

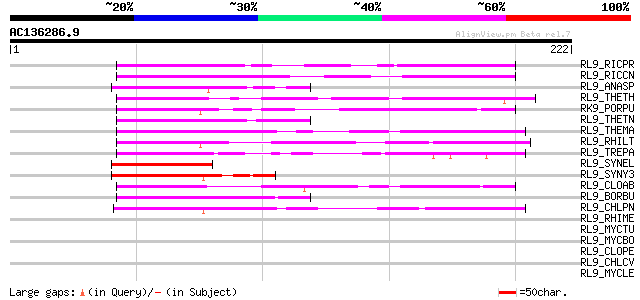
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.9 - phase: 0
(222 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
RL9_RICPR (Q9ZEA4) 50S ribosomal protein L9 53 5e-07
RL9_RICCN (Q92JK1) 50S ribosomal protein L9 52 8e-07
RL9_ANASP (Q8YZA0) 50S ribosomal protein L9 49 9e-06
RL9_THETH (P27151) 50S ribosomal protein L9 48 2e-05
RK9_PORPU (P51334) Chloroplast 50S ribosomal protein L9 48 2e-05
RL9_THETN (Q8R6M7) 50S ribosomal protein L9 47 3e-05
RL9_THEMA (Q9WZW7) 50S ribosomal protein L9 47 4e-05
RL9_RHILT (Q9F9K6) 50S ribosomal protein L9 47 5e-05
RL9_TREPA (O83099) 50S ribosomal protein L9 46 6e-05
RL9_SYNEL (P25904) 50S ribosomal protein L9 46 6e-05
RL9_SYNY3 (P42352) 50S ribosomal protein L9 44 3e-04
RL9_CLOAB (Q97CX9) 50S ribosomal protein L9 44 4e-04
RL9_BORBU (O51139) 50S ribosomal protein L9 44 4e-04
RL9_CHLPN (Q9Z6V3) 50S ribosomal protein L9 43 7e-04
RL9_RHIME (Q92QZ9) 50S ribosomal protein L9 41 0.003
RL9_MYCTU (P66315) 50S ribosomal protein L9 40 0.003
RL9_MYCBO (P66316) 50S ribosomal protein L9 40 0.003
RL9_CLOPE (Q8XH49) 50S ribosomal protein L9 40 0.003
RL9_CHLCV (Q821W9) 50S ribosomal protein L9 40 0.003
RL9_MYCLE (P46385) 50S ribosomal protein L9 40 0.006
>RL9_RICPR (Q9ZEA4) 50S ribosomal protein L9
Length = 171
Score = 53.1 bits (126), Expect = 5e-07
Identities = 43/158 (27%), Positives = 78/158 (49%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL + KLGK G+ +KVA GF RN+L+P+ LA+ L+ +Q+ Y+ E+
Sbjct: 1 MEVILIKPVRKLGKIGDILKVADGFGRNYLLPQKLAIRATKSNKELIVKQK--YEFEEKD 58
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
KQ + +E E+ I+ +LV R++ D+ +L V+
Sbjct: 59 KQ------------VREEVEKINTIIKDQQLVF----------IRQTSDD-CKLFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ ++++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KDIADKLSKNISYNISHSNIILDKQIKSTGIYTVEIRL 133
>RL9_RICCN (Q92JK1) 50S ribosomal protein L9
Length = 171
Score = 52.4 bits (124), Expect = 8e-07
Identities = 39/158 (24%), Positives = 76/158 (47%), Gaps = 25/158 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL + KLGK G+ +KVA GF RN+L+P+ LA+ + L+ +Q+ ++ ++
Sbjct: 1 MEIILIKPVRKLGKIGDILKVADGFGRNYLLPQKLAIRATEPNKELIVKQKHEFEAKDKQ 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
+E+V + ALI D+ + +R+ + K L V+
Sbjct: 61 IREEVEKIN-------------ALIKDQQLVFIRQTSNDGK------------LFGSVTN 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ +++ + NI+ N+ L + + G Y V +RL
Sbjct: 96 KEIADKLSENISYNISHSNIILDKQIKSTGIYTVEIRL 133
>RL9_ANASP (Q8YZA0) 50S ribosomal protein L9
Length = 152
Score = 48.9 bits (115), Expect = 9e-06
Identities = 31/82 (37%), Positives = 49/82 (58%), Gaps = 9/82 (10%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLL---AVPNIDKFAYLLTEQRKIYQ 97
+++Q++LT ++ KLG++G+ V VAPG+ RN+L+P+ L A P I K EQ + Q
Sbjct: 3 KRVQLVLTKDVSKLGRSGDLVDVAPGYARNYLIPQSLATHATPGILKQVERRREQER--Q 60
Query: 98 PTEEVKQEDVVVVTESKEDLMK 119
E++Q+ E KE L K
Sbjct: 61 RQLELRQQ----ALEQKESLEK 78
>RL9_THETH (P27151) 50S ribosomal protein L9
Length = 148
