

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136287.2 + phase: 0
(365 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
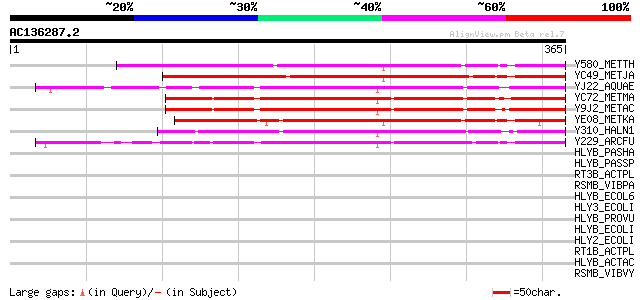
Score E
Sequences producing significant alignments: (bits) Value
Y580_METTH (O26680) Hypothetical UPF0245 protein MTH580 216 6e-56
YC49_METJA (Q58646) Hypothetical UPF0245 protein MJ1249 209 9e-54
YJ22_AQUAE (O67751) Hypothetical UPF0245 protein AQ_1922 201 2e-51
YC72_METMA (Q8PXE8) Hypothetical UPF0245 protein MM1272 198 2e-50
Y9J2_METAC (Q8THC5) Hypothetical UPF0245 protein MA4592 196 8e-50
YE08_METKA (Q8TVI1) Hypothetical UPF0245 protein MK1408 196 1e-49
Y310_HALN1 (Q9HSB6) Hypothetical UPF0245 protein Vng0310c 187 4e-47
Y229_ARCFU (O30010) Hypothetical UPF0245 protein AF0229 182 9e-46
HLYB_PASHA (P16532) Leukotoxin translocation ATP-binding protein... 41 0.004
HLYB_PASSP (P55122) Leukotoxin translocation ATP-binding protein... 39 0.021
RT3B_ACTPL (Q04473) Toxin RTX-III translocation ATP-binding prot... 37 0.060
RSMB_VIBPA (Q87KD3) Ribosomal RNA small subunit methyltransferas... 37 0.060
HLYB_ECOL6 (Q8FDZ8) Alpha-hemolysin translocation ATP-binding pr... 37 0.078
HLY3_ECOLI (Q47258) Alpha-hemolysin translocation ATP-binding pr... 37 0.078
HLYB_PROVU (P11599) Alpha-hemolysin translocation ATP-binding pr... 37 0.10
HLYB_ECOLI (P08716) Alpha-hemolysin translocation ATP-binding pr... 37 0.10
HLY2_ECOLI (P10089) Alpha-hemolysin translocation ATP-binding pr... 37 0.10
RT1B_ACTPL (P26760) Toxin RTX-I translocation ATP-binding protei... 36 0.17
HLYB_ACTAC (P23702) Leukotoxin translocation ATP-binding protein... 36 0.17
RSMB_VIBVY (Q7MGK4) Ribosomal RNA small subunit methyltransferas... 33 0.87
>Y580_METTH (O26680) Hypothetical UPF0245 protein MTH580
Length = 378
Score = 216 bits (551), Expect = 6e-56
Identities = 128/297 (43%), Positives = 178/297 (59%), Gaps = 12/297 (4%)
Query: 71 LDAQNKKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVF 130
L ++VA ++ + E R + +++ DW++IP ENIIA Q +
Sbjct: 92 LSESGRQVAAYVEIRSKAHEELARRLGRVVDYLILVGEDWKIIPLENIIADLQEEDVKLI 151
Query: 131 AISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQI 190
A + EA+ LE LEHG DG++++ D+ + ++ + ES L AT+T+I
Sbjct: 152 AAVADVDEARVALETLEHGTDGVLIEPADISQIKDIAALLENI--ESETYELKPATITRI 209
Query: 191 QVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTFSGEC--VHAY 248
+ G GDRVCVD CS+M GEG+LVGS+++GLFLVHSE LES+ + F V AY
Sbjct: 210 EPIGSGDRVCVDTCSIMGIGEGMLVGSYSQGLFLVHSESLESEYVASRPFRVNAGPVQAY 269
Query: 249 IAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISI 308
+ VPGGRT YLSEL++G EVI+VD+ GR R AIVGRVKIE RPL+LVEA + + +
