

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC136286.13 + phase: 0
(373 letters)
Database: sprot
164,201 sequences; 59,974,054 total letters
Searching..................................................done
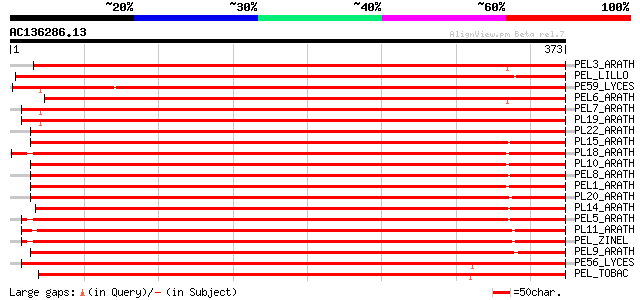
Score E
Sequences producing significant alignments: (bits) Value
PEL3_ARATH (Q9M9S2) Probable pectate lyase 3 precursor (EC 4.2.2... 501 e-141
PEL_LILLO (P40973) Pectate lyase precursor (EC 4.2.2.2) 483 e-136
PE59_LYCES (P15722) Probable pectate lyase P59 precursor (EC 4.2... 477 e-134
PEL6_ARATH (O64510) Probable pectate lyase 6 precursor (EC 4.2.2.2) 476 e-134
PEL7_ARATH (Q9SRH4) Probable pectate lyase 7 precursor (EC 4.2.2.2) 468 e-131
PL19_ARATH (Q9LFP5) Putative pectate lyase 19 precursor (EC 4.2.... 467 e-131
PL22_ARATH (Q93Z25) Probable pectate lyase 22 precursor (EC 4.2.... 451 e-126
PL15_ARATH (Q944R1) Probable pectate lyase 15 precursor (EC 4.2.... 451 e-126
PL18_ARATH (Q9C5M8) Probable pectate lyase 18 precursor (EC 4.2.... 449 e-126
PL10_ARATH (Q9LJ42) Probable pectate lyase 10 precursor (EC 4.2.... 446 e-125
PEL8_ARATH (Q9M8Z8) Probable pectate lyase 8 precursor (EC 4.2.2.2) 446 e-125
PEL1_ARATH (Q940Q1) Probable pectate lyase 1 precursor (EC 4.2.2... 445 e-124
PL20_ARATH (Q93WF1) Probable pectate lyase 20 precursor (EC 4.2.... 442 e-124
PL14_ARATH (Q9SVQ6) Putative pectate lyase 14 precursor (EC 4.2.... 440 e-123
PEL5_ARATH (Q9FXD8) Probable pectate lyase 5 precursor (EC 4.2.2.2) 440 e-123
PL11_ARATH (Q9LTZ0) Putative pectate lyase 11 precursor (EC 4.2.... 434 e-121
PEL_ZINEL (O24554) Pectate lyase precursor (EC 4.2.2.2) (ZePel) 434 e-121
PEL9_ARATH (Q9LRM5) Putative pectate lyase 9 precursor (EC 4.2.2.2) 415 e-115
PE56_LYCES (P15721) Probable pectate lyase P56 precursor (EC 4.2... 413 e-115
PEL_TOBAC (P40972) Pectate lyase precursor (EC 4.2.2.2) 410 e-114
>PEL3_ARATH (Q9M9S2) Probable pectate lyase 3 precursor (EC 4.2.2.2)
(Pectate lyase A2)
Length = 459
Score = 501 bits (1290), Expect = e-141
Identities = 239/362 (66%), Positives = 284/362 (78%), Gaps = 5/362 (1%)
Query: 17 NSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNP 76
+ C+A NPID CWRCD NWANNRKKLADCV GFGR+TTGGKDGPIYVV D SD+DL+NP
Sbjct: 98 SGKCLAYNPIDNCWRCDRNWANNRKKLADCVLGFGRRTTGGKDGPIYVVKDASDNDLINP 157
Query: 77 RPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIE 136
+PGTLRHAVTR+GPLWIIFARSM I+L QEL++ +KTIDGRGA V I GAG+T+QF+
Sbjct: 158 KPGTLRHAVTRDGPLWIIFARSMIIKLQQELMITSDKTIDGRGARVYIMEGAGLTLQFVN 217
Query: 137 NVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGL 196
NVIIH I + I+ G+GGL+RDSE H GLRT SDGDGIS+FG+++IWIDHVSM C DG+
Sbjct: 218 NVIIHNIYVKHIVPGNGGLIRDSEAHIGLRTKSDGDGISLFGATNIWIDHVSMTRCADGM 277
Query: 197 IDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRF 256
IDAI GSTA+TISNSHFTDH EVMLFGA D + D+KMQITV FN FGK+L QRMPRCR+
Sbjct: 278 IDAIDGSTAVTISNSHFTDHQEVMLFGARDEHVIDKKMQITVAFNHFGKRLEQRMPRCRY 337
Query: 257 GFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQW 316
G IHV+NN Y WEMYAIGG M+PTIIS+GN+FIAP N AK+ITKR P EWK+W W
Sbjct: 338 GTIHVVNNDYTHWEMYAIGGNMNPTIISQGNRFIAPPNEEAKQITKREYTPYGEWKSWNW 397
Query: 317 RSINDLYLNGAFFRQSG-----AELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGK 371
+S D +LNGA+F QSG + P NK I+ KPG+ V +LT + L C++G+
Sbjct: 398 QSEGDYFLNGAYFVQSGKANAWSSKPKTPLPNKFTIRPKPGTMVRKLTMDAGVLGCKLGE 457
Query: 372 PC 373
C
Sbjct: 458 AC 459
>PEL_LILLO (P40973) Pectate lyase precursor (EC 4.2.2.2)
Length = 434
Score = 483 bits (1244), Expect = e-136
Identities = 230/369 (62%), Positives = 281/369 (75%), Gaps = 1/369 (0%)
Query: 5 GINGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYV 64
G + RRNL C+ATNPIDRCWRC NW+ NRK L CV+GFGRKTTGG G IYV
Sbjct: 67 GNSTRRNLRTNKLGQCLATNPIDRCWRCKKNWSANRKDLVKCVKGFGRKTTGGAAGEIYV 126
Query: 65 VTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTI 124
VTDPSD L +P+ GTLR V ++ PLWIIF +SM IRL QELI+ +KTIDGRGA+V I