Score = 47.8 bits (112), Expect = 2e-05
Identities = 46/174 (26%), Positives = 78/174 (44%), Gaps = 34/174 (19%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL ++ LG G+ V V PG+ RN+L+P+ LAV L TE +
Sbjct: 1 MKVILLEPLENLGDVGQVVDVKPGYARNYLLPRGLAV--------LATE--------SNL 44
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
K + + ++K ++ E ++ K +L L +A E+K + V+
Sbjct: 45 KALEARIRAQAKRLAERKAE-----AERLKKILENLTLTIPVRAGETK-----IYGSVTA 94
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEY--------EVPLRLPRSIPLPE 208
+ ++RQ V I + L L P+ +GEY EVP++L S+ E
Sbjct: 95 KDIAEALSRQHGVTIDPKRLALEKPIKELGEYVLTYKPHPEVPIQLKVSVVAQE 148
>RK9_PORPU (P51334) Chloroplast 50S ribosomal protein L9
Length = 155
Score = 47.8 bits (112), Expect = 2e-05
Identities = 38/159 (23%), Positives = 76/159 (46%), Gaps = 27/159 (16%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMP-KLLAVPNIDKFAYLLTEQRKIYQPTEE 101
+QV+L NI KLGK+ + VKVA G+ RN L+P K+ +V + + Q+K+Y
Sbjct: 6 IQVVLKENIQKLGKSNDVVKVATGYARNFLIPNKMASVATVG-----MLNQQKLY---AA 57
Query: 102 VKQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVS 161
+K++ ++ E+ A+ + L +++K + + + V+
Sbjct: 58 IKEKKIIFAKEN-----------------ARKTQQLLEEIQKFSISKKTGDGETIFGSVT 100
Query: 162 KNALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
+ + + V+I +N+ +P + TIG Y + ++L
Sbjct: 101 EKEISQIIKNTTNVDIDKQNILIP-EIKTIGLYNIEIKL 138
>RL9_THETN (Q8R6M7) 50S ribosomal protein L9
Length = 147
Score = 47.4 bits (111), Expect = 3e-05
Identities = 26/77 (33%), Positives = 45/77 (57%), Gaps = 3/77 (3%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL ++ +GKAGE V V+ G+ RN+L+PK LA+ + +L E++K E
Sbjct: 1 MKVILVKDVKNVGKAGEVVNVSDGYGRNYLIPKGLAIEATESNLKMLNEKKK---AEERK 57
Query: 103 KQEDVVVVTESKEDLMK 119
+Q+++ E + L K
Sbjct: 58 RQQELEQAKELAQKLSK 74
>RL9_THEMA (Q9WZW7) 50S ribosomal protein L9
Length = 149
Score = 47.0 bits (110), Expect = 4e-05
Identities = 36/162 (22%), Positives = 75/162 (46%), Gaps = 25/162 (15%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL ++ K+GK GE +V+ G+ RN+L+P+ A + + +++I + +E
Sbjct: 1 MKVILLRDVPKIGKKGEIKEVSDGYARNYLIPRGFAKEYTEGLERAIKHEKEIEKRKKER 60
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
++E+ E ++KE L++ V K KA E ++ V+
Sbjct: 61 EREE-------SEKILKE--------------LKKRTHVVKVKAGEGG----KIFGAVTA 95
Query: 163 NALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRLPRSI 204
+ E+++ + + L P+ +GEY + + LP +
Sbjct: 96 ATVAEEISKTTGLKLDKRWFKLDKPIKELGEYSLEVSLPGGV 137
>RL9_RHILT (Q9F9K6) 50S ribosomal protein L9
Length = 192
Score = 46.6 bits (109), Expect = 5e-05
Identities = 44/166 (26%), Positives = 72/166 (42%), Gaps = 30/166 (18%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMP--KLLAVPNIDKFAYLLTEQRKIYQPTE 100
++VIL I KLG+ GETVKV GF RN+L+P K L +K +
Sbjct: 1 MEVILLERISKLGQMGETVKVRDGFARNYLLPLGKALRANAANKTRF------------- 47
Query: 101 EVKQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPV 160
+ + + + E ++ A +LD ++ R S E +L V
Sbjct: 48 ---EAERATLEARNLERKSEAQKVADVLDGKSFIVVR-----------SAGETGQLYGSV 93
Query: 161 SKNALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRLPRSIPL 206