Sbjct: 270 VMVPGGRTRYLSELETGDEVIIVDRDGRSRSAIVGRVKIEKRPLMLVEA--EYEGMKVRT 327
Query: 309 LLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
LLQNAET+ LV +G + V+ L GD VL+ ARH G+ I+E I+EK
Sbjct: 328 LLQNAETIRLVND-KGEPV-----SVSELGEGDRVLVYFDESARHFGMAIKETIIEK 378
>YC49_METJA (Q58646) Hypothetical UPF0245 protein MJ1249
Length = 361
Score = 209 bits (532), Expect = 9e-54
Identities = 118/267 (44%), Positives = 171/267 (63%), Gaps = 12/267 (4%)
Query: 101 ENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDV 160
+NI++ DW +IP EN+IA + + A ++ EA+ E LE G DG++L +++
Sbjct: 105 DNIILEGRDWTIIPLENLIADLFHRDVKIVASVNSVDEAKVAYEILEKGTDGVLLNPKNL 164
Query: 161 EPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFAR 220
E + EL + + +E L++ ATVT+++ G GDRVC+D CSLM+ GEG+L+GS++R
Sbjct: 165 EDIKELSKLIEEMNKEKVALDV--ATVTKVEPIGSGDRVCIDTCSLMKIGEGMLIGSYSR 222
Query: 221 GLFLVHSECLESKLHCKQTFSGEC--VHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQR 278
LFLVHSE +E+ + F VHAYI PG +T YLSELK+G +V++VD+ G R
Sbjct: 223 ALFLVHSETVENPYVATRPFRVNAGPVHAYILCPGNKTKYLSELKAGDKVLIVDKDGNTR 282
Query: 279 IAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLK 338
AIVGRVKIE RPL+L+EA+ D I +LQNAET+ LV +G + V LK
Sbjct: 283 EAIVGRVKIERRPLVLIEAEYKGD--IIRTILQNAETIRLV-NEKGEPI-----SVVDLK 334
Query: 339 VGDEVLLRVQGGARHTGIEIQEFIVEK 365
GD+VL++ + ARH G+ I+E I+EK
Sbjct: 335 PGDKVLIKPEEYARHFGMAIKETIIEK 361
>YJ22_AQUAE (O67751) Hypothetical UPF0245 protein AQ_1922
Length = 331
Score = 201 bits (512), Expect = 2e-51
Identities = 126/353 (35%), Positives = 194/353 (54%), Gaps = 28/353 (7%)
Query: 18 KKVWIWTK--NKQVMTAAVERGWNTFIFPSN-LPHLANDWSSIAVICPLFASEKEILDAQ 74
K+ W+W + ++++++ A+E G N + P I VI P + ++ +
Sbjct: 2 KEFWVWVEPFDRKIVSVALEAGANAVVIPEKGRVEEVKKVGRITVIAP----DGDLKLGE 57
Query: 75 NKKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISD 134
+ I E + P + ++V DW VIP EN+IA ++ ++A+
Sbjct: 58 DVVYVLIKGKEDEERAAKYPPNVK----VIVETTDWTVIPLENLIA----QREELYAVVK 109
Query: 135 NASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAG 194
NA EA+ L+ LE G+ G+VLK D+ E+K+ +E L L +T+I G
Sbjct: 110 NAQEAEVALQTLEKGVKGVVLKSRDIN---EIKKVGQIVSEHEEKLELVTIKITKILPLG 166
Query: 195 MGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVP 252
+GDRVCVD SL+ GEG+LVG+ + G+FLVH+E E+ + F + VH YI VP
Sbjct: 167 LGDRVCVDTISLLHRGEGMLVGNSSGGMFLVHAETEENPYVAARPFRVNAGAVHMYIRVP 226
Query: 253 GGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQN 312
RT YL ELK+G +V+V D +GR R+ VGR K+E RP++L+E + +N+ +S +LQN
Sbjct: 227 NNRTKYLCELKAGDKVMVYDYKGRGRVTYVGRAKVERRPMLLIEGR--YENKKLSCILQN 284
Query: 313 AETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
AET+ L P T V+ LK GDEVL V+ RH G++++E I+EK