Sbjct: 127 VTDPSDDSLTDPKFGTLRWGVIQDRPLWIIFGKSMVIRLKQELIINNDKTIDGRGANVQI 186
Query: 125 ANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWI 184
A GA +T+QF+ NVIIHGI I+DI G GGL+RDSE H G+RT SDGDGISI GSS+IWI
Sbjct: 187 AGGAQLTVQFVHNVIIHGIHIHDIKPGEGGLIRDSEKHSGIRTRSDGDGISIIGSSNIWI 246
Query: 185 DHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFG 244
DHVS+ C DGLID I+GSTAITISN H T+H++VML GASDTY D+ MQ+TV FN FG
Sbjct: 247 DHVSLARCSDGLIDVILGSTAITISNCHLTEHDDVMLLGASDTYTQDEIMQVTVAFNHFG 306
Query: 245 KKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRT 304
+ L+QRMPRCR+GF+HV+NN Y W MYA+GG+ HPTIIS+GN++IAP+ AKE+TKR
Sbjct: 307 RGLVQRMPRCRYGFVHVVNNDYTHWIMYAVGGSQHPTIISQGNRYIAPHIEAAKEVTKRD 366
Query: 305 LVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRS 364
AEW W W+S DL+++GAFF +SG N+ +S KD+IKAKPG++V RLTR+S +
Sbjct: 367 YAEPAEWSKWTWKSQGDLFVSGAFFVESGGPFENK-YSKKDLIKAKPGTFVQRLTRFSGA 425
Query: 365 LRCRVGKPC 373
L C+ C
Sbjct: 426 LNCKENMEC 434
>PE59_LYCES (P15722) Probable pectate lyase P59 precursor (EC
4.2.2.2)
Length = 449
Score = 477 bits (1227), Expect = e-134
Identities = 231/376 (61%), Positives = 280/376 (74%), Gaps = 6/376 (1%)
Query: 3 IGGINGRRNLVGLGNSS----CMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGK 58
+ G + GLG CM TNPID+CWRCDPNWA+NRKKLADC GFG K GGK
Sbjct: 75 VKGTHNNSTRRGLGTKKYTGPCMVTNPIDKCWRCDPNWADNRKKLADCAMGFGSKAIGGK 134
Query: 59 DGPIYVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGR 118
DG YVVTD SD D +P+PGTLRHAV + PLWIIF R M IRL+QE+IM +KTID R
Sbjct: 135 DGEFYVVTDNSD-DYNDPKPGTLRHAVIQKEPLWIIFKRGMNIRLHQEMIMQSDKTIDAR 193
Query: 119 GADVTIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFG 178
G +V I GAGIT+Q+I+NVIIHG+ I+DI+ G+GG+VRD+ DH G+RT SDGDGISIFG
Sbjct: 194 GVNVHITKGAGITLQYIKNVIIHGLHIHDIVEGNGGMVRDAVDHIGIRTKSDGDGISIFG 253
Query: 179 SSHIWIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITV 238
+S+IWIDHVSM+ C DGLIDA+ GST ITISN HFTDHNEVMLFGASD+ DQ MQIT+
Sbjct: 254 ASYIWIDHVSMQRCYDGLIDAVEGSTGITISNGHFTDHNEVMLFGASDSSSIDQVMQITL 313
Query: 239 VFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAK 298
FN FGK+LIQRMPRCR+G+IHV+NN Y W MYAIGG+MHPTII +GN+FIAP + K
Sbjct: 314 AFNHFGKRLIQRMPRCRWGYIHVVNNDYTHWNMYAIGGSMHPTIIHQGNRFIAPPDIFKK 373
Query: 299 EITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSG-AELINRPFSNKDMIKAKPGSYVGR 357
++TKR PE+ W W WRS +L++NGA+F +SG E ++ D I A P V
Sbjct: 374 QVTKREYNPESVWMQWTWRSEGNLFMNGAYFTESGDPEWSSKHKDLYDGISAAPAEDVTW 433
Query: 358 LTRYSRSLRCRVGKPC 373
+TR++ L C+ GKPC
Sbjct: 434 MTRFAGVLGCKPGKPC 449
>PEL6_ARATH (O64510) Probable pectate lyase 6 precursor (EC 4.2.2.2)
Length = 455
Score = 476 bits (1225), Expect = e-134
Identities = 227/355 (63%), Positives = 277/355 (77%), Gaps = 5/355 (1%)
Query: 24 NPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRPGTLRH 83
N ID+CWR D NW NRKKLADCV GFGRKTTGGK+GPIYVVTDPSD+DL+ P+PGT+RH
Sbjct: 101 NAIDKCWRGDKNWDKNRKKLADCVLGFGRKTTGGKNGPIYVVTDPSDNDLLKPKPGTIRH 160
Query: 84 AVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVIIHGI 143
AVTR+ PLWIIFARSM I+L QELI+ +KTIDGRGA + I GAG+T+QF+ NVIIH I
Sbjct: 161 AVTRDRPLWIIFARSMIIKLQQELIITNDKTIDGRGAKIYITGGAGLTLQFVRNVIIHNI 220
Query: 144 KIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDAIMGS 203
I I G+GGL+ DSE H+GLRT+SDGDGI+IFG++++WIDHVSM +C DG+IDAIMGS
Sbjct: 221 HIKQIKRGAGGLIIDSEQHFGLRTVSDGDGINIFGATNVWIDHVSMTDCSDGMIDAIMGS 280
Query: 204 TAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFIHVLN 263
TAITISNSHFTDH+EVMLFG ++ D+KMQITV FN FGK+L QRMPR RFG +HV+N
Sbjct: 281 TAITISNSHFTDHDEVMLFGGTNKDVIDKKMQITVAFNHFGKRLKQRMPRVRFGLVHVVN 340
Query: 264 NFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWRSINDLY 323
N Y WEMYAIGG M+PTIIS+GN+FIAP +K++TKR P EWK+W W+S D +
Sbjct: 341 NDYTHWEMYAIGGNMNPTIISQGNRFIAPPIEDSKQVTKREYTPYPEWKSWNWQSEKDYF 400
Query: 324 LNGAFFRQSG-----AELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
LNGA+F QSG + P K I+ +PG+ V RLT+ + +L C+ GK C
Sbjct: 401 LNGAYFVQSGKANAWSATPKNPIPRKFAIRPQPGTKVRRLTKDAGTLGCKPGKSC 455