+ +V E+ NI +HL TP+ +IG ++V L+L + +
Sbjct: 94 AARDVV-EILGAEGFNIGRNQVHLNTPIKSIGLHKVELQLHAEVEI 138
>RL9_TREPA (O83099) 50S ribosomal protein L9
Length = 156
Score = 46.2 bits (108), Expect = 6e-05
Identities = 45/172 (26%), Positives = 76/172 (44%), Gaps = 45/172 (26%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
+++IL ++ LG+ G+ +VA G+FRN+L P+ LAVP+ ++F +QR
Sbjct: 1 MKIILNQDVKILGEEGDVKEVAAGYFRNYLYPRNLAVPH-NRFTVARFKQR--------- 50
Query: 103 KQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQIPVSK 162
Q+D+ E ++ L ++ D KAR + +P+ + +P
Sbjct: 51 -QQDI----EMRKSLKRQ-------------------DAANLKAR-LEAQPVVIAMPAGT 85
Query: 163 NALV------AEVARQL-CVNITAENLHLPTP---LSTIGEYEVPLRLPRSI 204
N + VA QL C+ E + P L +G Y V +RL I
Sbjct: 86 NGKLYGAVTSHTVAEQLACMGFEVERKRVEVPGLTLKCVGNYHVTIRLYEEI 137
>RL9_SYNEL (P25904) 50S ribosomal protein L9
Length = 152
Score = 46.2 bits (108), Expect = 6e-05
Identities = 18/40 (45%), Positives = 31/40 (77%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVP 80
+++QV+L ++KLG+ G+ V+VAPG+ RN+L P+ +A P
Sbjct: 3 KRVQVVLNETVNKLGRMGQVVEVAPGYARNYLFPRGIAEP 42
>RL9_SYNY3 (P42352) 50S ribosomal protein L9
Length = 152
Score = 43.9 bits (102), Expect = 3e-04
Identities = 26/68 (38%), Positives = 44/68 (64%), Gaps = 8/68 (11%)
Query: 41 RKLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPK---LLAVPNIDKFAYLLTEQRKIYQ 97
++++V+L I+KLG G+ V+VAPG+ RN+L+PK ++A P I + EQR++ +
Sbjct: 3 KRVKVVLNETINKLGFTGDLVEVAPGYARNYLIPKGLGVVATPGILR----QVEQRRL-K 57
Query: 98 PTEEVKQE 105
E +K E
Sbjct: 58 ELERLKAE 65
>RL9_CLOAB (Q97CX9) 50S ribosomal protein L9
Length = 147
Score = 43.5 bits (101), Expect = 4e-04
Identities = 41/162 (25%), Positives = 74/162 (45%), Gaps = 34/162 (20%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL +I +GK GE + V+ G+ RN L P+ LA EE
Sbjct: 1 MKVILLKDIKSVGKKGEVINVSDGYARNFLFPRKLA---------------------EEA 39
Query: 103 KQEDVVVVTESKE----DLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQI 158
++ V+ K+ ++E E A + ++ K + +L AKA E+ L
Sbjct: 40 NNSNMRVLNLKKDAERKQKLQETEEAQKLANELKGKVLKL----SAKAGENG----RLFG 91
Query: 159 PVSKNALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRL 200
++ + AE+ +Q V+I + ++ T + +G YE+ L+L
Sbjct: 92 AITSKDIAAEIKKQFNVDIDKKKVNSET-IRKLGNYEIELKL 132
>RL9_BORBU (O51139) 50S ribosomal protein L9
Length = 173
Score = 43.5 bits (101), Expect = 4e-04
Identities = 27/77 (35%), Positives = 45/77 (58%), Gaps = 1/77 (1%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRKIYQPTEEV 102
++VIL + LGK G+TV+V GF RN+L+PK AV + + ++R+ +E
Sbjct: 1 MKVILKEDFINLGKEGDTVEVRDGFARNYLLPKGFAVFSNKHNIEIFNQKRRSILKKQEA 60
Query: 103 KQEDVVVVTESKEDLMK 119
K++ + +SK DL+K
Sbjct: 61 KKQ-MANDLKSKLDLVK 76
>RL9_CHLPN (Q9Z6V3) 50S ribosomal protein L9
Length = 169
Score = 42.7 bits (99), Expect = 7e-04
Identities = 33/166 (19%), Positives = 72/166 (42%), Gaps = 31/166 (18%)
Query: 42 KLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPK---LLAVPNIDKFAYLLTEQRKIYQP 98
K Q++L ++D LG++G+ + PG+ RN+L+PK ++A + L EQR I