Sbjct: 285 AETIRLTKPD------GTPISVSELKEGDEVLGYVEEAGRHFGMKVEETIIEK 331
>YC72_METMA (Q8PXE8) Hypothetical UPF0245 protein MM1272
Length = 380
Score = 198 bits (503), Expect = 2e-50
Identities = 120/267 (44%), Positives = 174/267 (64%), Gaps = 17/267 (6%)
Query: 103 IVVNLLDWQVIPAENIIAAFQNSQ-KTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVE 161
++V DW+VIP EN+IA Q + K +F + +A EA+ + LE G DG++L + +
Sbjct: 127 LLVTGTDWKVIPLENLIADLQREKVKIIFGVK-SAEEARLAFQTLETGADGVLLDSGNPQ 185
Query: 162 PMLELKEYFDRRTE-ESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFAR 220
E+K+ E ES L A VT+++ GMGDRVCVD C+LM+ GEG+L+GS A
Sbjct: 186 ---EIKDTIKAARELESESTELESAVVTRVEPLGMGDRVCVDTCNLMQRGEGMLIGSQAS 242
Query: 221 GLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQR 278
G+FLV+SE +S + F + VH+YI + G +T YLSEL++G V ++D +G+QR
Sbjct: 243 GMFLVNSESDDSPYVAARPFRVNAGAVHSYIKI-GDKTRYLSELQAGDSVTIIDSRGKQR 301
Query: 279 IAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLK 338
IVGR+KIESRPL+L+EAK + ++T++ +LQNAET+ LV +G T V LK
Sbjct: 302 EGIVGRIKIESRPLMLIEAK--AGDRTLTAILQNAETIKLV--GKGG----TPISVAKLK 353
Query: 339 VGDEVLLRVQGGARHTGIEIQEFIVEK 365
GDEVL+R++ GARH G +I+E I+EK
Sbjct: 354 KGDEVLVRLEEGARHFGKKIEETIIEK 380
>Y9J2_METAC (Q8THC5) Hypothetical UPF0245 protein MA4592
Length = 380
Score = 196 bits (498), Expect = 8e-50
Identities = 121/267 (45%), Positives = 174/267 (64%), Gaps = 17/267 (6%)
Query: 103 IVVNLLDWQVIPAENIIAAFQNSQ-KTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVE 161
++V DW+VIP EN+IA Q+ + K +F + +A EA+ + LE G DG++L + +
Sbjct: 127 LLVTGTDWKVIPLENLIADLQHQKVKIIFGVK-SAEEARLAFQTLEAGADGVLLDSGNPQ 185
Query: 162 PMLELKEYFDRRTE-ESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFAR 220
E+K+ E ES L A VT+++ GMGDRVCVD C+LM+ GEG+L+GS A
Sbjct: 186 ---EIKDTIKAARELESESAELEAAVVTRVEPLGMGDRVCVDTCNLMQRGEGMLIGSQAS 242
Query: 221 GLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQR 278
G+FLV+SE +S + F + VH+YI + G +T YLSEL++G V +VD +G+QR
Sbjct: 243 GMFLVNSESDDSPYVAARPFRVNAGAVHSYIKI-GEKTRYLSELRAGDPVTIVDSKGKQR 301
Query: 279 IAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLK 338
IVGRVKIESRPL+L+EAK + ++T++ +LQNAET+ LV G T V L+
Sbjct: 302 EGIVGRVKIESRPLMLIEAK--ARDRTLTAILQNAETIKLV----GKD--GTPISVAKLE 353
Query: 339 VGDEVLLRVQGGARHTGIEIQEFIVEK 365
GDEVL+R++ GARH G +I+E I+EK
Sbjct: 354 KGDEVLVRLEEGARHFGKKIEETIIEK 380
>YE08_METKA (Q8TVI1) Hypothetical UPF0245 protein MK1408
Length = 402
Score = 196 bits (497), Expect = 1e-49
Identities = 119/265 (44%), Positives = 163/265 (60%), Gaps = 19/265 (7%)
Query: 109 DWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELK- 167
DW++IP EN+IA Q + + A + +A EA+ E LE G DG++L E ++P E+K