>PEL7_ARATH (Q9SRH4) Probable pectate lyase 7 precursor (EC 4.2.2.2)
Length = 475
Score = 468 bits (1204), Expect = e-131
Identities = 220/377 (58%), Positives = 278/377 (73%), Gaps = 12/377 (3%)
Query: 9 RRNLVGLGNSS-----------CMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGG 57
RR+L G G C A+NPID+CWRC P+WA RKKL CV+GFG +TTGG
Sbjct: 99 RRSLTGRGKGKGKGKWSKLTGPCTASNPIDKCWRCQPDWARRRKKLVHCVRGFGYRTTGG 158
Query: 58 KDGPIYVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDG 117
K G IYVVT P D D+VNPRPGTLRHAV + PLWI+F M IRL+QEL++ +KTID
Sbjct: 159 KRGRIYVVTSPRDDDMVNPRPGTLRHAVIQKEPLWIVFKHDMSIRLSQELMITSDKTIDA 218
Query: 118 RGADVTIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIF 177
RGA+V IA GAGIT+Q++ N+IIHG+ ++ I+ SGGL+RDS +H+G R +DGDGISIF
Sbjct: 219 RGANVHIAYGAGITMQYVHNIIIHGLHVHHIVKSSGGLIRDSINHFGHRGEADGDGISIF 278
Query: 178 GSSHIWIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQIT 237
G+++IW+DH+SM C+DGLIDAIMGSTAITISNSHFT HN+VML GA + D+KMQ+T
Sbjct: 279 GATNIWLDHISMSKCQDGLIDAIMGSTAITISNSHFTHHNDVMLLGAQNNNMDDKKMQVT 338
Query: 238 VVFNRFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIA-PNNGH 296
V +N FGK L+QRMPR R+GF+HV+NN Y WE+YAIGG+ PTI+S GN+FIA P+ H
Sbjct: 339 VAYNHFGKGLVQRMPRVRWGFVHVVNNDYTHWELYAIGGSQGPTILSHGNRFIAPPHKQH 398
Query: 297 AKEITKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVG 356
+E+TKR E+EWK W WRS D+++N A+FRQSG S + MIK K G V
Sbjct: 399 YREVTKRDYASESEWKNWNWRSEKDVFMNNAYFRQSGNPHFKCSHSRQQMIKPKNGMAVS 458
Query: 357 RLTRYSRSLRCRVGKPC 373
+LT+Y+ +L CRVGK C
Sbjct: 459 KLTKYAGALDCRVGKAC 475
>PL19_ARATH (Q9LFP5) Putative pectate lyase 19 precursor (EC
4.2.2.2)
Length = 472
Score = 467 bits (1201), Expect = e-131
Identities = 219/373 (58%), Positives = 279/373 (74%), Gaps = 8/373 (2%)
Query: 9 RRNLVGLGNSS-------CMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGP 61
RR+L G G C A+NPID+CWRC +WA RKKL CV+GFG +TTGGK G
Sbjct: 100 RRSLRGKGKGKWSKLKGPCTASNPIDKCWRCRSDWAKRRKKLTRCVRGFGHRTTGGKRGR 159
Query: 62 IYVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGAD 121
IYVVT D D+VNP+PGTLRHAV + PLWIIF M IRLNQEL++ +KTID RGA+
Sbjct: 160 IYVVTSNLDEDMVNPKPGTLRHAVIQKEPLWIIFKNDMSIRLNQELLINSHKTIDARGAN 219
Query: 122 VTIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSH 181
V +A+GAGIT+QF++NVIIHG+ I+ I SGG++RDS DH+G+RT +DGDG+SI+GSS+
Sbjct: 220 VHVAHGAGITMQFVKNVIIHGLHIHHISESSGGMIRDSVDHFGMRTRADGDGLSIYGSSN 279
Query: 182 IWIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFN 241
IW+DH+SM C+DGLIDAI+GST ITISNSHFT HN+VML GA +T + D+ MQ+TV +N
Sbjct: 280 IWLDHISMSKCQDGLIDAIVGSTGITISNSHFTHHNDVMLLGAQNTNEADKHMQVTVAYN 339
Query: 242 RFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIA-PNNGHAKEI 300
FGK L+QRMPR R+GF+HV+NN Y WE+YAIGG+ PTI+S GN+FIA P+ H +E+
Sbjct: 340 HFGKGLVQRMPRIRWGFVHVVNNDYTHWELYAIGGSQGPTILSHGNRFIAPPHKPHYREV 399
Query: 301 TKRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTR 360
TKR E EWK W WRS D+++NGA+FRQSG + + MIK K G V +LT+
Sbjct: 400 TKRDYASEDEWKHWNWRSDKDVFMNGAYFRQSGNPQYKCAHTRQQMIKPKNGLAVSKLTK 459
Query: 361 YSRSLRCRVGKPC 373
Y+ +L CRVG+ C
Sbjct: 460 YAGALDCRVGRRC 472
>PL22_ARATH (Q93Z25) Probable pectate lyase 22 precursor (EC
4.2.2.2)
Length = 432
Score = 451 bits (1159), Expect = e-126
Identities = 210/360 (58%), Positives = 263/360 (72%), Gaps = 1/360 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG SC + NPID CWRCD +W NRK+LADC GFG+ GG+DG IYVVTDP + D V
Sbjct: 73 LGFFSCGSGNPIDDCWRCDKDWEKNRKRLADCGIGFGKNAIGGRDGEIYVVTDPGNDDPV 132
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NPRPGTLR+AV ++ PLWIIF R M I+L +ELIM KT+DGRGA V I+ G ITIQ+
Sbjct: 133 NPRPGTLRYAVIQDEPLWIIFKRDMTIQLKEELIMNSFKTLDGRGASVHISGGPCITIQY 192
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
+ N+IIHG+ I+D G VRDS +HYG RT+SDGDG+SIFG SH+W+DH S+ NC D
Sbjct: 193 VTNIIIHGLHIHDCKQGGNTYVRDSPEHYGYRTVSDGDGVSIFGGSHVWVDHCSLSNCND 252
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GLIDAI GSTAITISN++ T HN+VML G SDTY+ D+ MQ+T+ FN FG+ L+QRMPRC
