMEKILERYDRYLYSDKQLVGRDISQSENWVLEHAKLKARVEVLEKNKRNFMGE---D 116
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL 172
LD LS ++L+SLE ++++I IR RK + +KK + L+ N LL ++
Sbjct: 117 LDSLSLKELQSLEHQLHAAIKSIRSRKNQAMFESISALQKKDKVLQDHNNALLKKI 172
>AG_TOBAC (Q43585) Floral homeotic protein AGAMOUS (NAG1)
Length = 248
Score = 114 bits (286), Expect = 1e-25
Identities = 69/179 (38%), Positives = 105/179 (58%), Gaps = 9/179 (5%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
+GRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+LI+FS +++EY
Sbjct: 17 LGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYANN 76
Query: 61 GLSTKKIIDQYQKTLGDI----DLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
S K I++Y+K D + ++ + + KL+ L+ Q R+ +GE
Sbjct: 77 --SVKATIERYKKACSDSSNTGSISEANAQYYQQEASKLRAQIGNLQNQNRNMLGE---S 131
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLELVRT 175
L LS + L++LE+ + I+KIR +K ++ + +K+ L N L ++ T
Sbjct: 132 LAALSLRDLKNLEQKIEKGISKIRSKKNELLFAEIEYMQKREIDLHNNNQYLRAKIAET 190
>AG13_ARATH (Q38837) Agamous-like MADS box protein AGL13
Length = 244
Score = 114 bits (286), Expect = 1e-25
Identities = 74/208 (35%), Positives = 118/208 (56%), Gaps = 16/208 (7%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
MGRGK+E+K IEN RQVT+SKR++G+ KKA+ELSVLCDA+VSLI+FS K++E+
Sbjct: 1 MGRGKVEVKRIENKITRQVTFSKRKSGLLKKAYELSVLCDAEVSLIIFSTGGKLYEFSNV 60
Query: 61 GLSTKKIIDQYQK---TLGDIDLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGMEL 117
G+ + I++Y + L D D + + + + KLK L R R+ +GE +L
Sbjct: 61 GVG--RTIERYYRCKDNLLDNDTLED-TQGLRQEVTKLKCKYESLLRTHRNLVGE---DL 114
Query: 118 DDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLLEL----- 172
+ +S ++L++LE + +++ R++K V+ + + R+K R L +N L LE
Sbjct: 115 EGMSIKELQTLERQLEGALSATRKQKTQVMMEQMEELRRKERELGDINNKLKLETEDHDF 174
Query: 173 --VRTLFLSSYLYAFCQHHSHLNLPHHH 198
+ L L+ L A C L H +
Sbjct: 175 KGFQDLLLNPVLTAGCSTDFSLQSTHQN 202
>AGL5_ARATH (P29385) Agamous-like MADS box protein AGL5
Length = 246
Score = 114 bits (285), Expect = 2e-25
Identities = 65/174 (37%), Positives = 105/174 (59%), Gaps = 11/174 (6%)
Query: 1 MGRGKIEIKLIENPTNRQVTYSKRRNGIFKKAHELSVLCDAKVSLIMFSKNNKMHEYITP 60
+GRGKIEIK IEN TNRQVT+ KRRNG+ KKA+ELSVLCDA+V+L++FS +++EY
Sbjct: 16 IGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 GLSTKKIIDQYQKTLGDI----DLWRSHYEKMLENLKKLKDINHKLRRQIRHRIGEGGME 116
S + I++Y+K D + ++ + + KL+ ++ RH +GE
Sbjct: 76 --SVRGTIERYKKACSDAVNPPTITEANTQYYQQEASKLRRQIRDIQNLNRHILGE---S 130
Query: 117 LDDLSFQQLRSLEEDMNSSIAKIRERKFHVIKTRTDTCRKKVRSLEQMNGNLLL 170
L L+F++L++LE + I+++R +K ++ + +K R +E N N+ L
Sbjct: 131 LGSLNFKELKNLESRLEKGISRVRSKKHEMLVAEIEYMQK--REIELQNDNMYL 182
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.320 0.137 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,167,886
Number of Sequences: 164201
Number of extensions: 942532
Number of successful extensions: 3744
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 82
Number of HSP's successfully gapped in prelim test: 70
Number of HSP's that attempted gapping in prelim test: 3599
Number of HSP's gapped (non-prelim): 183
length of query: 210
length of database: 59,974,054
effective HSP length: 105
effective length of query: 105
effective length of database: 42,732,949
effective search space: 4486959645
effective search space used: 4486959645
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC136451.6