Sbjct: 149 DWKIIPLENLIAELQGEKTQLIAGARDAEEARIAFETLEVGSDGVLLDAERIDPS-EIKK 207
Query: 168 --EYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLV 225
E +R E L V +++ G GDRVCVD CSLM GEG+LVGS +RG+FL+
Sbjct: 208 TAEIAERAAAER--FELVAVEVKEVKPIGKGDRVCVDTCSLMSEGEGMLVGSTSRGMFLI 265
Query: 226 HSECLESKLHCKQTFSGEC--VHAYIAVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVG 283
HSE LE+ + F VHAYI VPGG+T YL+EL+ G EV++VD +GR R A+VG
Sbjct: 266 HSESLENPYVEPRPFRVNAGPVHAYIRVPGGKTKYLAELRPGDEVLIVDTEGRTRAAVVG 325
Query: 284 RVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEV 343
R+KIE RPL+L+ A + + I ++QNAET+ LV G + V LK GD+V
Sbjct: 326 RLKIERRPLMLIRA--EYEGVEIQTIVQNAETIHLV-REDGEPV-----SVVDLKPGDKV 377
Query: 344 LLRV---QGGARHTGIEIQEFIVEK 365
L V +G RH G+E++E IVEK
Sbjct: 378 LAYVETEEGKGRHFGMEVEETIVEK 402
>Y310_HALN1 (Q9HSB6) Hypothetical UPF0245 protein Vng0310c
Length = 387
Score = 187 bits (475), Expect = 4e-47
Identities = 118/270 (43%), Positives = 159/270 (58%), Gaps = 13/270 (4%)
Query: 98 EQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFAISDNASEAQTFLEALEHGLDGIVLKV 157
E A++ +V DW +IP EN+IA + T+ A ++A+EA+T E L+ G D ++L
Sbjct: 129 EVADHTIVVGDDWTIIPLENLIARI-GEETTLVAGVESAAEAETAFETLDIGADAVLLDS 187
Query: 158 EDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQVAGMGDRVCVDLCSLMRPGEGLLVGS 217
+D + + D E L L+ AT+T I+ AG DRVCVD SL+ EG+LVGS
Sbjct: 188 DDPDEIRRTVSVRDAADREH--LALSTATITTIEEAGSADRVCVDTGSLLADDEGMLVGS 245
Query: 218 FARGLFLVHSECLESKLHCKQTF--SGECVHAYIAVPGGRTSYLSELKSGKEVIVVDQQG 275
+RGLF VH+E +S + F + VHAY+ P G T YL+EL SG EV VVD G
Sbjct: 246 MSRGLFFVHAETAQSPYVAARPFRVNAGAVHAYVRTPDGGTKYLAELGSGDEVQVVDGDG 305
Query: 276 RQRIAIVGRVKIESRPLILVEAKTDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVT 335
R R A+VGR KIE RP+ VEA+TD D I LLQNAET+ + P +TA VT
Sbjct: 306 RTRSAVVGRAKIEKRPMFRVEAETD-DGDRIETLLQNAETIKVATPNG-----RTA--VT 357
Query: 336 SLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
L VGD++ + +Q G RH G I E I+E+
Sbjct: 358 DLSVGDDLHVFLQDGGRHFGEAIDERIIEQ 387
>Y229_ARCFU (O30010) Hypothetical UPF0245 protein AF0229
Length = 323
Score = 182 bits (463), Expect = 9e-46
Identities = 130/356 (36%), Positives = 197/356 (54%), Gaps = 42/356 (11%)
Query: 18 KKVWI------WTKNKQVMTAAVERGWNTFIFPSNLPHLANDWSSIAVICPLFASEKEIL 71
K++W+ W + K+++ A+E G++ + + A + ++ I
Sbjct: 2 KEIWLLAESESWDEAKEMLKDAIEIGFDGALVRRDFLERAEKLGRMKIV--------PIE 53
Query: 72 DAQNKKVATIFDVSTPEELERLRPEDEQAENIVVNLLDWQVIPAENIIAAFQNSQKTVFA 131
DA + +S+ E+ ER Q E +V+ DW+VIP ENI+A + S K + A
Sbjct: 54 DA-------VVKISSAEDQERAL----QREVVVLKFEDWKVIPLENIVA-MKKSGKVIAA 101
Query: 132 ISDNASEAQTFLEALEHGLDGIVLKVEDVEPMLELKEYFDRRTEESNVLNLTKATVTQIQ 191