Sbjct: 253 GLIDAIRGSTAITISNNYLTHHNKVMLLGHSDTYEQDKNMQVTIAFNHFGEGLVQRMPRC 312
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y WEMYAIGG+ +PTI S+GN+F+AP++ +KE+TK PE EW+ W
Sbjct: 313 RHGYFHVVNNDYTHWEMYAIGGSANPTINSQGNRFLAPDDSSSKEVTKHEDAPEDEWRNW 372
Query: 315 QWRSINDLYLNGAFFRQSGA-ELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL LNGAFF SGA + +S + A+P S+VG +T S +L C+ G C
Sbjct: 373 NWRSEGDLLLNGAFFTYSGAGPAKSSSYSKASSLAARPSSHVGEITIASGALSCKRGSHC 432
>PL15_ARATH (Q944R1) Probable pectate lyase 15 precursor (EC
4.2.2.2) (Pectate lyase A11)
Length = 470
Score = 451 bits (1159), Expect = e-126
Identities = 213/359 (59%), Positives = 259/359 (71%), Gaps = 1/359 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG SC NPID CWRCD NW NRK+LADC GFGR GG+DG Y+VTDP+D D+V
Sbjct: 113 LGFFSCGTGNPIDDCWRCDRNWHKNRKRLADCGIGFGRNAIGGRDGRFYIVTDPTDEDVV 172
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NP+PGTLRHAV + PLWI+F R M I L QELIM KTID RG++V IANGA ITIQF
Sbjct: 173 NPKPGTLRHAVIQEEPLWIVFKRDMVIELKQELIMNSFKTIDARGSNVHIANGACITIQF 232
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
I NVIIHG+ I+D +VR S H+G RT++DGD +SIFGSSHIWIDH S+ +C D
Sbjct: 233 ITNVIIHGLHIHDCKPTGNAMVRSSPSHFGWRTMADGDAVSIFGSSHIWIDHNSLSHCAD 292
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GL+DA+MGSTAIT+SN+HFT HNEVML G SD+Y D+ MQ+T+ +N FG+ L+QRMPRC
Sbjct: 293 GLVDAVMGSTAITVSNNHFTHHNEVMLLGHSDSYTKDKLMQVTIAYNHFGEGLVQRMPRC 352
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y WEMYAIGG+ PTI S+GN++ AP + AKE+TKR +EWK W
Sbjct: 353 RHGYFHVVNNDYTHWEMYAIGGSAEPTINSQGNRYAAPMDRFAKEVTKRVETDASEWKKW 412
Query: 315 QWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL LNGAFFR SGA + + + AKP S V +T + +L CR G+PC
Sbjct: 413 NWRSEGDLLLNGAFFRPSGAG-ASASYGRASSLAAKPSSMVDTITSTAGALGCRKGRPC 470
>PL18_ARATH (Q9C5M8) Probable pectate lyase 18 precursor (EC
4.2.2.2) (Pectate lyase A10)
Length = 408
Score = 449 bits (1155), Expect = e-126
Identities = 214/372 (57%), Positives = 263/372 (70%), Gaps = 4/372 (1%)
Query: 2 IIGGINGRRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGP 61
I + GRR L G SC NPID CWRCDP+W +R++LADC GFG+ GG+DG
Sbjct: 41 INASVAGRRKL---GYLSCTTGNPIDDCWRCDPHWEQHRQRLADCAIGFGKNAIGGRDGR 97
Query: 62 IYVVTDPSDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGAD 121
IYVVTD + + V+P+PGTLRHAV ++ PLWIIF R M I+L +ELIM KTIDGRGA
Sbjct: 98 IYVVTDSGNDNPVSPKPGTLRHAVVQDEPLWIIFQRDMTIQLKEELIMNSFKTIDGRGAS 157
Query: 122 VTIANGAGITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSH 181
V I+ G ITIQ++ N+IIHGI I+D G +VR S H+G RTISDGDG+SIFG SH
Sbjct: 158 VHISGGPCITIQYVTNIIIHGIHIHDCKQGGNAMVRSSPRHFGWRTISDGDGVSIFGGSH 217
Query: 182 IWIDHVSMRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFN 241
+W+DH S NC DGLIDAIMGSTAIT+SN+H T H++VML G SDTY D+ MQ+T+ FN
Sbjct: 218 VWVDHCSFSNCEDGLIDAIMGSTAITLSNNHMTHHDKVMLLGHSDTYSRDKNMQVTIAFN 277
Query: 242 RFGKKLIQRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEIT 301
FG+ L+QRMPRCR G+ HV+NN Y WEMYAIGG+ +PTI S+GN+F+APN +KE+T
Sbjct: 278 HFGEGLVQRMPRCRHGYFHVVNNDYTHWEMYAIGGSANPTINSQGNRFLAPNIRFSKEVT 337
Query: 302 KRTLVPEAEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRY 361
K PE+EWK W WRS DL LNGAFF SG + ++ + AKP S VG LT
Sbjct: 338 KHEDAPESEWKRWNWRSSGDLLLNGAFFTPSGG-AASSSYAKASSLGAKPSSLVGPLTST 396
Query: 362 SRSLRCRVGKPC 373
S +L CR G C
Sbjct: 397 SGALNCRKGSRC 408
>PL10_ARATH (Q9LJ42) Probable pectate lyase 10 precursor (EC
4.2.2.2)
Length = 440
Score = 446 bits (1148), Expect = e-125
Identities = 216/359 (60%), Positives = 257/359 (71%), Gaps = 1/359 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG SC NPID CWRCD NW RK+LA+C GFGR GG+DG YVVTDPSD D V
Sbjct: 83 LGFFSCATGNPIDDCWRCDRNWHLRRKRLANCAIGFGRNAIGGRDGRYYVVTDPSDHDAV 142
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NPRPGTLRHAV ++ PLWI+F R M I L QELIM KTIDGRG +V IA GA ITIQ+
Sbjct: 143 NPRPGTLRHAVIQDRPLWIVFKRDMVITLTQELIMNSFKTIDGRGVNVAIAGGACITIQY 202