Sbjct: 2 KQQLLLLEDVDGLGRSGDLITARPGYVRNYLIPKKKAVIAGAGTLRLQAKLKEQRLIQAA 61
Query: 99 TEEVKQEDVVVVTESKEDLMKEYERAALILDKAKLVLRRLIDVKKAKARESKDEPLELQI 158
++ E + ++ +D++ E++ R D + +
Sbjct: 62 ADKADSER---IAQALKDIVLEFQ-----------------------VRVDPDNNMYGSV 95
Query: 159 PVSKNALVAEVARQLCVNITAENLHLPTPLSTIGEYEVPLRLPRSI 204
++ ++AE A++ + H + +G+ +PL+L +
Sbjct: 96 TIAD--IIAEAAKKNIFLVRKNFPHAHYAIKNLGKKNIPLKLKEEV 139
>RL9_RHIME (Q92QZ9) 50S ribosomal protein L9
Length = 191
Score = 40.8 bits (94), Expect = 0.003
Identities = 19/32 (59%), Positives = 24/32 (74%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMP 74
++VIL I KLG+ GETVKV GF RN+L+P
Sbjct: 1 MEVILLERIAKLGQMGETVKVRDGFARNYLLP 32
>RL9_MYCTU (P66315) 50S ribosomal protein L9
Length = 152
Score = 40.4 bits (93), Expect = 0.003
Identities = 17/37 (45%), Positives = 28/37 (74%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAV 79
+++ILT ++D LG G+TV+V G+ RN L+P+ LA+
Sbjct: 1 MKLILTADVDHLGSIGDTVEVKDGYGRNFLLPRGLAI 37
>RL9_MYCBO (P66316) 50S ribosomal protein L9
Length = 152
Score = 40.4 bits (93), Expect = 0.003
Identities = 17/37 (45%), Positives = 28/37 (74%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAV 79
+++ILT ++D LG G+TV+V G+ RN L+P+ LA+
Sbjct: 1 MKLILTADVDHLGSIGDTVEVKDGYGRNFLLPRGLAI 37
>RL9_CLOPE (Q8XH49) 50S ribosomal protein L9
Length = 148
Score = 40.4 bits (93), Expect = 0.003
Identities = 18/52 (34%), Positives = 31/52 (59%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAVPNIDKFAYLLTEQRK 94
++VIL ++ KLGK G+ + + G+ RN L PK LA D ++L +++
Sbjct: 1 MKVILLQDVKKLGKKGDVINASDGYARNFLFPKKLAQEATDNNLHILNNKKE 52
>RL9_CHLCV (Q821W9) 50S ribosomal protein L9
Length = 172
Score = 40.4 bits (93), Expect = 0.003
Identities = 23/84 (27%), Positives = 46/84 (54%), Gaps = 6/84 (7%)
Query: 42 KLQVILTTNIDKLGKAGETVKVAPGFFRNHLMPK---LLAVPNIDKFAYLLTEQRKIYQP 98
K Q++L ++D LG++G+ V PG+ RN+LMP+ ++A + L E+R +
Sbjct: 2 KQQLLLLEDVDGLGRSGDIVTARPGYVRNYLMPQKKAVIAGAGTLRLQAKLKEERLLRAA 61
Query: 99 TEEVKQEDVVVVTESKEDLMKEYE 122
+ + E + E+ D++ E++
Sbjct: 62 EDRAESEK---LAEALRDVILEFQ 82
>RL9_MYCLE (P46385) 50S ribosomal protein L9
Length = 152
Score = 39.7 bits (91), Expect = 0.006
Identities = 17/37 (45%), Positives = 27/37 (72%)
Query: 43 LQVILTTNIDKLGKAGETVKVAPGFFRNHLMPKLLAV 79
+++ILT ++D LG G+TV+V G+ RN L+P LA+
Sbjct: 1 MKLILTADVDHLGSVGDTVEVKDGYGRNFLLPHGLAI 37
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.318 0.137 0.378
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 24,758,327
Number of Sequences: 164201
Number of extensions: 1007231
Number of successful extensions: 3307
Number of sequences better than 10.0: 110
Number of HSP's better than 10.0 without gapping: 77
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 3211
Number of HSP's gapped (non-prelim): 126
length of query: 222
length of database: 59,974,054
effective HSP length: 106
effective length of query: 116
effective length of database: 42,568,748
effective search space: 4937974768
effective search space used: 4937974768
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC136286.9