+ D +A+ L LE G DGI + + L+++++ EE + L +A V +I+
Sbjct: 102 V-DTIEDAKLALTTLERGADGIAVSGDRET----LRKFYEVVKEEGERVELVRARVKEIR 156
Query: 192 VAGMGDRVCVDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTF--SGECVHAYI 249
G+G+RVC+D +LM PGEG+LVG+ A +FLV SE ES+ + F + V+AY+
Sbjct: 157 PLGVGERVCIDTVTLMTPGEGMLVGNQASFMFLVASESEESEYVASRPFRVNAGSVNAYL 216
Query: 250 AVPGGRTSYLSELKSGKEVIVVDQQGRQRIAIVGRVKIESRPLILVEAKTDSDNQTISIL 309
V G +T YL+ELK+G EV VV G R + VGRVKIE RPLIL+ A+ D S++
Sbjct: 217 KV-GDKTRYLAELKAGDEVEVVKFDGAVRKSYVGRVKIERRPLILIRAEVDGVEG--SVI 273
Query: 310 LQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQGGARHTGIEIQEFIVEK 365
LQNAET+ LV P G + V LK GDE+L+ + ARH G+E+ EFIVE+
Sbjct: 274 LQNAETIKLVAP-DGKHV-----SVAELKPGDEILVWLGKKARHFGVEVDEFIVER 323
>HLYB_PASHA (P16532) Leukotoxin translocation ATP-binding protein
lktB
Length = 708
Score = 41.2 bits (95), Expect = 0.004
Identities = 30/108 (27%), Positives = 53/108 (48%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV +Q RQRIAI + + LI EA +
Sbjct: 578 YAAKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 637
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ I++QN + + QG ++ A ++++K D +++ +G
Sbjct: 638 DYESEHIIMQNMQKIC-----QGRTVILIAHRLSTVKNADRIIVMEKG 680
>HLYB_PASSP (P55122) Leukotoxin translocation ATP-binding protein
lktB
Length = 708
Score = 38.9 bits (89), Expect = 0.021
Identities = 28/108 (25%), Positives = 53/108 (48%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++S+++ G IV +Q RQRIAI + + LI EA +
Sbjct: 578 YAAKLAGAHDFISDVREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 637
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ I++QN + + QG ++ A ++++K D +++ +G
Sbjct: 638 DYESEHIIMQNMQKIC-----QGRTVILIAHRLSTVKNADRIIVMEKG 680
>RT3B_ACTPL (Q04473) Toxin RTX-III translocation ATP-binding protein
(RTX-III toxin determinant B) (APX-IIIB) (Cytolysin
IIIB) (CLY-IIIB)
Length = 711
Score = 37.4 bits (85), Expect = 0.060
Identities = 29/108 (26%), Positives = 51/108 (46%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV + RQRIAI + R LI EA +
Sbjct: 581 YAAKLAGAHDFISELREGYNTIVGELGAGLSGGQRQRIAIARALVNNPRILIFDEATSAL 640
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ I++QN + + G ++ A ++++K D +++ +G
Sbjct: 641 DYESEHIIMQNMQKIC-----HGRTVIIIAHRLSTVKNADRIIVMEKG 683
>RSMB_VIBPA (Q87KD3) Ribosomal RNA small subunit methyltransferase B
(EC 2.1.1.-) (rRNA (cytosine-C(5)-)-methyltransferase)
(16S rRNA m5C967 methyltransferase)
Length = 427
Score = 37.4 bits (85), Expect = 0.060
Identities = 19/72 (26%), Positives = 35/72 (48%)
Query: 201 VDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTFSGECVHAYIAVPGGRTSYLS 260