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
+ N+IIHGI ++D +VR S HYG RT++DGD ISIFGSSHIWIDH S+ NC D
Sbjct: 203 VTNIIIHGINVHDCRRTGNAMVRSSPSHYGWRTMADGDAISIFGSSHIWIDHNSLSNCAD 262
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GLIDAIMGSTAITISN++ T HNEVML G SD+Y D+ MQ+T+ +N FG+ LIQRMPRC
Sbjct: 263 GLIDAIMGSTAITISNNYMTHHNEVMLMGHSDSYTRDKLMQVTIAYNHFGEGLIQRMPRC 322
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y W MYAIGG+ +PTI S+GN+F+AP N AKE+TKR + EWK W
Sbjct: 323 RHGYFHVVNNDYTHWVMYAIGGSANPTINSQGNRFLAPGNPFAKEVTKRVGSWQGEWKQW 382
Query: 315 QWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL LNGA+F +SGA ++ + AKP S V LT S +L+CR+G C
Sbjct: 383 NWRSQGDLMLNGAYFTKSGA-AAPASYARASSLGAKPASVVSMLTYSSGALKCRIGMRC 440
>PEL8_ARATH (Q9M8Z8) Probable pectate lyase 8 precursor (EC 4.2.2.2)
Length = 416
Score = 446 bits (1148), Expect = e-125
Identities = 212/359 (59%), Positives = 256/359 (71%), Gaps = 1/359 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG SC NPID CWRCD W RK+LADC GFGR GG+DG YVVTDP D D V
Sbjct: 59 LGYFSCATGNPIDDCWRCDRKWQLRRKRLADCSIGFGRNAIGGRDGRFYVVTDPGDDDPV 118
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NP PGTLRHAV ++ PLWIIF R M I L QELIM KTIDGRG +V IANGA +TIQ+
Sbjct: 119 NPIPGTLRHAVIQDEPLWIIFKRDMVITLKQELIMNSFKTIDGRGVNVHIANGACLTIQY 178
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
+ N+I+HGI ++D +VR S HYG R+++DGD ISIFGSSHIWIDH S+ NC D
Sbjct: 179 VTNIIVHGIHVHDCKPTGNAMVRSSPSHYGFRSMADGDAISIFGSSHIWIDHNSLSNCAD 238
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GL+DA+M STAIT+SN+ FT HNEVML G SD+Y D+ MQ+T+ +N FG+ LIQRMPRC
Sbjct: 239 GLVDAVMSSTAITVSNNFFTHHNEVMLLGHSDSYTRDKVMQVTIAYNHFGEGLIQRMPRC 298
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y WEMYAIGG+ PTI S+GN+F+AP N AKE+TKR E++WK W
Sbjct: 299 RHGYFHVVNNDYTHWEMYAIGGSAGPTINSQGNRFLAPVNPFAKEVTKREYTGESKWKHW 358
Query: 315 QWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL+LNGAFF +SGA ++ + AK S VG +T YS +L CR G+ C
Sbjct: 359 NWRSEGDLFLNGAFFTRSGAG-AGANYARASSLSAKSSSLVGTMTSYSGALNCRAGRRC 416
>PEL1_ARATH (Q940Q1) Probable pectate lyase 1 precursor (EC 4.2.2.2)
(Pectate lyase A1)
Length = 431
Score = 445 bits (1144), Expect = e-124
Identities = 211/359 (58%), Positives = 258/359 (71%), Gaps = 1/359 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG +C NPID CWRCDPNW NRK+LADC GFGR GG+DG YVVTDP D + V
Sbjct: 72 LGYFTCGTGNPIDDCWRCDPNWHKNRKRLADCGIGFGRNAIGGRDGRFYVVTDPRDDNPV 131
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NPRPGTLRHAV ++ PLWI+F R M I+L QELI+ KTIDGRGA+V IANG ITIQF
Sbjct: 132 NPRPGTLRHAVIQDRPLWIVFKRDMVIQLKQELIVNSFKTIDGRGANVHIANGGCITIQF 191
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
+ NVI+HG+ I+D +VR SE H+G RT++DGD ISIFGSSH+WIDH S+ +C D
Sbjct: 192 VTNVIVHGLHIHDCKPTGNAMVRSSETHFGWRTMADGDAISIFGSSHVWIDHNSLSHCAD 251
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GL+DA+MGSTAITISN+H T HNEVML G SD+Y D+ MQ+T+ +N FG LIQRMPRC
Sbjct: 252 GLVDAVMGSTAITISNNHLTHHNEVMLLGHSDSYMRDKAMQVTIAYNHFGVGLIQRMPRC 311
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y WEMYAIGG+ +PTI S+GN++ AP N AKE+TKR P + WK W
Sbjct: 312 RHGYFHVVNNDYTHWEMYAIGGSANPTINSQGNRYAAPKNPFAKEVTKRVDTPASHWKGW 371
Query: 315 QWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL NGA+F SGA + ++ + AK S VG +T + +L CR G+ C
Sbjct: 372 NWRSEGDLLQNGAYFTSSGA-AASGSYARASSLSAKSSSLVGHITSDAGALPCRRGRQC 429
>PL20_ARATH (Q93WF1) Probable pectate lyase 20 precursor (EC
4.2.2.2)
Length = 417
Score = 442 bits (1137), Expect = e-124
Identities = 211/359 (58%), Positives = 258/359 (71%), Gaps = 1/359 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG SC NPID CWRCD W + RK LA+C GFGR GG+DG YVV+DP+D + V
Sbjct: 60 LGYFSCSTGNPIDDCWRCDRRWQSRRKHLANCAIGFGRNAIGGRDGRYYVVSDPNDDNPV 119
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NP+PGTLRHAV + PLWI+F R M I L +ELIM KTIDGRG +V IANGA ITIQF
Sbjct: 120 NPKPGTLRHAVIQEEPLWIVFKRDMVITLKEELIMNSFKTIDGRGVNVHIANGACITIQF 179
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
+ N+IIHGI I+D +VR S HYG RT++DGDGISIFGSSHIWIDH S+ NC D
Sbjct: 180 VTNIIIHGIHIHDCRPTGNAMVRSSPSHYGWRTMADGDGISIFGSSHIWIDHNSLSNCAD 239
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GLIDA+M STAITISN++FT HNEVML G SDTY D+ MQ+T+ +N FG+ LIQRMPRC
Sbjct: 240 GLIDAVMASTAITISNNYFTHHNEVMLLGHSDTYTRDKVMQVTIAYNHFGEGLIQRMPRC 299
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y WEMYAIGG+ PTI S+GN+++AP N AKE+TKR + +W+ W
Sbjct: 300 RHGYFHVVNNDYTHWEMYAIGGSASPTINSQGNRYLAPRNRFAKEVTKRDYAGQWQWRHW 359
Query: 315 QWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL+LNGAFF +SG+ L ++ + AK S VG +T + +L CR G+ C
Sbjct: 360 NWRSEGDLFLNGAFFTRSGSGL-GASYARASSLAAKSSSLVGVITYNAGALNCRGGRRC 417
>PL14_ARATH (Q9SVQ6) Putative pectate lyase 14 precursor (EC
4.2.2.2)
Length = 418
Score = 440 bits (1132), Expect = e-123
Identities = 208/356 (58%), Positives = 256/356 (71%), Gaps = 1/356 (0%)
Query: 18 SSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPR 77
SSC NPID CWRCD W RK+LADC GFGR GG+DG Y+VTDPSD D V P+
Sbjct: 64 SSCSTGNPIDDCWRCDKKWHRRRKRLADCAIGFGRNAVGGRDGRYYIVTDPSDHDPVTPK 123
Query: 78 PGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIEN 137
PGTLR+AV ++ PLWI+F R M I L+QELIM KTIDGRG +V IA GA +T+Q++ N
Sbjct: 124 PGTLRYAVIQDEPLWIVFKRDMVITLSQELIMNSFKTIDGRGVNVHIAGGACLTVQYVTN 183
Query: 138 VIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLI 197
+IIHGI I+D +VR SE HYG RT++DGDGISIFGSSHIWIDH S+ +C DGLI
Sbjct: 184 IIIHGINIHDCKRTGNAMVRSSESHYGWRTMADGDGISIFGSSHIWIDHNSLSSCADGLI 243
Query: 198 DAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFG 257
DAIMGSTAITISN++ T HNE +L G +D+Y D+ MQ+T+ +N FG+ LIQRMPRCR G
Sbjct: 244 DAIMGSTAITISNNYLTHHNEAILLGHTDSYTRDKMMQVTIAYNHFGEGLIQRMPRCRHG 303
Query: 258 FIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTWQWR 317
+ HV+NN Y WEMYAIGG+ +PTI S+GN+F+AP N AKE+TKR + EW W WR
Sbjct: 304 YFHVVNNDYTHWEMYAIGGSANPTINSQGNRFLAPGNRFAKEVTKRVGAGKGEWNNWNWR 363
Query: 318 SINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
S DL LNGA+F SGA + ++ + AK S VG LT S +L+CR+G C
Sbjct: 364 SQGDLMLNGAYFTSSGAG-ASANYARASSLAAKSSSLVGMLTSSSGALKCRIGTLC 418
>PEL5_ARATH (Q9FXD8) Probable pectate lyase 5 precursor (EC 4.2.2.2)
Length = 408
Score = 440 bits (1132), Expect = e-123
Identities = 209/365 (57%), Positives = 259/365 (70%), Gaps = 4/365 (1%)
Query: 9 RRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDP 68
RRNL G SC NPID CWRCDP W NR++LADC GFG+ GG+DG IYVVTD
Sbjct: 48 RRNL---GVLSCGTGNPIDDCWRCDPKWEKNRQRLADCAIGFGKHAIGGRDGKIYVVTDS 104
Query: 69 SDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGA 128
SD D+VNP+PGTLRHAV ++ PLWIIFAR M I+L +ELIM KTIDGRGA V IA GA
Sbjct: 105 SDKDVVNPKPGTLRHAVIQDEPLWIIFARDMVIKLKEELIMNSFKTIDGRGASVHIAGGA 164
Query: 129 GITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVS 188
IT+Q++ N+IIHG+ I+D VRDS HYG RT SDGD +SIFG SH+W+DH S
Sbjct: 165 CITVQYVTNIIIHGVNIHDCKRKGNAYVRDSPSHYGWRTASDGDAVSIFGGSHVWVDHCS 224
Query: 189 MRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLI 248
+ NC DGLIDAI GSTAITISN++ + HN+VML G SD+Y D+ MQ+T+ FN FG+ L+
Sbjct: 225 LSNCADGLIDAIHGSTAITISNNYLSHHNKVMLLGHSDSYTRDKNMQVTIAFNHFGEGLV 284
Query: 249 QRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPE 308
QRMPRCR G+ HV+NN Y W+MYAIGG+ PTI S+GN+F+APN+ KE+TK P
Sbjct: 285 QRMPRCRHGYFHVVNNDYTHWQMYAIGGSAAPTINSQGNRFLAPNDHVFKEVTKYEDAPR 344
Query: 309 AEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCR 368
++WK W WRS DL+LNGAFF SG + ++ + A+P S V +T + +L CR
Sbjct: 345 SKWKKWNWRSEGDLFLNGAFFTPSGGG-ASSSYAKASSLSARPSSLVASVTSNAGALFCR 403
Query: 369 VGKPC 373
G C
Sbjct: 404 KGSRC 408
>PL11_ARATH (Q9LTZ0) Putative pectate lyase 11 precursor (EC
4.2.2.2)
Length = 409
Score = 434 bits (1117), Expect = e-121
Identities = 204/365 (55%), Positives = 259/365 (70%), Gaps = 4/365 (1%)
Query: 9 RRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDP 68
RR+L L SC NPID CWRCDPNW NR++LADC GFG+ GG+ G IYVVTDP