+D L P + + + F +G V + ++ + GE + A PGG+T+++
Sbjct: 203 MDAIKLAAPCDVMKLPGFDKGWVSVQDAAAQLSINYLKPQDGELILDCCAAPGGKTAHIL 262
Query: 261 ELKSGKEVIVVD 272
E SG EV+ +D
Sbjct: 263 ERTSGSEVVAID 274
>HLYB_ECOL6 (Q8FDZ8) Alpha-hemolysin translocation ATP-binding
protein hlyB
Length = 707
Score = 37.0 bits (84), Expect = 0.078
Identities = 28/108 (25%), Positives = 52/108 (47%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV +Q RQRIAI + + LI EA +
Sbjct: 577 YAAKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 636
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ I+++N + +G ++ A ++++K D +++ +G
Sbjct: 637 DYESEHIIMRNMHKIC-----KGRTVIIIAHRLSTVKNADRIIVMEKG 679
>HLY3_ECOLI (Q47258) Alpha-hemolysin translocation ATP-binding
protein hlyB
Length = 707
Score = 37.0 bits (84), Expect = 0.078
Identities = 28/108 (25%), Positives = 52/108 (47%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV +Q RQRIAI + + LI EA +
Sbjct: 577 YAAKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 636
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ I+++N + +G ++ A ++++K D +++ +G
Sbjct: 637 DYESEHIIMRNMHKIC-----KGRTVIIIAHRLSTVKNADRIIVMEKG 679
>HLYB_PROVU (P11599) Alpha-hemolysin translocation ATP-binding
protein hlyB
Length = 707
Score = 36.6 bits (83), Expect = 0.10
Identities = 29/111 (26%), Positives = 54/111 (48%), Gaps = 13/111 (11%)
Query: 245 VHAYIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAK 298
+HA A G ++SEL+ G IV +Q RQRIAI + + LI EA
Sbjct: 576 IHA--AKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEAT 633
Query: 299 TDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
+ D ++ ++++N + QG ++ A ++++K D +++ +G
Sbjct: 634 SALDYESEHVIMRNMHKIC-----QGRTVIIIAHRLSTVKNADRIIVMEKG 679
>HLYB_ECOLI (P08716) Alpha-hemolysin translocation ATP-binding
protein hlyB
Length = 707
Score = 36.6 bits (83), Expect = 0.10
Identities = 27/108 (25%), Positives = 52/108 (48%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV +Q RQRIAI + + LI EA +
Sbjct: 577 YAAKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 636
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ ++++N + +G ++ A ++++K D +++ +G
Sbjct: 637 DYESEHVIMRNMHKIC-----KGRTVIIIAHRLSTVKNADRIIVMEKG 679
>HLY2_ECOLI (P10089) Alpha-hemolysin translocation ATP-binding
protein hlyB
Length = 707
Score = 36.6 bits (83), Expect = 0.10
Identities = 27/108 (25%), Positives = 52/108 (48%), Gaps = 11/108 (10%)
Query: 248 YIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDS 301
Y A G ++SEL+ G IV +Q RQRIAI + + LI EA +
Sbjct: 577 YAAKLAGAHDFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEATSAL 636
Query: 302 DNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