Sbjct: 49 RRSLAYL---SCRTGNPIDDCWRCDPNWETNRQRLADCAIGFGKNAIGGRKGRIYVVTDP 105
Query: 69 SDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGA 128
++ D VNPRPGTLR+AVT+ PLWIIF R M IRL +ELI+ KTIDGRG+ V I +G
Sbjct: 106 ANDDPVNPRPGTLRYAVTQEEPLWIIFKRDMVIRLKKELIITSFKTIDGRGSSVHITDGP 165
Query: 129 GITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVS 188
+ I + N+IIHGI I+D GSGG+++D H G SDGD ++IFG H+WIDH S
Sbjct: 166 CLKIHYATNIIIHGINIHDCKPGSGGMIKDGPHHTGWWMQSDGDAVAIFGGKHVWIDHCS 225
Query: 189 MRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLI 248
+ NC DGLIDAI GSTAITISN+H T H++VML G SD+Y D+ MQ+T+ FN FG+ L+
Sbjct: 226 LSNCDDGLIDAIHGSTAITISNNHMTHHDKVMLLGHSDSYTQDKNMQVTIAFNHFGEGLV 285
Query: 249 QRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPE 308
QRMPRCR G+ HV+NN Y WEMYAIGG+ PTI S+GN+F+APN KE+TK PE
Sbjct: 286 QRMPRCRHGYFHVVNNDYTHWEMYAIGGSASPTIYSQGNRFLAPNTRFNKEVTKHEDAPE 345
Query: 309 AEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCR 368
++W+ W WRS D+ LNGA+FR+SGAE + ++ + A+P S VG +T + +L CR
Sbjct: 346 SKWRDWNWRSEGDMLLNGAYFRESGAEAPS-TYARASSLSARPSSLVGSITTTAGTLSCR 404
Query: 369 VGKPC 373
G+ C
Sbjct: 405 RGRRC 409
>PEL_ZINEL (O24554) Pectate lyase precursor (EC 4.2.2.2) (ZePel)
Length = 401
Score = 434 bits (1116), Expect = e-121
Identities = 209/365 (57%), Positives = 256/365 (69%), Gaps = 4/365 (1%)
Query: 9 RRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDP 68
RRNL G SC NPID CWRCDPNWANNR++LADC GFG+ GG++G IYVVTDP
Sbjct: 41 RRNL---GYLSCGTGNPIDDCWRCDPNWANNRQRLADCAIGFGKNAMGGRNGRIYVVTDP 97
Query: 69 SDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGA 128
+ D VNP PGTLR+AV ++ PLWIIF R M I+L QEL+M +KTIDGRG +V I NG
Sbjct: 98 GNDDPVNPVPGTLRYAVIQDEPLWIIFKRDMVIQLRQELVMNSHKTIDGRGVNVHIGNGP 157
Query: 129 GITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVS 188
ITI + N+IIHGI I+D G +R+S H G T SDGDGISIF S IWIDH S
Sbjct: 158 CITIHYASNIIIHGIHIHDCKQAGNGNIRNSPHHSGWWTQSDGDGISIFASKDIWIDHNS 217
Query: 189 MRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLI 248
+ NC DGLIDAI GSTAITISN++ T H++VML G SD+Y D+ MQ+T+ FN FG+ L+
Sbjct: 218 LSNCHDGLIDAIHGSTAITISNNYMTHHDKVMLLGHSDSYTQDKNMQVTIAFNHFGEGLV 277
Query: 249 QRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPE 308
QRMPRCR G+ HV+NN Y WEMYAIGG+ PTI S+GN+F+APN KE+TK PE
Sbjct: 278 QRMPRCRHGYFHVVNNDYTHWEMYAIGGSASPTIYSQGNRFLAPNTRFDKEVTKHENAPE 337
Query: 309 AEWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCR 368
+EWK W WRS DL LNGA+FR+SG + F+ + +P + V +TR + +L CR
Sbjct: 338 SEWKNWNWRSEGDLMLNGAYFRESGGRAAS-SFARASSLSGRPSTLVASMTRSAGALVCR 396
Query: 369 VGKPC 373
G C
Sbjct: 397 KGSRC 401
>PEL9_ARATH (Q9LRM5) Putative pectate lyase 9 precursor (EC 4.2.2.2)
Length = 452
Score = 415 bits (1066), Expect = e-115
Identities = 209/359 (58%), Positives = 245/359 (68%), Gaps = 2/359 (0%)
Query: 15 LGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLV 74
LG SC N ID CWRCD NW NRK LADC GFG K GG++G YVVTD SD D+V
Sbjct: 95 LGFFSCGNGNLIDDCWRCDRNWNKNRKHLADCGMGFGSKAFGGRNGSYYVVTDHSDDDVV 154
Query: 75 NPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQF 134
NP+PGTLRHAV + PLWIIF R M I+L QELIM KTID RGA+V IANGA ITIQ
Sbjct: 155 NPKPGTLRHAVIQVEPLWIIFKRDMVIKLKQELIMNSFKTIDARGANVHIANGACITIQN 214
Query: 135 IENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRD 194
I NVI+HG+ I+D VR S G R +DGD I+IFGSSHIWIDH S+ NC D
Sbjct: 215 ITNVIVHGLHIHDCKRTGNVTVRSSPSQAGFRGTADGDAINIFGSSHIWIDHNSLSNCTD 274
Query: 195 GLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRC 254
GL+D + GSTAITISN+HFT H+EVML G +D+Y D+ MQ+TV +N FG+ LIQRMPRC
Sbjct: 275 GLVDVVNGSTAITISNNHFTHHDEVMLLGHNDSYTRDKMMQVTVAYNHFGEGLIQRMPRC 334
Query: 255 RFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPEAEWKTW 314
R G+ HV+NN Y W+MYAIGG+ +PTI S+GN+F AP N AKE+TKR EW W
Sbjct: 335 RHGYFHVVNNDYTHWKMYAIGGSANPTINSQGNRFAAPKNHSAKEVTKRLDTKGNEWMEW 394
Query: 315 QWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
WRS DL +NGAFF SG S + AKP S V +T + +L CR GKPC
Sbjct: 395 NWRSEKDLLVNGAFFTPSGEGASGD--SQTLSLPAKPASMVDAITASAGALSCRRGKPC 451
>PE56_LYCES (P15721) Probable pectate lyase P56 precursor (EC
4.2.2.2)
Length = 398
Score = 413 bits (1061), Expect = e-115
Identities = 201/367 (54%), Positives = 257/367 (69%), Gaps = 2/367 (0%)
Query: 9 RRNLVGLGNSSCMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDP 68
RR L CMA N ID+CWRCDP WA +R+K+ADC GFG GGK GP Y+VTD
Sbjct: 32 RRKLTKKYRGPCMAVNSIDKCWRCDPFWAEDRQKMADCALGFGINAMGGKYGPYYIVTDN 91
Query: 69 SDSDLVNPRPGTLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGA 128
SD D+V+P+PGTLR V + GPLWI FARSM+IRL +ELI++ NKTIDGRG V IANGA
Sbjct: 92 SDDDVVDPKPGTLRFGVIQKGPLWITFARSMRIRLTRELIVSSNKTIDGRGKYVHIANGA 151
Query: 129 GITIQFIENVIIHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVS 188
GI IQ NVII ++I++I+ +GGL+R+S+DH GLR +GD ISIF S IWIDH+S
Sbjct: 152 GIKIQSASNVIISNLRIHNIVPTAGGLLRESDDHLGLRGADEGDAISIFNSHDIWIDHIS 211
Query: 189 MRNCRDGLIDAIMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLI 248
M DGLIDA+ GST ITISN HFTDH +VMLFGA+D + D+ M+IT+ +N FGK+L
Sbjct: 212 MSRATDGLIDAVAGSTNITISNCHFTDHEKVMLFGANDHAEEDRGMKITLAYNHFGKRLD 271
Query: 249 QRMPRCRFGFIHVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPE 308
QRMPRCRFGF H++NN Y WE YAIGG+ TIIS+GN+FIA + KE+T R
Sbjct: 272 QRMPRCRFGFFHLVNNDYTHWERYAIGGSSGATIISQGNRFIAEDKLLVKEVTYREKSTS 331
Query: 309 A--EWKTWQWRSINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLR 366
+ EW W W + D + NGA F SG + + + ++I+ +P S VG LT++S +L
Sbjct: 332 SVEEWMKWTWITDGDDFENGATFTPSGDQNLLSKIDHLNLIQPEPSSKVGLLTKFSGALS 391
Query: 367 CRVGKPC 373
C++ +PC
Sbjct: 392 CKIRRPC 398
>PEL_TOBAC (P40972) Pectate lyase precursor (EC 4.2.2.2)
Length = 397
Score = 410 bits (1054), Expect = e-114
Identities = 201/356 (56%), Positives = 252/356 (70%), Gaps = 2/356 (0%)
Query: 20 CMATNPIDRCWRCDPNWANNRKKLADCVQGFGRKTTGGKDGPIYVVTDPSDSDLVNPRPG 79
C A N ID+CWRCDPNWA NR+K+ADC GFG GGK G IYVVTD SD D+V+P+PG
Sbjct: 42 CRAENAIDKCWRCDPNWAENRQKMADCALGFGSNAIGGKLGRIYVVTDNSDDDVVDPKPG 101
Query: 80 TLRHAVTRNGPLWIIFARSMKIRLNQELIMAGNKTIDGRGADVTIANGAGITIQFIENVI 139
TLR+ V + PLWIIF ++MKI+L++ELI+ NKTIDGRG +V I NGAGI IQ N+I
Sbjct: 102 TLRYGVIQKEPLWIIFGKNMKIKLSRELIVTSNKTIDGRGFNVHIQNGAGIKIQSASNII 161
Query: 140 IHGIKIYDIMVGSGGLVRDSEDHYGLRTISDGDGISIFGSSHIWIDHVSMRNCRDGLIDA 199
I ++I++I+ GGL+R+SEDH GLR +GDGISIF S IWIDH+SM DGLIDA
Sbjct: 162 ISNLRIHNIVPTPGGLLRESEDHVGLRGSDEGDGISIFSSHDIWIDHISMSRATDGLIDA 221
Query: 200 IMGSTAITISNSHFTDHNEVMLFGASDTYDGDQKMQITVVFNRFGKKLIQRMPRCRFGFI 259
+ ST ITISN HFTDH +VMLFGA+D Y D+ M+IT+ +N FGK+L QRMPRCRFGF
Sbjct: 222 VAASTNITISNCHFTDHEKVMLFGANDHYVLDKDMKITLAYNHFGKRLDQRMPRCRFGFF 281
Query: 260 HVLNNFYNRWEMYAIGGTMHPTIISEGNKFIAPNNGHAKEITKRTLVPE--AEWKTWQWR 317
H++NN Y WE YAIGG+ TIIS+GN+FIA + KE+T R + AEW W W
Sbjct: 282 HLVNNDYTHWERYAIGGSSGATIISQGNRFIAEDELLVKEVTYREKLTASVAEWMKWTWI 341
Query: 318 SINDLYLNGAFFRQSGAELINRPFSNKDMIKAKPGSYVGRLTRYSRSLRCRVGKPC 373
S D NGA F SG + + + ++IK +P S VG LT++S +L C G+PC
Sbjct: 342 SDGDDMENGATFTPSGDQNLLDKIDHLNLIKPEPSSKVGILTKFSGALSCVKGRPC 397
Database: sprot
Posted date: Nov 25, 2004 10:54 AM
Number of letters in database: 59,974,054
Number of sequences in database: 164,201
Lambda K H
0.322 0.140 0.442
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 48,197,007
Number of Sequences: 164201
Number of extensions: 2152208
Number of successful extensions: 4178
Number of sequences better than 10.0: 68
Number of HSP's better than 10.0 without gapping: 52
Number of HSP's successfully gapped in prelim test: 16
Number of HSP's that attempted gapping in prelim test: 4021
Number of HSP's gapped (non-prelim): 70
length of query: 373
length of database: 59,974,054
effective HSP length: 112
effective length of query: 261
effective length of database: 41,583,542
effective search space: 10853304462
effective search space used: 10853304462
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC136286.13