D ++ ++++N + +G ++ A ++++K D +++ +G
Sbjct: 637 DYESEHVIMRNMHKIC-----KGRTVIIIAHRLSTVKNADRIIVMEKG 679
>RT1B_ACTPL (P26760) Toxin RTX-I translocation ATP-binding protein
(RTX-I toxin determinant B) (APX-IB) (HLY-IB) (Cytolysin
IB) (CLY-IB)
Length = 707
Score = 35.8 bits (81), Expect = 0.17
Identities = 30/111 (27%), Positives = 54/111 (48%), Gaps = 13/111 (11%)
Query: 245 VHAYIAVPGGRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAK 298
VHA A G ++SEL+ G IV +Q RQRIAI + + LI EA
Sbjct: 576 VHA--AKLAGAHEFISELREGYNTIVGEQGAGLSGGQRQRIAIARALVNNPKILIFDEAT 633
Query: 299 TDSDNQTISILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
+ D ++ I+++N + +G ++ A ++++K D +++ +G
Sbjct: 634 SALDYESEHIIMRNMHQIC-----KGRTVIIIAHRLSTVKNADRIIVMEKG 679
>HLYB_ACTAC (P23702) Leukotoxin translocation ATP-binding protein
lktB
Length = 707
Score = 35.8 bits (81), Expect = 0.17
Identities = 27/102 (26%), Positives = 49/102 (47%), Gaps = 11/102 (10%)
Query: 254 GRTSYLSELKSGKEVIVVDQQG------RQRIAIVGRVKIESRPLILVEAKTDSDNQTIS 307
G ++SEL+ G +V +Q RQRIAI + R LI EA + D ++ +
Sbjct: 583 GAHDFISELREGYNTVVGEQGAGLSGGQRQRIAIARALVNNPRILIFDEATSALDYESEN 642
Query: 308 ILLQNAETVALVCPVQGNKLLKTAFPVTSLKVGDEVLLRVQG 349
I++ N + Q +L A ++++K D +++ +G
Sbjct: 643 IIMHNMHKIC-----QNRTVLIIAHRLSTVKNADRIIVMDKG 679
>RSMB_VIBVY (Q7MGK4) Ribosomal RNA small subunit methyltransferase B
(EC 2.1.1.-) (rRNA (cytosine-C(5)-)-methyltransferase)
(16S rRNA m5C967 methyltransferase)
Length = 426
Score = 33.5 bits (75), Expect = 0.87
Identities = 19/72 (26%), Positives = 33/72 (45%)
Query: 201 VDLCSLMRPGEGLLVGSFARGLFLVHSECLESKLHCKQTFSGECVHAYIAVPGGRTSYLS 260
+D L P + + F +G V + + Q GE + A PGG+T+++
Sbjct: 203 MDAIKLAAPCDVHSLPGFEQGWVSVQDAAAQLSFNYLQPKDGELILDCCAAPGGKTAHIL 262
Query: 261 ELKSGKEVIVVD 272
E S +EV+ +D
Sbjct: 263 ERVSAREVVALD 274
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.317 0.133 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 40,974,061
Number of Sequences: 164201
Number of extensions: 1649558
Number of successful extensions: 5076
Number of sequences better than 10.0: 46
Number of HSP's better than 10.0 without gapping: 15
Number of HSP's successfully gapped in prelim test: 31
Number of HSP's that attempted gapping in prelim test: 5017
Number of HSP's gapped (non-prelim): 50
length of query: 365
length of database: 59,974,054
effective HSP length: 112
effective length of query: 253
effective length of database: 41,583,542
effective search space: 10520636126
effective search space used: 10520636126
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 66 (30.0 bits)
Medicago: description of AC136287.2


